Characterization of Oligopeptides in Solanum lycopersicum Xylem Exudates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Materials and Xylem Exudates Harvesting
2.2. Protein and Peptide Purification Using a Gel-Free System
2.3. Data-Dependent MS/MS Analysis
2.4. Exploration of the Tomato CLE-like Sequence
2.5. Quantitative PCR Analyses
2.6. Statistical Analyses
3. Results
3.1. Characterization of Small Protein- or Peptide Fragment-Containing Fractions of Tomato Xylem Exudates
3.2. Identification of Tomato Xylem Exudate-Associated Oligopeptides
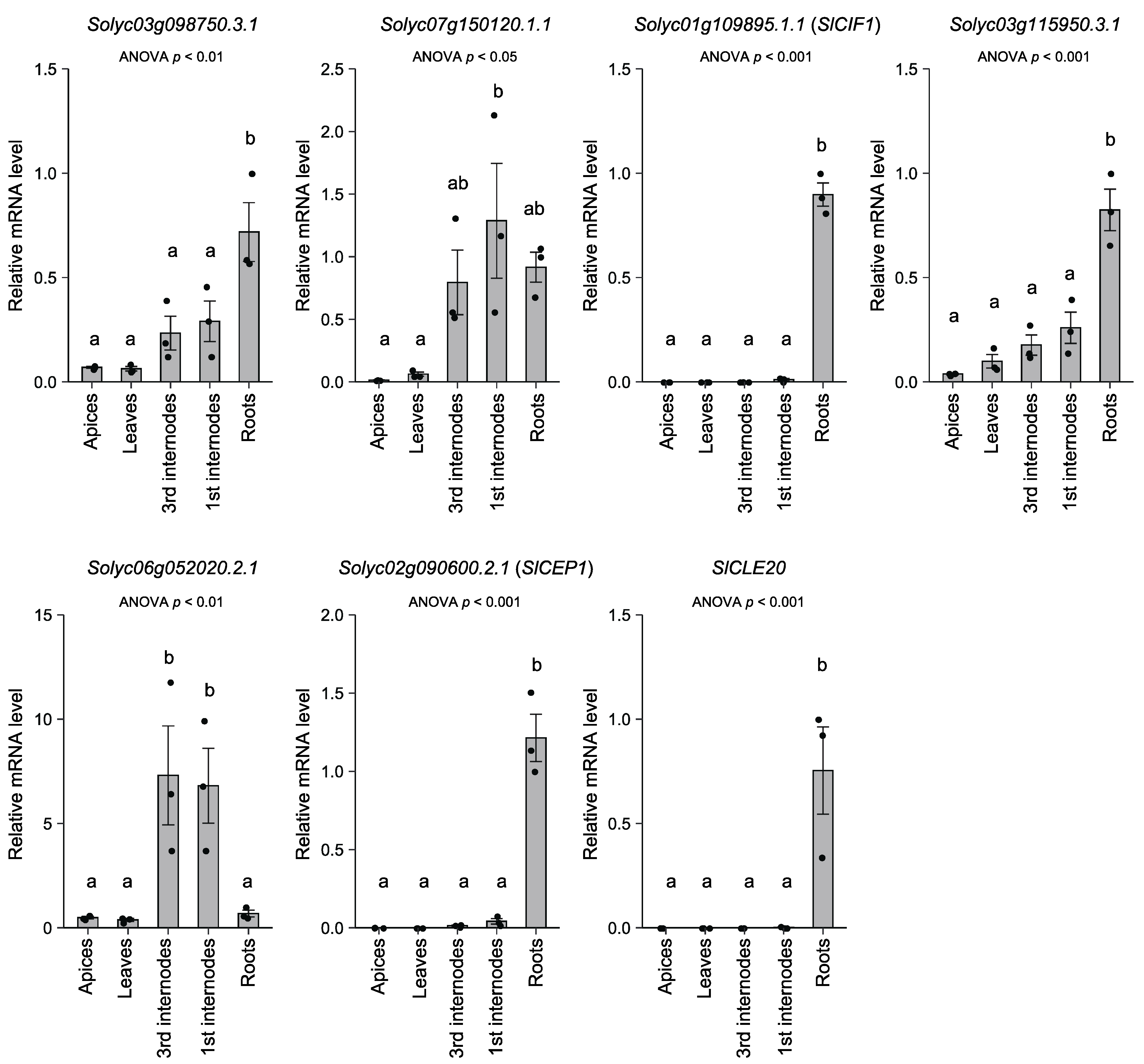
3.3. Expression Analysis of the Oligopeptide-Encoding Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biles, C.L.; Abeles, F.B. Xylem Sap Proteins. Plant Physiol. 1991, 96, 597–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwai, H.; Usui, M.; Hoshino, H.; Kamada, H.; Matsunaga, T.; Kakegawa, K.; Ishii, T.; Satoh, S. Analysis of Sugars in Squash Xylem Sap. Plant Cell Physiol. 2003, 44, 582–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buhtz, A.; Kolasa, A.; Arlt, K.; Walz, C.; Kehr, J. Xylem Sap Protein Composition Is Conserved among Different Plant Species. Planta 2004, 219, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.; Buhtz, A.; Giavalisco, P. Analysis of Xylem Sap Proteins from Brassica Napus. BMC Plant Biol. 2005, 5, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, S.; Goodger, J.Q.D.; Marsh, E.L.; Chen, S.; Asirvatham, V.S.; Schachtman, D.P. Characterization of the Maize Xylem Sap Proteome. J. Proteome Res. 2006, 5, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, M.A.; Oakes, M.; Li, D.X.; Hwang, C.H.; Hocart, C.H.; Gresshoff, P.M. The Glycine Max Xylem Sap and Apoplast Proteome. J. Proteome Res. 2007, 6, 3771–3779. [Google Scholar] [CrossRef] [PubMed]
- Ligat, L.; Lauber, E.; Albenne, C.; San Clemente, H.; Valot, B.; Zivy, M.; Pont-Lezica, R.; Arlat, M.; Jamet, E. Analysis of the Xylem Sap Proteome of Brassica Oleracea Reveals a High Content in Secreted Proteins. Proteomics 2011, 11, 1798–1813. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, S.; Suzuki, T.; Kawaguchi, M.; Higashiyama, T.; Matsubayashi, Y. A Comprehensive Strategy for Identifying Long-Distance Mobile Peptides in Xylem Sap. Plant J. 2015, 84, 611–620. [Google Scholar] [CrossRef]
- Luo, J.-S.; Zhang, Z. Proteomic Changes in the Xylem Sap of Brassica Napus under Cadmium Stress and Functional Validation. BMC Plant Biol. 2019, 19, 280. [Google Scholar] [CrossRef]
- Young, S.A.; Guo, A.; Guikema, J.A.; White, F.F.; Leach, J.E. Rice Cationic Peroxidase Accumulates in Xylem Vessels during Incompatible Interactions with Xanthomonas Oryzae Pv Oryzae. Plant Physiol. 1995, 107, 1333–1341. [Google Scholar] [CrossRef]
- Marjamaa, K.; Kukkola, E.M.; Fagerstedt, K.V. The Role of Xylem Class III Peroxidases in Lignification. J. Exp. Bot. 2009, 60, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, A.M.; Doerner, P.; Dixon, R.A.; Lamb, C.J.; Cameron, R.K. A Putative Lipid Transfer Protein Involved in Systemic Resistance Signalling in Arabidopsis. Nature 2002, 419, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Chanda, B.; Xia, Y.; Mandal, M.K.; Yu, K.; Sekine, K.; Gao, Q.; Selote, D.; Hu, Y.; Stromberg, A.; Navarre, D.; et al. Glycerol-3-Phosphate Is a Critical Mobile Inducer of Systemic Immunity in Plants. Nat. Genet. 2011, 43, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, Y. Post-Translational Modifications in Secreted Peptide Hormones in Plants. Plant Cell Physiol. 2011, 52, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, S.; Shinohara, H.; Mori, T.; Matsubayashi, Y.; Kawaguchi, M. Root-Derived CLE Glycopeptides Control Nodulation by Direct Binding to HAR1 Receptor Kinase. Nat. Commun. 2013, 4, 2191. [Google Scholar] [CrossRef]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of Root-Derived Peptides by Shoot LRR-RKs Mediates Systemic N-Demand Signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef]
- Okamoto, S.; Ohnishi, E.; Sato, S.; Takahashi, H.; Nakazono, M.; Tabata, S.; Kawaguchi, M. Nod Factor/Nitrate-Induced CLE Genes That Drive HAR1-Mediated Systemic Regulation of Nodulation. Plant Cell Physiol. 2009, 50, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A Small Peptide Modulates Stomatal Control via Abscisic Acid in Long-Distance Signalling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef]
- Ohyama, K.; Shinohara, H.; Ogawa-Ohnishi, M.; Matsubayashi, Y. A Glycopeptide Regulating Stem Cell Fate in Arabidopsis Thaliana. Nat. Chem. Biol. 2009, 5, 578–580. [Google Scholar] [CrossRef]
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S.; et al. An Improved de Novo Assembly and Annotation of the Tomato Reference Genome Using Single-Molecule Sequencing, Hi-C Proximity Ligation and Optical Maps. bioRxiv 2019, 767764. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Cock, J.M.; McCormick, S. A Large Family of Genes That Share Homology WithCLAVATA3. Plant Physiol. 2001, 126, 939–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yang, S.; Song, Y.; Wang, J. Genome-Wide Characterization, Expression and Functional Analysis of CLV3/ESR Gene Family in Tomato. BMC Genom. 2014, 15, 827. [Google Scholar] [CrossRef] [Green Version]
- Dyrløv Bendtsen, J.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved Prediction of Signal Peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B.P.A. The Plant Peptidome: An Expanding Repertoire of Structural Features and Biological Functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef] [Green Version]
- Notaguchi, M.; Okamoto, S. Dynamics of Long-Distance Signaling via Plant Vascular Tissues. Front. Plant Sci. 2015, 6, 160. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, S.; Tabata, R.; Matsubayashi, Y. Long-Distance Peptide Signaling Essential for Nutrient Homeostasis in Plants. Curr. Opin. Plant Biol. 2016, 34, 35–40. [Google Scholar] [CrossRef]
- Takahashi, F.; Hanada, K.; Kondo, T.; Shinozaki, K. Hormone-like Peptides and Small Coding Genes in Plant Stress Signaling and Development. Curr. Opin. Plant Biol. 2019, 51, 88–95. [Google Scholar] [CrossRef]
- Nakayama, T.; Shinohara, H.; Tanaka, M.; Baba, K.; Ogawa-Ohnishi, M.; Matsubayashi, Y. A Peptide Hormone Required for Casparian Strip Diffusion Barrier Formation in Arabidopsis Roots. Science 2017, 355, 284–286. [Google Scholar] [CrossRef] [PubMed]
- Doblas Verónica, G.; Smakowska-Luzan, E.; Fujita, S.; Alassimone, J.; Barberon, M.; Madalinski, M.; Belkhadir, Y.; Geldner, N. Root Diffusion Barrier Control by a Vasculature-Derived Peptide Binding to the SGN3 Receptor. Science 2017, 355, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Penel, C.; Dunand, C. Performing the Paradoxical: How Plant Peroxidases Modify the Cell Wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Yadeta, K.; Thomma, B. The Xylem as Battleground for Plant Hosts and Vascular Wilt Pathogens. Front. Plant Sci. 2013, 4, 97. [Google Scholar] [CrossRef] [Green Version]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. Null 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Chukhchin, D.G.; Bolotova, K.; Sinelnikov, I.; Churilov, D.; Novozhilov, E. Exosomes in the Phloem and Xylem of Woody Plants. Planta 2019, 251, 12. [Google Scholar] [CrossRef]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating Protein-Coding and Noncoding RNA: Challenges and Ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef] [Green Version]
- Hanada, K.; Zhang, X.; Borevitz, J.O.; Li, W.-H.; Shiu, S.-H. A Large Number of Novel Coding Small Open Reading Frames in the Intergenic Regions of the Arabidopsis Thaliana Genome Are Transcribed and/or under Purifying Selection. Genome Res. 2007, 17, 632–640. [Google Scholar] [CrossRef] [Green Version]
- Hanada, K.; Higuchi-Takeuchi, M.; Okamoto, M.; Yoshizumi, T.; Shimizu, M.; Nakaminami, K.; Nishi, R.; Ohashi, C.; Iida, K.; Tanaka, M.; et al. Small Open Reading Frames Associated with Morphogenesis Are Hidden in Plant Genomes. Proc. Natl. Acad. Sci. USA 2013, 110, 2395–2400. [Google Scholar] [CrossRef] [Green Version]
- Araya, T.; Miyamoto, M.; Wibowo, J.; Suzuki, A.; Kojima, S.; Tsuchiya, Y.N.; Sawa, S.; Fukuda, H.; von Wirén, N.; Takahashi, H. CLE-CLAVATA1 Peptide-Receptor Signaling Module Regulates the Expansion of Plant Root Systems in a Nitrogen-Dependent Manner. Proc. Natl. Acad. Sci. USA 2014, 111, 2029–2034. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Wang, Y.; Takahashi, H. CLE-CLAVATA1 Signaling Pathway Modulates Lateral Root Development under Sulfur Deficiency. Plants 2019, 8, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.; Endo, S.; Betsuyaku, S.; Shimotohno, A.; Fukuda, H. CLE2 Regulates Light-Dependent Carbohydrate Metabolism in Arabidopsis Shoots. Plant Mol. Biol. 2020, 104, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Kumar, A.; Jain, M.; Sudan, J.; Singh, K.; Kumari, S.; Mustafiz, A. C-Terminally Encoded Peptides (CEPs) Are Potential Mediators of Abiotic Stress Response in Plants. Physiol. Mol. Biol. Plants 2020, 26, 2019–2033. [Google Scholar] [CrossRef] [PubMed]
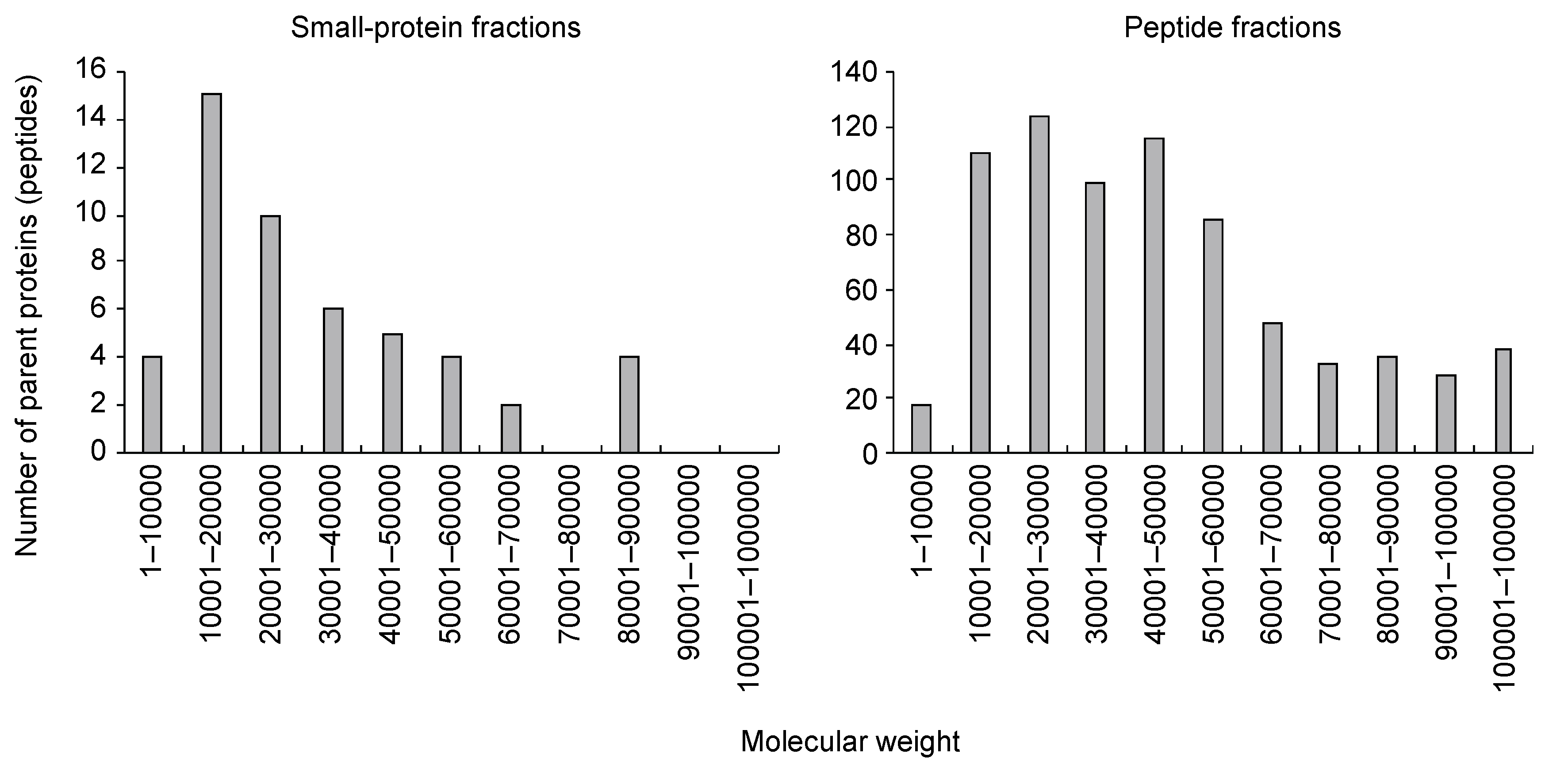
| Secreted Proteins | Nonsecreted Proteins | |
|---|---|---|
| Small-protein fractions | 33 (66.0%) | 17 (34.0%) |
| Peptide fractions | 287 (39.4%) | 442 (60.6%) |
| Tomato protein database (ITAG 4.0) | 5563 (16.3%) | 28,511 (83.7%) |
| Accession Number in ITAG4.0 | MW of Precursor Protein | Amino Acid Sequence | Note |
|---|---|---|---|
| Solyc03g098750.3.1 | 11,668 | DyAGTGANNHHDPKPpGSQW † | Homolog of GmXAP1 |
| Solyc07g150120.1.1 | 9252 | DyPSSGANNRHTpSHP † | --- |
| Solyc01g109895.1.1 (SlCIF1) | 13,357 | DyGRYDPTpALSKPPFKLIPN † | Homolog of GmXAP3 |
| Solyc03g115950.3.1 | 10,409 | DyPGSGANNRHTp † | --- |
| Solyc06g052020.2.1 | 8649 | DYDNAGpNTKHD | Homolog of GmXAP5 |
| Solyc02g090600.2.1 (SlCEP1) | 18,882 | YApKQTGNSpGIGHSS | Homolog of GmXAP6 |
| (SlCLE20) | 8402 | RVSpGGp*DPHHH | Homolog of GmXAP4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okamoto, S.; Kawasaki, A.; Makino, Y. Characterization of Oligopeptides in Solanum lycopersicum Xylem Exudates. Life 2022, 12, 592. https://doi.org/10.3390/life12040592
Okamoto S, Kawasaki A, Makino Y. Characterization of Oligopeptides in Solanum lycopersicum Xylem Exudates. Life. 2022; 12(4):592. https://doi.org/10.3390/life12040592
Chicago/Turabian StyleOkamoto, Satoru, Azusa Kawasaki, and Yumiko Makino. 2022. "Characterization of Oligopeptides in Solanum lycopersicum Xylem Exudates" Life 12, no. 4: 592. https://doi.org/10.3390/life12040592
APA StyleOkamoto, S., Kawasaki, A., & Makino, Y. (2022). Characterization of Oligopeptides in Solanum lycopersicum Xylem Exudates. Life, 12(4), 592. https://doi.org/10.3390/life12040592






