Do Ty3/Gypsy Transposable Elements Play Preferential Roles in Sex Chromosome Differentiation?
Abstract
1. Introduction
2. Roles of TEs during Sex Chromosome Differentiation
3. Non-Random Distribution of TEs on Sex Chromosomes across Different Lineages
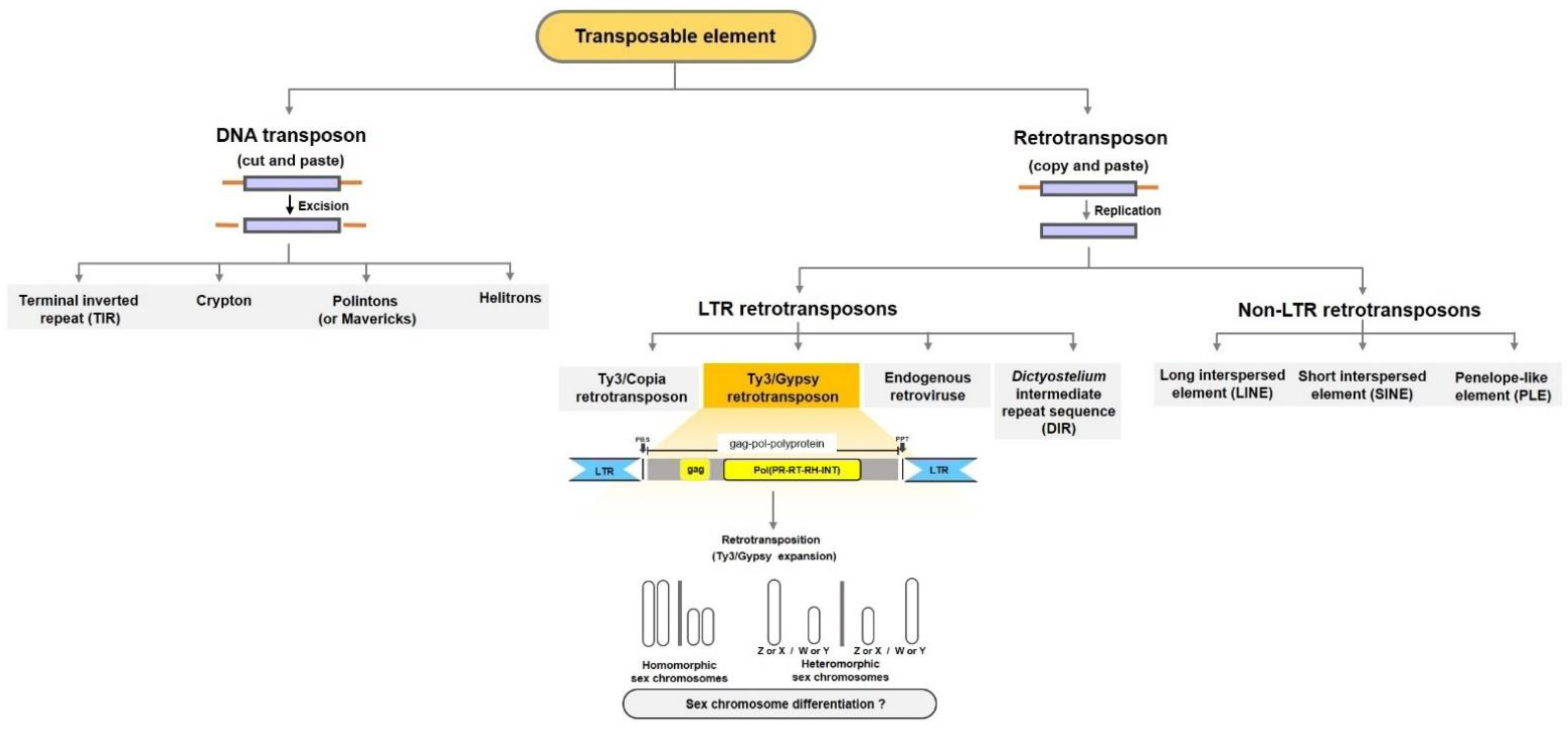
4. Do Ty3/Gypsy TEs Facilitate Sex Chromosome Differentiation?
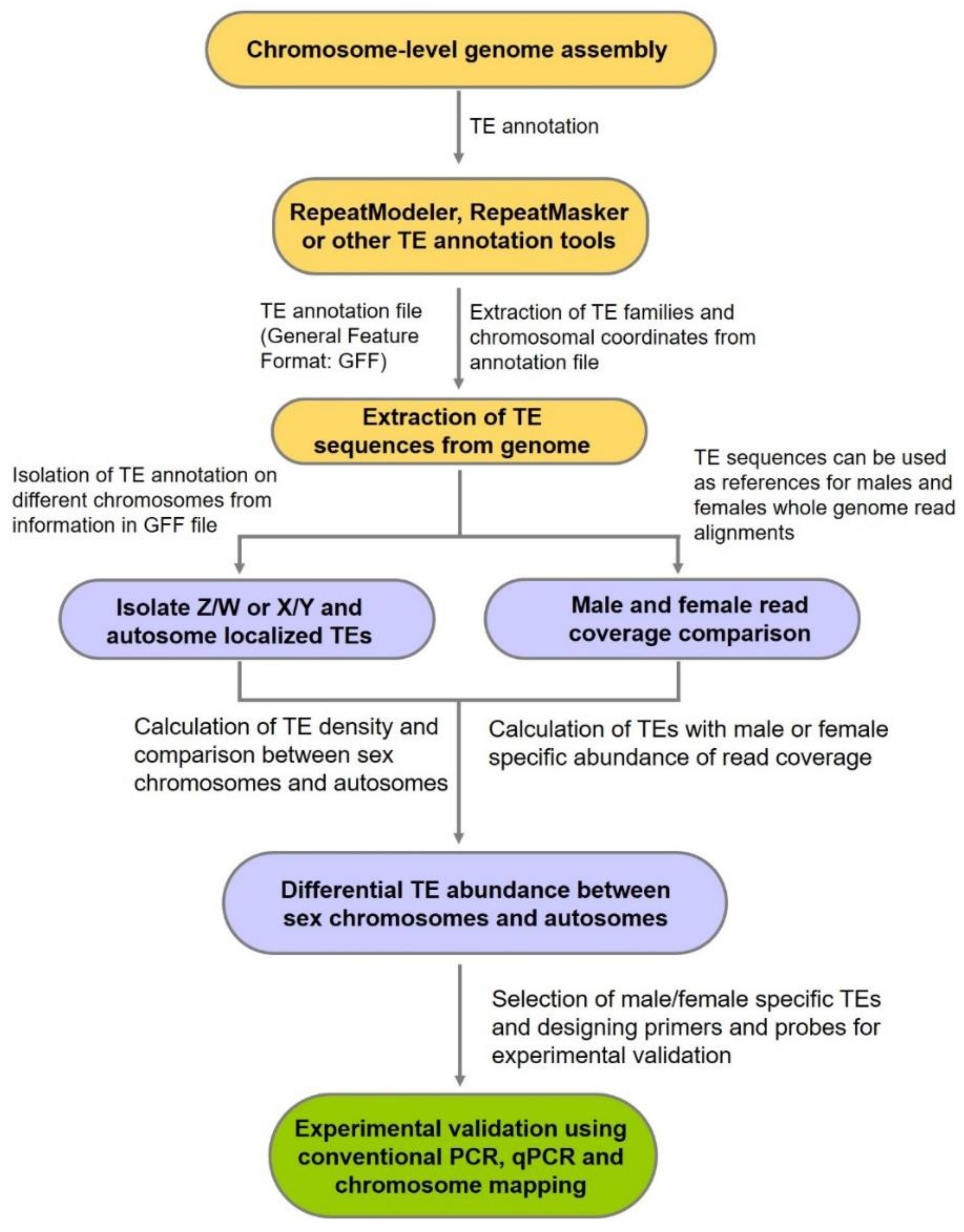
5. Determination of the Crucial Role of Ty3/Gypsy in Sex Chromosome Evolution
6. Final Considerations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bull, J.J. Sex determination in reptiles. Q. Rev. Biol. 1980, 55, 3–21. [Google Scholar] [CrossRef]
- Rhen, T.; Schroeder, A. Molecular mechanisms of sex determination in reptiles. Sex. Dev. 2010, 4, 16–28. [Google Scholar] [CrossRef]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef] [PubMed]
- Ezaz, T.; Srikulnath, K.; Graves, J.A.M. Origin of amniote sex chromosomes: An ancestral super-sex chromosome, or common requirements? J. Hered. 2017, 108, 94–105. [Google Scholar] [CrossRef]
- Li, S.; Ajimura, M.; Chen, Z.; Liu, J.; Chen, E.; Guo, H.; Tadapatri, V.; Reddy, C.G.; Zhang, J.; Kishino, H.; et al. A new approach for comprehensively describing heterogametic sex chromosomes. DNA Res. 2018, 25, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Furman, B.L.; Metzger, D.C.; Darolti, I.; Wright, A.E.; Sandkam, B.A.; Almeida, P.; Shu, J.J.; Mank, J.E. Sex chromosome evolution: So many exceptions to the rules. Genome Biol. Evol. 2020, 12, 750–763. [Google Scholar] [CrossRef] [PubMed]
- Stöck, M.; Kratochvíl, L.; Kuhl, H.; Rovatsos, M.; Evans, B.J.; Suh, A.; Valenzuela, N.; Veyrunes, F.; Zhou, Q.; Guiguen, Y.; et al. A brief review of vertebrate sex evolution with a pledge for integrative research: Towards ‘sexomics’. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200426. [Google Scholar] [CrossRef]
- Sinclair, A.H.; Berta, P.; Palmer, M.S.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.M.; Lovell-Badge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990, 346, 240–244. [Google Scholar] [CrossRef]
- Koopman, P.; Gubbay, J.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. Male development of chromosomally female mice transgenic for Sry. Nature 1991, 351, 117–121. [Google Scholar] [CrossRef]
- Temminck, C.J.; Schlegel, H. Oryzias latipes (Temminck & Schlegel, 1846). Available online: https://www.gbif.org/species/2368377 (accessed on 5 January 2022).
- Matsuda, M. Sex determination in the teleost medaka, Oryzias latipes. Annu. Rev. Genet. 2005, 39, 293–307. [Google Scholar] [CrossRef]
- Nanda, I.; Kondo, M.; Hornung, U.; Asakawa, S.; Winkler, C.; Shimizu, A.; Shan, Z.; Haaf, T.; Shimizu, N.; Shima, A.; et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 2002, 99, 11778–11783. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A. Sex chromosome specialization and degeneration in mammals. Cell 2006, 124, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Graves, A.; Fernández, S.; Gomez, F.; Schmidhuber, J. Connectionist temporal classification: Labelling unsegmented sequence data with recurrent neural networks. In Proceedings of the 23rd International Conference on Machine Learning, Pittsburgh, PA, USA, 25–29 June 2006; pp. 369–376. [Google Scholar]
- Ellegren, H. Sex-chromosome evolution: Recent progress and the influence of male and female heterogamety. Nat. Rev. Genet. 2011, 12, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Abbott, J.K.; Nordén, A.K.; Hansson, B. Sex chromosome evolution: Historical insights and future perspectives. Proc. Biol. Sci. 2017, 284, 20162806. [Google Scholar] [CrossRef]
- Singchat, W.; O’Connor, R.E.; Tawichasri, P.; Suntronpong, A.; Sillapaprayoon, S.; Suntrarachun, S.; Muangmai, N.; Baicharoen, S.; Peyachoknagul, S.; Chanhome, L.; et al. Chromosome map of the Siamese cobra: Did partial synteny of sex chromosomes in the amniote represent “a hypothetical ancestral super-sex chromosome” or random distribution? BMC Genom. 2018, 19, 939. [Google Scholar] [CrossRef]
- Singchat, W.; Ahmad, S.F.; Sillapaprayoon, S.; Muangmai, N.; Duengkae, P.; Peyachoknagul, S.; O’Connor, R.E.; Griffin, D.K.; Srikulnath, K. Partial amniote sex chromosomal linkage homologies shared on snake W sex chromosomes support the ancestral super-sex chromosome evolution in amniotes. Front. Genet. 2020, 11, 948. [Google Scholar] [CrossRef]
- Singchat, W.; Sillapaprayoon, S.; Muangmai, N.; Baicharoen, S.; Indananda, C.; Duengkae, P.; Peyachoknagul, S.; O’Connor, R.E.; Griffin, D.K.; Srikulnath, K. Do sex chromosomes of snakes, monitor lizards, and iguanian lizards result from multiple fission of an “ancestral amniote super-sex chromosome”? Chromosome Res. 2020, 28, 209–228. [Google Scholar] [CrossRef]
- Singchat, W.; Ahmad, S.F.; Laopichienpong, N.; Suntronpong, A.; Panthum, T.; Griffin, D.K.; Srikulnath, K. Snake W sex chromosome: The shadow of ancestral amniote super-sex chromosome. Cells 2020, 9, 2386. [Google Scholar] [CrossRef]
- Singchat, W.; Panthum, T.; Ahmad, S.F.; Baicharoen, S.; Muangmai, N.; Duengkae, P.; Griffin, D.K.; Srikulnath, K. Remnant of unrelated amniote sex chromosomal linkage sharing on the same chromosome in house gecko lizards, providing a better understanding of the ancestral super-sex chromosome. Cells 2021, 10, 2969. [Google Scholar] [CrossRef]
- Charlesworth, D.; Charlesworth, B.; Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity 2005, 95, 118–128. [Google Scholar] [CrossRef]
- Linnaeus [family CARICACEAE], Sp. Pl., 2: 1036, 1753. Available online: https://www.gbif.org/species/2874484 (accessed on 5 January 2022).
- Ma, H.; Moore, P.H.; Liu, Z.; Kim, M.S.; Yu, Q.; Fitch, M.M.; Sekioka, T.; Paterson, A.H.; Ming, R. High-density linkage mapping revealed suppression of recombination at the sex determination locus in papaya. Genetics 2004, 166, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Linnaeus [Spinacia oleracea], Sp. Pl.: 1027, 1753. Available online: https://www.gbif.org/species/3083647 (accessed on 5 January 2022).
- Arumuganathan, K.; Slattery, J.P.; Tanksley, S.D.; Earle, E.D. Preparation and flow cytometric analysis of metaphase chromosomes of tomato. Theor. Appl. Genet. 1991, 82, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Takahata, S.; Yago, T.; Iwabuchi, K.; Hirakawa, H.; Suzuki, Y.; Onodera, Y. Comparison of spinach sex chromosomes with sugar beet autosomes reveals extensive synteny and low recombination at the male-determining locus. J. Hered. 2016, 107, 679–685. [Google Scholar] [CrossRef]
- Xu, C.; Jiao, C.; Sun, H.; Cai, X.; Wang, X.; Ge, C.; Zheng, Y.; Liu, W.; Sun, X.; Xu, Y.; et al. Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat. Commun. 2017, 8, e15275. [Google Scholar] [CrossRef] [PubMed]
- Kudoh, T.; Takahashi, M.; Osabe, T.; Toyoda, A.; Hirakawa, H.; Suzuki, Y.; Ohmido, N.; Onodera, Y. Molecular insights into the non-recombining nature of the spinach male-determining region. Mol. Genet. Genom. 2018, 293, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Sayres, M.A.W. Genetic diversity on the sex chromosomes. Genome Biol. Evol. 2018, 10, 1064. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, J.; Bachtrog, D.; An, N.; Huang, Q.; Jarvis, E.D.; Gilbert, M.T.P.; Zhang, G. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 2014, 346, 1246338. [Google Scholar] [CrossRef]
- Bellott, D.W.; Skaletsky, H.; Cho, T.J.; Brown, L.; Locke, D.; Chen, N.; Page, D.C. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat. Genet. 2017, 49, 387–394. [Google Scholar] [CrossRef]
- Sigeman, H.; Ponnikas, S.; Chauhan, P.; Dierickx, E.; Brooke, M.D.L.; Hansson, B. Repeated sex chromosome evolution in vertebrates supported by expanded avian sex chromosomes. Proc. R. Soc. B 2019, 286, 20192051. [Google Scholar] [CrossRef]
- Kratochvíl, L.; Stöck, M.; Rovatsos, M.; Bullejos, M.; Herpin, A.; Jeffries, D.L.; Peichel, C.L.; Perrin, N.; Valenzuela, N.; Pokorná, M.J. Expanding the classical paradigm: What we have learnt from vertebrates about sex chromosome evolution. Philos. Trans. R. Soc. 2021, 376, 20200097. [Google Scholar] [CrossRef]
- Charlesworth, B.; Charlesworth, D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Steinemann, S.; Steinemann, M. Y chromosomes: Born to be destroyed. BioEssays 2005, 27, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Malone, J.H.; Oliver, B. The sex chromosome that refused to die. BioEssays 2008, 30, 409–411. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Yamana, Y.; Usui, T.; Ogawa, H.I.; Yamamoto, M.T.; Kusano, K. Homologous recombination via synthesis-dependent strand annealing in yeast requires the Irc20 and Srs2 DNA helicases. Genetics 2012, 191, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Blaser, O.; Grossen, C.; Neuenschwander, S.; Perrin, N. Sex-chromosome turnovers induced by deleterious mutation load. Evolution 2013, 67, 635–645. [Google Scholar] [CrossRef]
- Charlesworth, B.; Jarne, P.; Assimacopoulos, S. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. III. element abundances in heterochromatin. Genet. Res. 1994, 64, 183–197. [Google Scholar] [CrossRef][Green Version]
- Peichel, C.L.; Ross, J.A.; Matson, C.K.; Dickson, M.; Grimwood, J.; Schmutz, J.; Myers, R.M.; Mori, S.; Schluter, D.; Kingsley, D.M. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 2004, 14, 1416–1424. [Google Scholar] [CrossRef]
- Bachtrog, D. The temporal dynamics of processes underlying Y-chromosome degeneration. Genetics 2008, 179, 1513–1525. [Google Scholar] [CrossRef]
- Matsubara, K.; Uno, Y.; Srikulnath, K.; Seki, R.; Nishida, C.; Matsuda, Y. Molecular cloning and characterization of satellite DNA sequences from constitutive heterochromatin of the habu snake (Protobothrops flavoviridis, Viperidae) and the Burmese python (Python bivittatus, Pythonidae). Chromosoma 2015, 124, 529–539. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Panthum, T.; Srikulnath, K. Consequence of paradigm shift with repeat landscapes in reptiles: Powerful facilitators of chromosomal rearrangements for diversity and evolution. Genes 2020, 11, 827. [Google Scholar] [CrossRef]
- Carvalho, B.; Koerich, L.B.; Clark, A.G. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009, 25, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Rifkin, J.L.; Beaudry, F.E.; Humphries, Z.; Choudhury, B.I.; Barrett, S.C.; Wright, S.I. Widespread recombination suppression facilitates plant sex chromosome evolution. Mol. Biol. Evol. 2021, 38, 1018–1030. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.F. Transposable Elements and Evolution; Kluver: Dodrect, The Netherlands, 1993. [Google Scholar]
- Hua-Van, A.; Le Rouzic, A.; Maisonhaute, C.; Capy, P. Abundance, distribution and dynamics of retrotransposable elements and transposons: Similarities and differences. Cytogenet. Genome Res. 2005, 110, 426–440. [Google Scholar] [CrossRef]
- Erlandsson, R.; Wilson, J.F.; Pääbo, S. Sex chromosomal transposable element accumulation and male-driven substitutional evolution in humans. Mol. Biol. Evol. 2000, 17, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Ezaz, T.; Deakin, J.E. Repetitive sequence and sex chromosome evolution in vertebrates. Adv. Evol. Biol. 2014, 2014, 104683. [Google Scholar] [CrossRef]
- Chalopin, D.; Volff, J.; Galiana, D.; Anderson, J.L.; Schartl, M. Transposable elements and early evolution of sex chromosomes in fish. Chromosome Res. 2015, 23, 545–560. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Sotero-Caio, C.G.; Platt, R.N.; Suh, A.; Ray, D.A. Evolution and diversity of transposable elements in vertebrate genomes. Genome Biol. Evol. 2017, 9, 161–177. [Google Scholar] [CrossRef]
- Almojil, D.; Bourgeois, Y.; Falis, M.; Hariyani, I.; Wilcox, J.; Boissinot, S. The structural, functional and evolutionary impact of transposable elements in eukaryotes. Genes 2021, 12, 918. [Google Scholar] [CrossRef]
- Callinan, P.A.; Batzer, M.A. Retrotransposable elements and human disease. Genome Dyn. 2006, 1, 104–115. [Google Scholar]
- Eickbush, T.H.; Malik, H.S. Evolution of retrotransposons. In Mobile DNA II; Craig, N., Craigie, R., Gellert, M., Lambowitz, A., Eds.; ASM Press: Washington, DC, USA, 2002; pp. 1111–1144. [Google Scholar]
- Kent, T.V.; Uzunović, J.; Wright, S.I. Coevolution between transposable elements and recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160458. [Google Scholar] [CrossRef] [PubMed]
- Drost, H.G.; Sanchez, D.H. Becoming a selfish clan: Recombination associated to reverse-transcription in LTR retrotransposons. Genome Biol. Evol. 2019, 11, 3382–3392. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Hou, S.; Hobza, R.; Feltus, F.A.; Wang, X.; Jin, W.; Skelton, R.L.; Blas, A.; Lemke, C.; Saw, J.H.; et al. Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Mol. Genet. Genom. 2007, 278, 177–185. [Google Scholar] [CrossRef]
- Ming, R.; Hou, S.; Feng, Y.; Yu, Q.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.; et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 2008, 452, 991–996. [Google Scholar] [CrossRef]
- Na, J.K.; Wang, J.; Ming, R. Accumulation of interspersed and sex-specific repeats in the non-recombining region of papaya sex chromosomes. BMC Genom. 2014, 15, 335. [Google Scholar] [CrossRef]
- VanBuren, R.; Zeng, F.C.; Chen, C.X.; Zhang, J.S.; Wai, C.M.; Han, J.; Aryal, R.; Gschwend, A.R.; Wang, J.P.; Na, J.K.; et al. Originanddomestication of papaya Yh chromosome. Genome Res. 2015, 25, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Linnaeus [Rumex acetosa], Sp. Pl. 338, 1753. Available online: https://www.gbif.org/species/2888951 (accessed on 5 January 2022).
- Steflova, P.; Tokan, V.; Vogel, I.; Lexa, M.; Macas, J.; Novak, P.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Contrasting patterns of transposable element and satellite distribution on sex chromosomes (XY1Y2) in the dioecious plant Rumex acetosa. Genome Biol. Evol. 2013, 5, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Herpin, A.; Braasch, I.; Kraeussling, M.; Schmidt, C.; Thoma, E.C.; Nakamura, S.; Tanaka, M.; Schartl, M. Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements. PLoS Genet. 2010, 6, e1000844. [Google Scholar] [CrossRef] [PubMed]
- Regan, C.T. The Asiatic fishes of the family Anabantidae. J. Zool. 1910, 1909, 767–787. [Google Scholar]
- Burchell [Clarias gariepinus]. 1822. Available online: https://www.gbif.org/species/5202793 (accessed on 5 January 2022).
- Günther [Clarias macrocephalus]. 1864. Available online: https://www.gbif.org/species/5202728 (accessed on 5 January 2022).
- McCulloch, A.R.; Waite, E.R. Results of the South Australian Museum expedition to Strzelecki and Coopers Creeks. Trans. R. Soc. S. Aust. 1917, 41, 472–475. [Google Scholar]
- Lesson, R.P. Catalogue des Reptiles qui Font Partie d’une Collection Zoologique Recueille Dans l’Inde Continentale ou en Afrique, et Apportée en France par M. Lamare-Piquot; Bulletin des Sciences Naturelles et de Géologie: Paris, France, 1831; Volume 25, pp. 119–123. [Google Scholar]
- Kotov, A.A. Priority of Carl Linnaeus as the author of the oldest species of Cladocera (Crustacea: Branchiopoda): Daphnia pulex (Linnaeus, 1758) and Polyphemus pediculus (Linnaeus, 1758). Zootaxa 2020, 4803, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Koomgun, T.; Laopichienpong, N.; Singchat, W.; Panthum, T.; Phatcharakullawarawat, R.; Kraichak, E.; Sillapaprayoon, S.; Ahmad, S.F.; Muangmai, N.; Peyachoknagul, S.; et al. Genome complexity reduction high-throughput genome sequencing of green iguana (Iguana iguana) reveal a paradigm shift in understanding sex-chromosomal linkages on homomorphic X and Y sex chromosomes. Front. Genet. 2020, 11, 1217. [Google Scholar] [CrossRef] [PubMed]
- Laopichienpong, N.; Kraichak, E.; Singchat, W.; Sillapaprayoon, S.; Muangmai, N.; Suntrarachun, S.; Baicharoen, S.; Peyachok-nagul, S.; Chanhome, L.; Ezaz, T.; et al. Genome-wide SNP analysis of Siamese cobra (Naja kaouthia) reveals the molecular basis of transitions between Z and W sex chromosomes and supports the presence of an ancestral super-sex chromosome in amniotes. Genomics 2021, 113, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.H.M.; Panthum, T.; Ponjarat, J.; Laopichiengpong, N.; Kraichak, E.; Singchat, W.; Muangmai, N.; Peyachoknagul, S.; Na-Nakorn, U.; Srikulnath, K. An investigation of ZZ/ZW and XX/XY sex determination systems in North African catfish (Clarias gariepinus, Burchell 1822). Front. Genet. 2021, 11, 1719. [Google Scholar] [CrossRef]
- Nguyen, D.H.M.; Ponjarat, J.; Laopichienpong, N.; Kraichak, E.; Panthum, T.; Singchat, W.; Ahmad, S.F.; Muangmai, N.; Duengkae, P.; Peyachoknagul, S.; et al. Genome-wide SNP analysis suggests male heterogamety in bighead catfish (Clarias macrocephalus). Aquaculture 2021, 545, 737005. [Google Scholar] [CrossRef]
- Nguyen, D.H.M.; Ponjarat, J.; Laopichienpong, N.; Panthum, T.; Singchat, W.; Ahmad, S.F.; Kraichak, E.; Muangmai, N.; Duengkae, P.; Peyachoknagul, S.; et al. Genome-wide SNP analysis of hybrid clariid fish reflects the existence of polygenic sex-determination in the lineage. Front. Genet. 2022, 13, 789573. [Google Scholar] [CrossRef]
- Panthum, T.; Laopichienpong, N.; Kraichak, E.; Singchat, W.; Ho My Nguyen, D.; Ariyaraphong, N.; Ahmad, S.F.; Muangmai, N.; Duengkae, P.; Peyachoknagul, S.; et al. The snakeskin gourami (Trichopodus pectoralis) tends to exhibit XX/XY sex determination. Fishes 2021, 6, 43. [Google Scholar] [CrossRef]
- Suntronpong, A.; Panthum, T.; Laopichienpong, N.; Nguyen, D.H.M.; Kraichak, E.; Singchat, W.; Ariyaraphong, N.; Ahmad, S.F.; Muangmai, N.; Duengkae, P.; et al. Implications of genome-wide single nucleotide polymorphisms in jade perch (Scortum barcoo) reveals the putative XX/XY sex-determination system, facilitating a new chapter of sex control in aquaculture. Aquaculture 2022, 548, 737587. [Google Scholar] [CrossRef]
- Arkhipova, I.R. Neutral theory, transposable elements, and eukaryotic genome evolution. Mol. Biol. Evol. 2018, 35, 1332–1337. [Google Scholar] [CrossRef]
- Makałowski, W.; Gotea, V.; Pande, A.; Makałowska, I. Transposable elements: Classification, identification, and their use as a tool for comparative genomics. Evol. Genom. 2019, 1910, 177–207. [Google Scholar]
- Dechaud, C.; Volff, J.N.; Schartl, M.; Naville, M. Sex and the TEs: Transposable elements in sexual development and function in animals. Mob. DNA 2019, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Hollister, J.D.; Gaut, B.S. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009, 19, 1419–1428. [Google Scholar] [CrossRef]
- Hobza, R.; Cegan, R.; Jesionek, W.; Kejnovsky, E.; Vyskot, B.; Kubat, Z. Impact of repetitive elements on the Y chromosome formation in plants. Genes 2017, 8, 302. [Google Scholar] [CrossRef] [PubMed]
- Muyle, A.; Bachtrog, D.; Marais, G.; Turner, J. Epigenetics drive the evolution of sex chromosomes in animals and plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200124. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Tarui, H.; Toriba, M.; Yamada, K.; Nishida, C.; Agata, K.; Matsuda, Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 18190–18195. [Google Scholar] [CrossRef]
- Bellott, D.W.; Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Cho, T.J.; Koutseva, N.; Zaghlul, S.; Graves, T.; Rock, S.; et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 2014, 508, 494–499. [Google Scholar] [CrossRef]
- Hughes, J.F.; Skaletsky, H.; Pyntikova, T.; Graves, T.A.; van Daalen, S.K.; Minx, P.J.; Fulton, R.S.; McGrath, S.D.; Locke, D.P.; Friedman, C.; et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 2010, 463, 536–539. [Google Scholar] [CrossRef]
- Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Graves, T.; Fulton, R.S.; Dugan, S.; Ding, Y.; Buhay, C.J.; Kremitzki, C.; et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 2012, 482, 82–86. [Google Scholar] [CrossRef]
- Soh, Y.S.; Alföldi, J.; Pyntikova, T.; Brown, L.G.; Graves, T.; Minx, P.J.; Page, D.C. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 2014, 159, 800–813. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Panthum, T.; Srikulnath, K. Impact of repetitive DNA elements on snake genome biology and evolution. Cells 2021, 10, 1707. [Google Scholar] [CrossRef]
- Kondo, M.; Hornung, U.; Nanda, I.; Imai, S.; Sasaki, T.; Shimizu, A.; Asakawa, S.; Hori, H.; Schmid, M.; Shimizu, N.; et al. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 2006, 16, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Günther [Xiphophorus maculatus]. 1866. Available online: https://www.gbif.org/species/2350164 (accessed on 5 January 2022).
- Kallman, K.D.; Schreibman, M.P. A sex-linked gene controlling gonadotrop differentiation and its significance in determining the age of sexual maturation and size of the platyfish, Xiphophorus maculatus. Gen. Comp. Endocrinol. 1973, 21, 287–304. [Google Scholar] [CrossRef]
- Volff, J.N.; Schartl, M. Variability of genetic sex determination in poeciliid fishes. Genetica 2001, 111, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Śliwińska, E.B.; Martyka, R.; Tryjanowski, P. Evolutionary interaction between W/Y chromosome and transposable elements. Genetica 2016, 144, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Günther [Cynoglossus semilaevis]. 1873. Available online: https://www.gbif.org/species/2409626 (accessed on 6 January 2022).
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.N.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef]
- Blavet, N.; Blavet, H.; Čegan, R.; Zemp, N.; Zdanska, J.; Janoušek, B.; Hobza, R.; Widmer, A. Comparative analysis of a plant pseudoautosomal region (PAR) in Silene latifolia with the corresponding S. vulgaris autosome. BMC Genom. 2012, 13, 226. [Google Scholar] [CrossRef]
- Ishii, K.; Nishiyama, R.; Shibata, F.; Kazama, Y.; Abe, T.; Kawano, S. Rapid degeneration of noncoding DNA regions surrounding SlAP3X/Y after recombination suppression in the dioecious plant Silene latifolia. G3 2013, 3, 2121–2130. [Google Scholar] [CrossRef]
- Daudin [Xenopus laevis]. 1802. Available online: https://www.gbif.org/species/5217334 (accessed on 6 January 2022).
- Mawaribuchi, S.; Takahashi, S.; Wada, M.; Uno, Y.; Matsuda, Y.; Kondo, M.; Fukui, A.; Takamatsu, N.; Taira, M.; Ito, M. Sex chromosome differentiation and the W-and Z-specific loci in Xenopus laevis. Dev. Biol. 2017, 426, 393–400. [Google Scholar] [CrossRef]
- Bachtrog, D. Adaptation shapes patterns of genome evolution in sexual and asexual genomes in Drosophila. Nat. Genet. 2003, 34, 215–219. [Google Scholar] [CrossRef]
- Hoehn, K.B.; Gall, A.; Bashford-Rogers, R.; Fidler, S.J.; Kaye, S.; Weber, J.N.; McClure, M.O.; Kellam, P.; Pybus, O.G. Dynamics of immunoglobulin sequence diversity in HIV-1 infected individuals. Philos. Trans. R. Soc. B 2015, 370, 20140241. [Google Scholar] [CrossRef][Green Version]
- Schemberger, M.O.; Nascimento, V.D.; Coan, R.; Ramos, É.; Nogaroto, V.; Ziemniczak, K.; Valente, G.T.; Moreira-Filho, O.; Martins, C.; Vicari, M.R. DNA transposon invasion and microsatellite accumulation guide W chromosome differentiation in a Neotropical fish genome. Chromosoma 2019, 128, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.A.; Clark, F.E.; Roberts, R.B.; Xu, L.; Tao, W.; Zhou, Q.; Wang, D.; Kocher, T.D. Origin of a giant sex chromosome. Mol. Biol. Evol. 2021, 38, 1554–1569. [Google Scholar] [CrossRef] [PubMed]
- Pourrajab, F.; Hekmatimoghaddam, S. Transposable elements, contributors in the evolution of organisms (from an arms race to a source of raw materials). Heliyon 2021, 7, e06029. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M.; Peichel, C.L. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 2010, 11, 205. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Takashi, T.; Nagamatsu, S.; Kojima, M.; Sakakibara, H.; Kitano, H.; Matsuoka, M.; Aya, K. Efficacy of microarray profiling data combined with QTL mapping for the identification of a QTL gene controlling the initial growth rate in rice. Plant Cell Physiol. 2012, 53, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Yano, A.; Nicol, B.; Jouanno, E.; Quillet, E.; Fostier, A.; Guyomard, R.; Guiguen, Y. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol. Appl. 2013, 6, 486–496. [Google Scholar] [CrossRef]
- Faber-Hammond, J.J.; Phillips, R.B.; Brown, K.H. Comparative analysis of the shared sex-determination region (SDR) among salmonid fishes. Genome Biol. Evol. 2015, 7, 1972–1987. [Google Scholar] [CrossRef]
- Eisbrenner, W.D.; Botwright, N.; Cook, M.; Davidson, E.A.; Dominik, S.; Elliott, N.G.; Henshall, J.; Jones, S.L.; Kube, P.D.; Lubieniecki, K.P.; et al. Evidence for multiple sex-determining loci in Tasmanian Atlantic salmon (Salmo salar). Heredity 2014, 113, 86–92. [Google Scholar] [CrossRef]
- Lubieniecki, K.P.; Lin, S.; Cabana, E.I.; Li, J.; Lai, Y.Y.; Davidson, W.S. Genomic instability of the sex-determining locus in Atlantic salmon (Salmo salar). G3 2015, 5, 2513–2522. [Google Scholar] [CrossRef]
- Larson, J.; Mattu, S.; Kirchner, L.; Angwin, J. How We Analyzed the COMPAS Recidivism Algorithm; ProPublica: New York, NY, USA, 2016; Volume 9, p. 3. [Google Scholar]
- Hornung, E.; Vilisics, F.; Szlávecz, K. Szárazföldi ászkarák (Isopoda, Oniscidea) fajok tipizálása hazai előfordulási adatok alapján (különös tekintettel a sikeres megtelepedőkre). Természetvédelmi Közlemények 2007, 13, 47–57. [Google Scholar]
- Chaumeil, J.; Waters, P.D.; Koina, E.; Gilbert, C.; Robinson, T.J.; Graves, J.A.M. Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: Epigenetic modifications in distantly related mammals. PLoS ONE 2011, 6, e19040. [Google Scholar] [CrossRef] [PubMed]
- Branco, A.T.; Tao, Y.; Hartl, D.L.; Lemos, B. Natural variation of the Y chromosome suppresses sex ratio distortion and modulates testis-specific gene expression in Drosophila simulans. Heredity 2013, 111, 8–15. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ellison, C.E.; Bachtrog, D. Dosage compensation via transposable element mediated rewiring of a regulatory network. Science 2013, 342, 846–850. [Google Scholar] [CrossRef] [PubMed]
- Sackton, T.B.; Hartl, D.L. Meta-analysis reveals that genes regulated by the Y chromosome in Drosophila melanogaster are preferentially localized to repressive chromatin. Genome Biol. Evol. 2013, 5, 255–266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vicoso, B.; Emerson, J.J.; Zektser, Y.; Mahajan, S.; Bachtrog, D. Comparative sex chromosome genomics in snakes: Differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013, 11, 1001643. [Google Scholar] [CrossRef]
- Zhou, Q.; Ellison, C.E.; Kaiser, V.B.; Alekseyenko, A.A.; Gorchakov, A.A.; Bachtrog, D. The epigenome of evolving Drosophila neo-sex chromosomes: Dosage compensation and heterochromatin formation. PLoS Biol. 2013, 11, e1001711. [Google Scholar] [CrossRef]
- White, M.A.; Kitano, J.; Peichel, C.L. Purifying selection maintains dosage-sensitive genes during degeneration of the threespine stickleback Y chromosome. Mol. Biol. Evol. 2015, 32, 1981–1995. [Google Scholar] [CrossRef]
- Lyon, M.F. The Lyon and the LINE hypothesis. Semin. Cell Dev. Biol. 2003, 14, 313–318. [Google Scholar] [CrossRef]
- Roberts, N.B.; Juntti, S.A.; Coyle, K.P.; Dumont, B.L.; Stanley, M.K.; Ryan, A.Q.; Fernald, R.D.; Roberts, R.B. Polygenic sex determination in the cichlid fish Astatotilapia burtoni. BMC Genom. 2016, 17, 835. [Google Scholar] [CrossRef]
- Shevchenko, A.I.; Zakharova, I.S.; Zakian, S.M. The evolutionary pathway of X chromosome inactivation in mammals. Acta Nat. 2013, 5, 40–53. [Google Scholar] [CrossRef]
- Abrusán, G.; Giordano, J.; Warburton, P.E. Analysis of transposon interruptions suggests selection for L1 elements on the X chromosome. PLoS Genet. 2008, 4, e1000172. [Google Scholar] [CrossRef]
- Lyon, M.F. LINE-1 elements and X chromosome inactivation: A function for “junk” DNA? Proc. Natl. Acad. Sci. USA 2000, 97, 6248–6249. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y. On the Amami Oshima spiny rat. Bot. Zool. 1934, 3, 936–942. [Google Scholar]
- Honda, T.; Suzuki, H.; Ithu, M. An unusual sex chromosome constitution found in the amami spinous country-rat, Tokudaia osimensis osimensis. Jpn. Genet. 1977, 52, 247–249. [Google Scholar] [CrossRef]
- Kobayashi, T.; Yamada, F.; Hashimoto, A.S.; Matsuda, Y.; Kuroiwa, A. Centromere repositioning in the X chromosome of XO/XO mammals, Ryukyu spiny rat. Chromosome Res. 2008, 16, 587. [Google Scholar] [CrossRef]
- Hansen, R.S. X inactivation-specific methylation of LINE-1 elements by DNMT3B: Implications for the Lyon repeat hypothesis. Hum. Mol. Genet. 2003, 12, 2559–2567. [Google Scholar] [CrossRef]
- Regan [Betta splendens]. 1910. Available online: https://www.gbif.org/species/2393998 (accessed on 7 January 2022).
- Wang, L.; Sun, F.; Wan, Z.Y.; Yang, Z.; Tay, Y.X.; Lee, M.; Ye, B.; Wen, Y.; Meng, Z.; Fan, B.; et al. Transposon-induced epigenetic silencing in the X chromosome as a novel form of dmrt1 expression regulation during sex determination in the fighting fish. BMC Biol. 2022, 20, 5. [Google Scholar] [CrossRef]
- Martin, A.; Troadec, C.; Boualem, A.; Rajab, M.; Fernandez, R.; Morin, H.; Pitrat, M.; Dogimont, C.; Bendahmane, A. A transposon-induced epigenetic change leads to sex determination in melon. Nature 2009, 461, 1135–1138. [Google Scholar] [CrossRef]
- Tomaszkiewicz, M.; Chalopin, D.; Schartl, M.; Galiana, D.; Volff, J.N. A multicopy Y-chromosomal SGNH hydrolase gene expressed in the testis of the platyfish has been captured and mobilized by a Helitron transposon. BMC Genet. 2014, 15, 44. [Google Scholar] [CrossRef]
- Britten, R.J.; McCormack, T.J.; Mears, T.L.; Davidson, E.H. Gypsy/Ty3-class retrotransposons integrated in the DNA of herring, tunicate, and echinoderms. J. Mol. Evol. 1995, 40, 13–24. [Google Scholar] [CrossRef]
- Feschotte, C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Meigen [Drosophila melanogaster]. 1830. Available online: https://www.gbif.org/species/11195063 (accessed on 5 January 2022).
- Lawlor, M.A.; Cao, W.; Ellison, C.E. A transposon expression burst accompanies the activation of Y-chromosome fertility genes during Drosophila spermatogenesis. Nat Commun. 2021, 12, 6854. [Google Scholar] [CrossRef] [PubMed]
- Linnaeus [Bombyx mori]. 1758. Available online: https://www.gbif.org/species/1868664 (accessed on 5 January 2022).
- Hara, K.; Fujii, T.; Suzuki, Y.; Sugano, S.; Shimada, T.; Katsuma, S.; Kawaoka, S. Altered expression of testis-specific genes, piRNAs, and transposons in the silkworm ovary masculinized by a W chromosome mutation. BMC Genom. 2012, 13, 119. [Google Scholar] [CrossRef] [PubMed]
- Zamudio, N.; Bourc’his, D. Transposable elements in the mammalian germline: A comfortable niche or a deadly trap? Heredity 2010, 105, 92–104. [Google Scholar] [CrossRef]
- Pimpinelli, S.; Berloco, M.; Fanti, L.; Dimitri, P.; Bonaccorsi, S.; Marchetti, E.; Caizzi, R.; Caggese, C.; Gatti, M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc. Natl. Acad. Sci. USA 1995, 92, 3804–3808. [Google Scholar] [CrossRef]
- Abe, H.; Seki, M.; Ohbayashi, F.; Tanaka, N.; Yamashita, J.; Fujii, T.; Yokoyama, T.; Takahashi, M.; Banno, Y.; Sahara, K.; et al. Partial deletions of the W chromosome due to reciprocal translocation in the silkworm Bombyx mori. Insect Mol. Biol. 2005, 14, 339–352. [Google Scholar] [CrossRef]
- Matzke, M.; Kanno, T.; Daxinger, L.; Huettel, B.; Matzke, A.J.M. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell. Biol. 2009, 21, 357–376. [Google Scholar] [CrossRef]
- Piergentili, R. Multiple roles of the Y chromosome in the biology of Drosophila melanogaster. Sci. World J. 2010, 10, 1749–1767. [Google Scholar] [CrossRef]
- Kharchenko, P.V.; Xi, R.; Park, P.J. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat. Genet. 2011, 43, 1167–1169. [Google Scholar] [CrossRef]
- Bertocchi, N.A.; de Oliveira, T.D.; del Valle Garnero, A.; Coan, R.L.B.; Gunski, R.J.; Martins, C.; Torres, F.P. Distribution of CR1-like transposable element in woodpeckers (Aves Piciformes): Z sex chromosomes can act as a refuge for transposable elements. Chromosome Res. 2018, 26, 333–343. [Google Scholar] [CrossRef]
- García, E.; Cross, I.; Portela-Bens, S.; Rodríguez, M.E.; García-Angulo, A.; Molina, B.; Cuadrado, A.; Liehr, T.; Rebordinos, L. Integrative genetic map of repetitive DNA in the sole Solea senegalensis genome shows a Rex transposon located in a proto-sex chromosome. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Eigenmann [Apareiodon spp]. 1916. Available online: https://www.gbif.org/species/2354811 (accessed on 5 January 2022).
- Lönnberg [Chionodraco hamatus]. 1905. Available online: https://www.gbif.org/species/2389952 (accessed on 5 January 2022).
- Capriglione, T.; Odierna, G.; Caputo, V.; Canapa, A.; Olmo, E. Characterization of a Tc1-like transposon in the Antarctic ice-fish, Chionodraco hamatus. Gene 2002, 295, 193–198. [Google Scholar] [CrossRef]
- Furano, A.V.; Duvernell, D.D.; Boissinot, S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004, 20, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Cocca, E.; Petraccioli, A.; Morescalchi, M.A.; Odierna, G.; Capriglione, T. Laser microdissection-based analysis of the Y sex chromosome of the Antarctic fish Chionodraco hamatus (Notothenioidei, Channichthyidae). Comp. Cytogenet. 2015, 9, 1–15. [Google Scholar] [CrossRef]
- Gschwend, A.R.; Weingartner, L.A.; Moore, R.C.; Ming, R. The sex-specific region of sex chromosomes in animals and plants. Chromosome Res. 2012, 20, 57–69. [Google Scholar] [CrossRef]
- Linnaeus, C. Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis, 10th ed.; Typis Ioannis Thomae: Vindobonae, Vienna, 1758; Volume 1, p. 824. [Google Scholar] [CrossRef]
- Latham [Casuarius novaehollandiae]. 1790. Available online: https://www.gbif.org/species/9293251 (accessed on 5 January 2022).
- Lesson, R. [Ornismya anna]. 1829. Available online: https://www.gbif.org/species/11331860 (accessed on 5 January 2022).
- Bonaparte [Lycocorax pyrrhopterus]. 1850. Available online: https://www.gbif.org/species/2494874 (accessed on 5 January 2022).
- Puinongpo, W.; Singchat, W.; Petpradub, S.; Kraichak, E.; Nunome, M.; Laopichienpong, N.; Thongchum, R.; Intarasorn, T.; Sillapaprayoon, S.; Indananda, C.; et al. Existence of Bov-B LINE retrotransposons in snake lineages reveals recent multiple horizontal gene transfers with copy number variation. Genes 2020, 11, 1241. [Google Scholar] [CrossRef]
- Kordis, D. Transposable elements in reptilian and avian (Sauropsida) genomes. Cytogenet. Genome Res. 2010, 127, 94–111. [Google Scholar] [CrossRef]
- Voigt [Anolis carolinensis]. 1832. Available online: https://www.gbif.org/species/2466939 (accessed on 5 January 2022).
- Tanaka, Y.; Oyama, S.; Hori, S.; Ushio, K.; Shioi, N.; Terada, S.; Deshimaru, M. Accelerated evolution of fetuin family proteins in Protobothrops flavoviridis (habu snake) serum and the discovery of an L1-like genomic element in the intronic sequence of a fetuin-encoding gene. Biosci. Biotech. Biochem. 2013, 77, 582–590. [Google Scholar] [CrossRef][Green Version]
- Mezzasalma, M.; Dall’Asta, A.; Loy, A.; Cheylan, M.; Lymberakis, P.; Zuffi, M.A.L.; Tomovic, L.; Odierna, G.; Guarino, F.M. A sisters’ story: Comparative phylogeography and taxonomy of Hierophis viridiflavus and H. gemonensis (Serpentes, Colubridae). Zoolog. Scr. 2015, 44, 495–508. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Visone, V.; Petraccioli, A.; Odierna, G.; Capriglione, T.; Guarino, F.M. Non-random accumulation of LINE1-like sequences on differentiated snake W chromosomes. J. Zool. 2016, 300, 67–75. [Google Scholar] [CrossRef]
- Lacépède [Hierophis carbonarius]. 1789. Available online: https://reptile-database.reptarium.cz/species?genus=Hierophis&species=viridiflavus (accessed on 5 January 2022).
- Lacépède [Elaphe quatuorlineata]. 1789. Available online: https://www.gbif.org/species/9099408 (accessed on 5 January 2022).
- Aprea, G.; Gentilli, A.; Zuffi, M.A.L.; Odierna, G. The karyology of Vipera aspis, V. atra, V. hugyi, and Cerastes vipera. Amphib-reptil 2006, 27, 113–119. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Branch, W.R.; Glaw, F.; Guarino, F.M.; Nagy, Z.T.; Odierna, G.; Aprea, G. Chromosome evolution in pseudoxyrhophiine snakes from Madagascar: A wide range of karyotypic variability. Biol. J. Linn. Soc. 2014, 112, 450–460. [Google Scholar] [CrossRef]
- Marchal, J.A.; Acosta, M.J.; Bullejos, M.; Puerma, E.; Dıaz de la Guardia, R.; Sanchez, A. Distribution of L1- retroposons on the giant sex chromosomes of Microtus cabrerae (Arvicolidae, Rodentia): Functional and evolutionary implications. Chromosome Res. 2006, 14, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Melamed, E.; Arnold, A.P. The role of LINEs and CpG islands in dosage compensation on the chicken Z chromosome. Chromosome Res. 2009, 17, 726–736. [Google Scholar] [CrossRef][Green Version]
- Deininger, P.L.; Batzer, D.A. Mammalian retroelements. Genome Res. 2002, 12, 1455–1465. [Google Scholar] [CrossRef]
- Sellis, D.; Provata, A.; Almirantis, Y. Alu and LINE1 distributions in the human chromosomes: Evidence of global genomic organization expressed in the form of power laws. Mol. Biol. Evol. 2007, 24, 2385–2399. [Google Scholar] [CrossRef]
- Boissinot, S.; Davis, J.; Entezam, A.; Petrov, D.; Furano, A.V. Fitness cost of LINE-1 (L1) activity in humans. Proc. Natl. Acad. Sci. USA 2006, 103, 9590–9594. [Google Scholar] [CrossRef]
- Pontius, J.U.; Mullikin, J.C.; Smith, D.R.; Team, A.S.; LindbladToh, K.; Gnerre, S.; Clamp, M.; Chang, J.; Stephens, R.; Neelam, B.; et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007, 17, 1675–1689. [Google Scholar] [CrossRef]
- Novick, P.A.; Basta, H.; Floumanhaft, M.; McClure, M.A.; Boissinot, S. The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals. Mol. Biol. Evol. 2009, 26, 1811–1822. [Google Scholar] [CrossRef]
- Tollis, M.; Boissinot, S. The transposable element profile of the Anolis genome: How a lizard can provide insights into the evolution of vertebrate genome size and structure. Mob. Genet. Elem. 2011, 1, 107–111. [Google Scholar] [CrossRef][Green Version]
- Tollis, M.; Boissinot, S. Lizards and LINEs: Selection and demography affect the fate of L1 retrotransposons in the genome of the green anole (Anolis carolinensis). Genome Biol. Evol. 2013, 5, 1754–1768. [Google Scholar] [CrossRef] [PubMed]
- Sultana, T.; van Essen, D.; Siol, O.; Bailly-Bechet, M.; Philippe, C.; Aabidine, A.Z.E.; Pioger, L.; Nigumann, P.; Saccani, S.; Andrau, J.C. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol. Cell 2019, 74, 555–570. [Google Scholar] [CrossRef]
- Bailey, J.A.; Carrel, L.; Chakravarti, A.; Eichler, E.E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA 2000, 97, 6634–6639. [Google Scholar] [CrossRef]
- Presting, G.G.; Malysheva, L.; Fuchs, J.; Schubert, I. A TY3/GYPSY retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 1998, 16, 721–728. [Google Scholar] [CrossRef]
- Tubio, J.M.C.; Tojo, M.; Bassaganyas, L.; Escaramis, G.; Sharakhov, I.V.; Sharakhova, M.V.; Tornador, C.; Unger, M.F.; Naveira, H.; Costas, J.; et al. Evolutionary dynamics of the Ty3/gypsy LTR retrotransposons in the genome of Anopheles gambiae. PLoS ONE 2011, 6, e16328. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kralova, T.; Cegan, R.; Kubat, Z.; Vrana, J.; Vyskot, B.; Vogel, I.; Kejnovsky, E.; Hobza, R. Identification of a novel retrotransposon with sex chromosome-specific distribution in Silene latifolia. Cytogenet. Genome Res. 2014, 143, 87–95. [Google Scholar] [CrossRef]
- Li, N.; Li, X.; Zhou, J.; Yu, L.A.; Li, S.; Zhang, Y.; Qin, R.; Gao, W.; Deng, C. Genome-wide analysis of transposable elements and satellite DNAs in Spinacia species to shed light on their roles in sex chromosome evolution. Front. Plant Sci. 2021, 11, 575462. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Eickbush, T.H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990, 9, 3353–3362. [Google Scholar] [CrossRef]
- Lerat, E.; Capy, P. Retrotransposons and retroviruses: Analysis of the envelope gene. Mol. Biol. Evol. 1999, 16, 1198–1207. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pantazidis, A.; Labrador, M.; Fontdevila, A. The retrotransposon Osvaldo from D. buzzatii displays all structural features of functional retrovirus. Mol. Biol. Evol. 1999, 16, 909–921. [Google Scholar] [CrossRef][Green Version]
- Peterson-Burch, B.D.; Wright, D.A.; Laten, H.M.; Voytas, D.F. Retroviruses in plants? Trends Genet. 2000, 16, 151–152. [Google Scholar] [CrossRef]
- Terzian, C.; Ferraz, C.; Demaille, J.; Bucheton, A. Evolution of the gypsy endogenous retrovirus in the Drosophila melanogaster subgroup. Mol. Biol. Evol. 2000, 17, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, P.; Lessios, H.A. Evolution of sea urchin retroviral-like (SURL) elements: Evidence from 40 echinoid species. Mol. Biol. Evol. 1999, 16, 938–952. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Eickbush, T.H. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 1999, 73, 5186–5189. [Google Scholar] [CrossRef]
- Temminck and Schlegel [Fugu rubripes]. 1850. Available online: https://www.gbif.org/species/2407607 (accessed on 5 January 2022).
- Poulter, R.; Butler, M.; Ormandy, J. A LINE element from the pufferfish (fugu) Fugu rubripes which shows similarities to the CR1 family of non-LTR retrotransposons. Gene 1998, 227, 169–179. [Google Scholar] [CrossRef]
- Volff, J.N.; Körting, C.; Altschmied, J.; Duschl, J.; Sweeney, K.; Wichert, K.; Froschauer, A.; Schartl, M. Jule from the fish Xiphophorus is the first complete vertebrate Ty3/Gypsy retrotransposon from the Mag family. Mol. Biol. Evol. 2001, 18, 101–111. [Google Scholar] [CrossRef][Green Version]
- Tristem, M.; Kabat, P.; Herniou, E.H.; Karpas, A.; Hill, F. Ease1, a Gypsy LTR retrotransposon in the Salmonidae. Mol. Gen. Genet. 1995, 249, 229–236. [Google Scholar] [CrossRef]
- Marracci, S.; Batistoni, R.; Pesole, G.; Citti, L.; Nardi, I. Gypsy/Ty3-like elements in the genome of the terrestrial salamander Hydromantes (Amphibia, Urodela). J. Mol. Evol. 1996, 43, 584–593. [Google Scholar] [CrossRef]
- Miller, K.; Lynch, C.; Martin, J.; Herniou, E.; Tristem, M. Identification of multiple Gypsy LTR retrotransposons lineages in vertebrate genomes. J. Mol. Evol. 1999, 49, 358–366. [Google Scholar] [CrossRef]
- Marín, I.; Lloréns, C. Ty3/Gypsy retrotransposons: Description of new Arabidopsis thaliana elements and evolutionary perspectives derived from comparative genomic data. Mol. Biol. Evol. 2000, 17, 1040–1049. [Google Scholar] [CrossRef]
- Herédia, F.; Loreto, E.L.S.; Valente, V.L.S. Complex evolution of gypsy in Drosophilid species. Mol. Biol. Evol. 2004, 21, 1831–1842. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Willd. [Sagittaria latifolia] In: Sp. Pl. 4: 409, 1805. Available online: https://www.gbif.org/species/5328909 (accessed on 5 January 2022).
- Puterova, J.; Kubat, Z.; Kejnovsky, E.; Jesionek, W.; Cizkova, J.; Vyskot, B.; Hobza, R. The slowdown of Y chromosome expansion in dioecious Silene latifolia due to DNA loss and male-specific silencing of retrotransposons. BMC Genom. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Mourier, T.; Willerslev, E. Large-scale transcriptome data reveals transcriptional activity of fission yeast LTR retrotransposons. BMC Genom. 2010, 11, 167. [Google Scholar] [CrossRef] [PubMed]
- Nuzhdin, S.V.; Mackay, T. The genomic rate of transposable element movement in Drosophila melanogaster. Mol. Biol. Evol. 1995, 12, 180–181. [Google Scholar] [CrossRef] [PubMed]
- Rhie, A.; McCarthy, S.A.; Fedrigo, O.; Damas, J.; Formenti, G.; Koren, S.; Uliano-Silva, M.; Chow, W.; Fungtammasan, A.; Kim, J.; et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 2021, 592, 737–746. [Google Scholar] [CrossRef]
- Froschauer, A.; Körting, C.; Katagiri, T.; Aoki, T.; Asakawa, S.; Shimizu, N.; Schartl, M.; Volff, J.N. Construction and initial analysis of bacterial artificial chromosome (BAC) contigs from the sex-determining region of the platyfish Xiphophorus maculatus. Gene 2002, 295, 247–254. [Google Scholar] [CrossRef]
- Deakin, J.E.; Potter, S.; O’Neill, R.; Ruiz-Herrera, A.; Cioffi, M.B.; Eldridge, M.D.; Fukui, K.; Graves, J.A.M.; Griffin, D.; Grutzner, F.; et al. Chromosomics: Bridging the gap between genomes and chromosomes. Genes 2019, 10, 627. [Google Scholar] [CrossRef]
- Bao, Z.; Eddy, S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002, 12, 1269–1276. [Google Scholar] [CrossRef]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21, i351–i358. [Google Scholar] [CrossRef]
- Ellinghaus, D.; Kurtz, S.; Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 2008, 9, 18. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. 2013. Available online: http://www.repeatmasker.org (accessed on 5 January 2022).
- Ou, S.; Su, W.; Liao, Y.; Chougule, K.; Agda, J.R.A.; Hellinga, A.J.; Lugo, C.S.B.; Elliott, T.A.; Ware, D.; Peterson, T.; et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019, 20, 275. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Böhne, A.; Brunet, F.; Galiana-Arnoux, D.; Schultheis, C.; Volff, J.N. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008, 16, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Lerat, E.; Casacuberta, J.; Chaparro, C.; Vieira, C. On the Importance to Acknowledge Transposable Elements in Epigenomic Analyses. Genes 2019, 10, 258. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srikulnath, K.; Ahmad, S.F.; Singchat, W.; Panthum, T. Do Ty3/Gypsy Transposable Elements Play Preferential Roles in Sex Chromosome Differentiation? Life 2022, 12, 522. https://doi.org/10.3390/life12040522
Srikulnath K, Ahmad SF, Singchat W, Panthum T. Do Ty3/Gypsy Transposable Elements Play Preferential Roles in Sex Chromosome Differentiation? Life. 2022; 12(4):522. https://doi.org/10.3390/life12040522
Chicago/Turabian StyleSrikulnath, Kornsorn, Syed Farhan Ahmad, Worapong Singchat, and Thitipong Panthum. 2022. "Do Ty3/Gypsy Transposable Elements Play Preferential Roles in Sex Chromosome Differentiation?" Life 12, no. 4: 522. https://doi.org/10.3390/life12040522
APA StyleSrikulnath, K., Ahmad, S. F., Singchat, W., & Panthum, T. (2022). Do Ty3/Gypsy Transposable Elements Play Preferential Roles in Sex Chromosome Differentiation? Life, 12(4), 522. https://doi.org/10.3390/life12040522








