SIRT1: A Key Player in Male Reproduction
Abstract
1. Introduction
2. Sirtuin 1 (SIRT1)
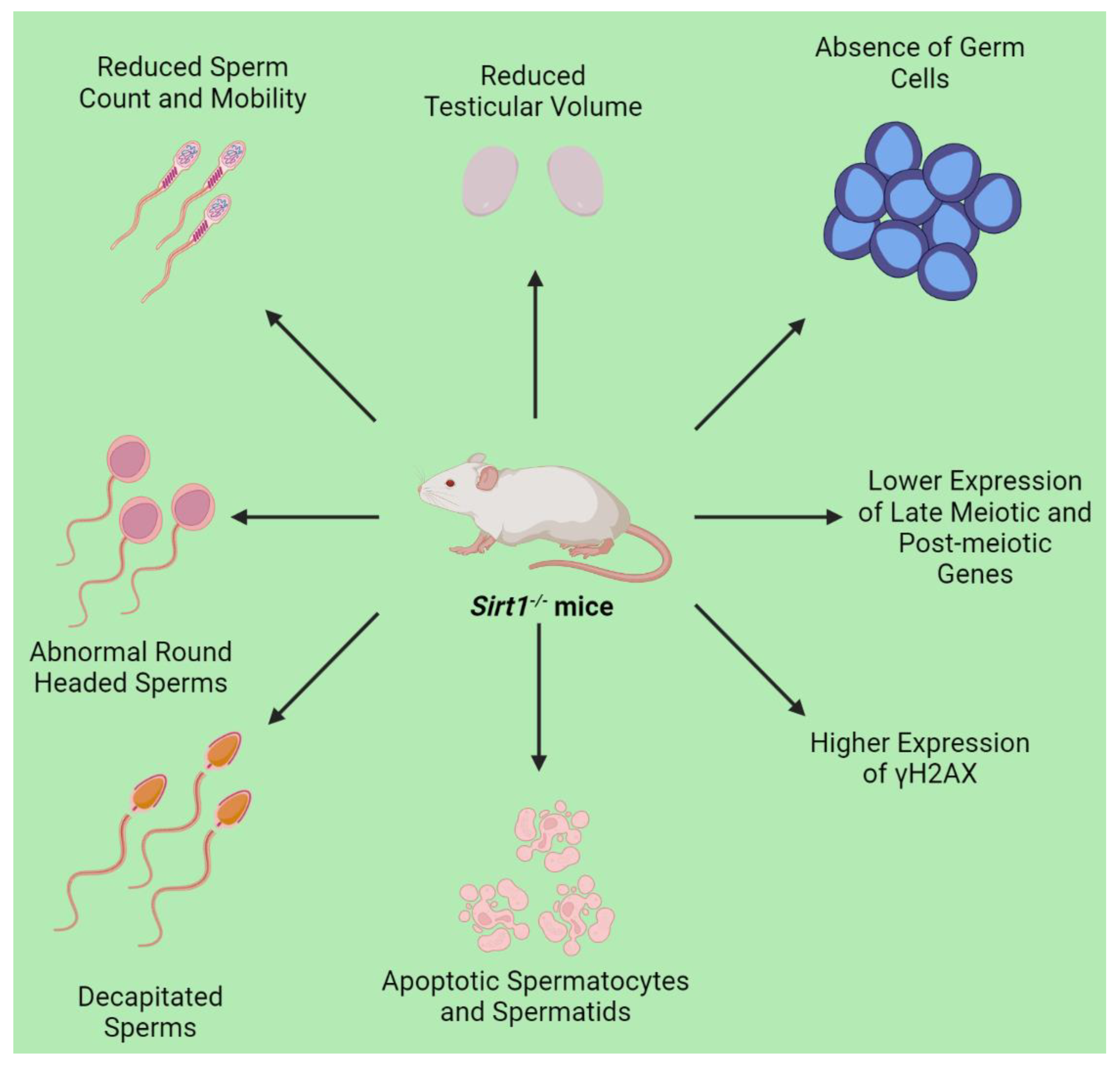
3. SIRT1 in Male Germ Cells
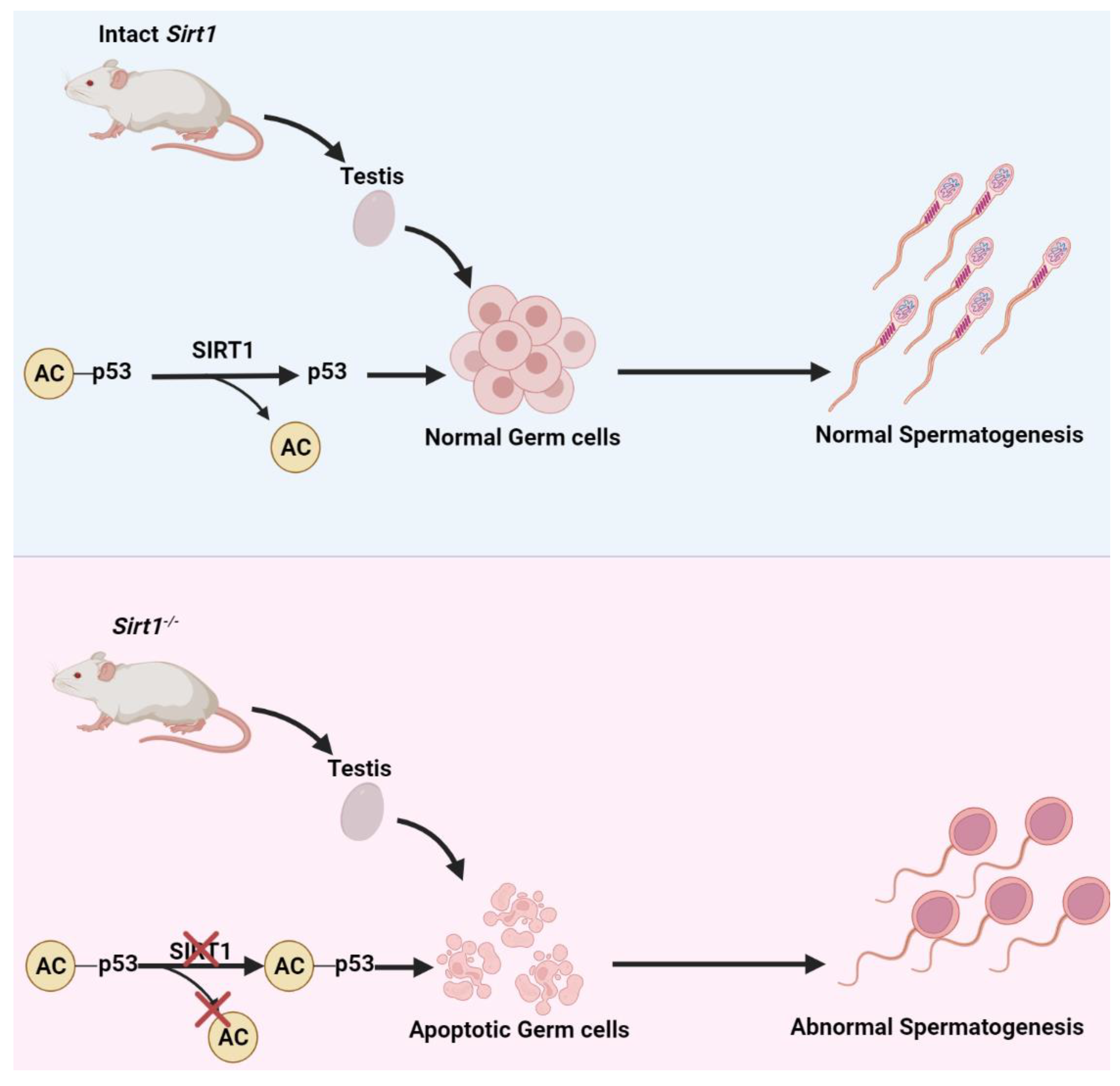
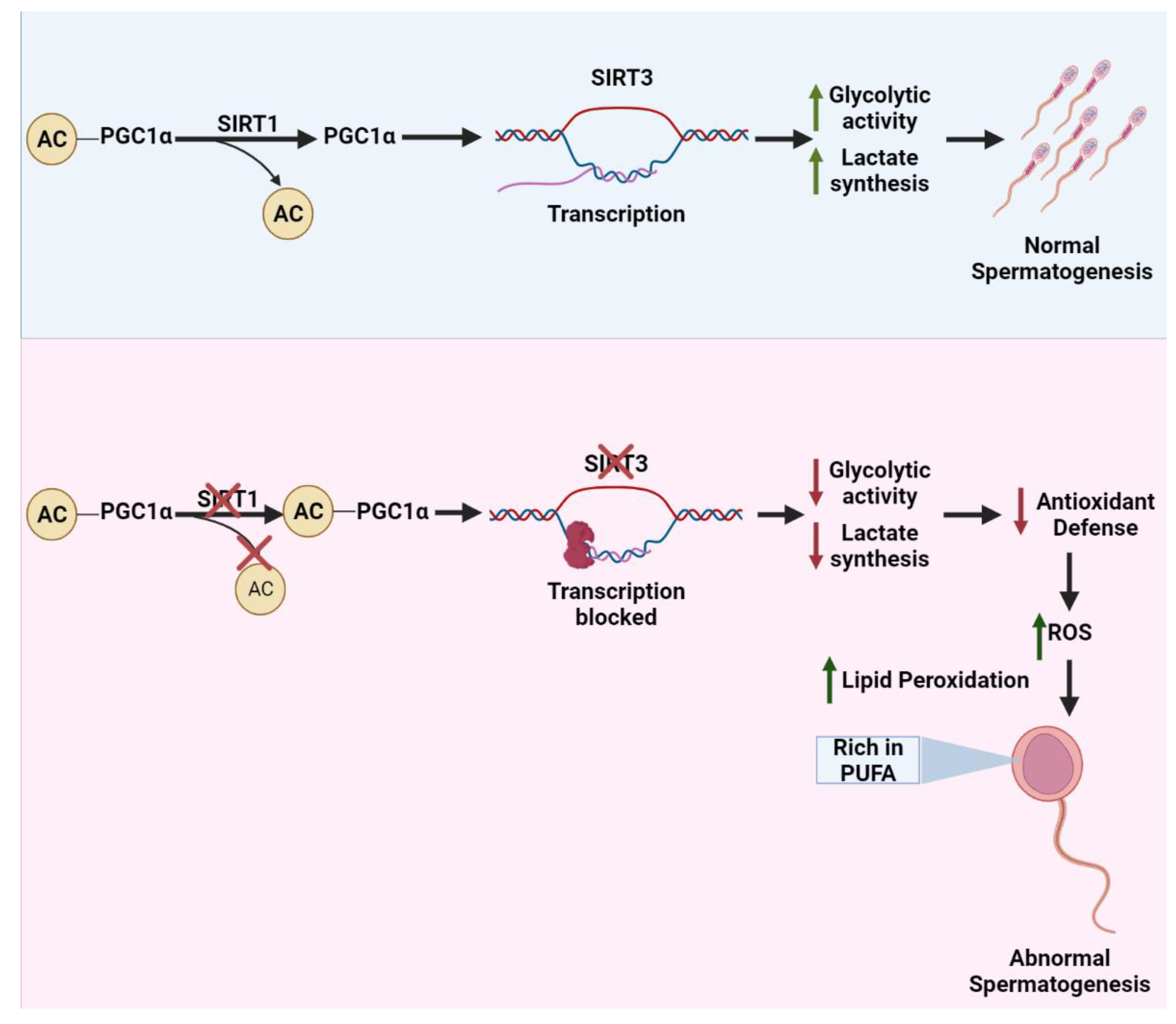
3.1. Mechanism Underlying SIRT1-Related Germ Cell Death
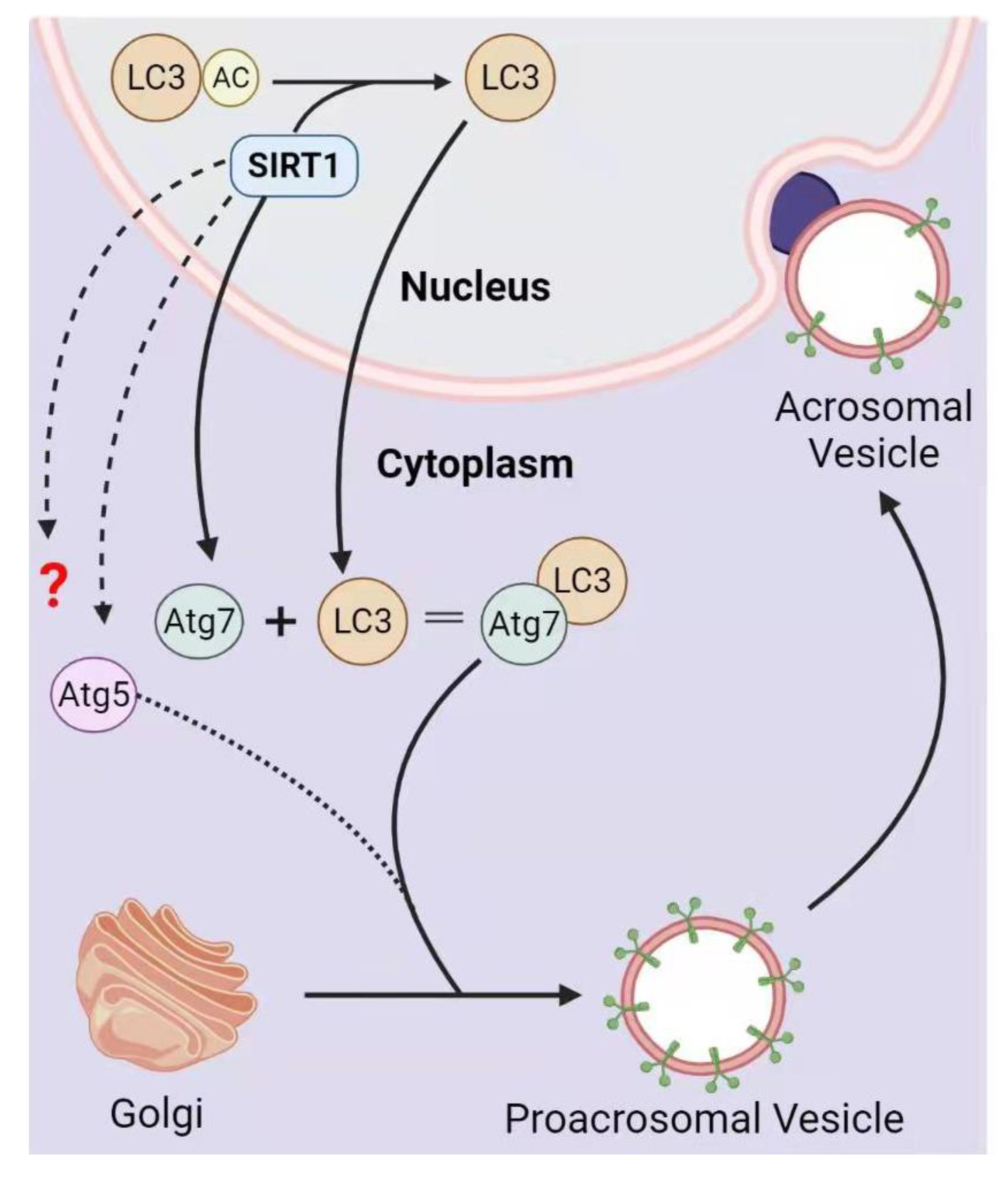
3.2. SIRT1 in Acrosome Biogenesis
3.3. SIRT1-Mediated Protamine Replacement
4. SIRT1 Functions in the Hypothalamic-Pituitary-Gonadal (HPG) Axis
5. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vaquero, A. The conserved role of sirtuins in chromatin regulation. Int. J. Dev. Biol. 2009, 53, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Li, X. Sirtuins in metabolic and epigenetic regulation of stem cells. Sirtuin Biol. Cancer Metab. Dis. 2021, 25–37. [Google Scholar] [CrossRef]
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 119–141. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Xiong, Z.; Gu, J.; Sun, Z.; Shuai, J.; Fan, D.; Li, W. Sirtuins: Potential Therapeutic Targets for Defense against Oxidative Stress in Spinal Cord Injury. Oxidative Med. Cell. Longev. 2021, 2021, 7207692. [Google Scholar] [CrossRef] [PubMed]
- Donlon, T.A.; Morris, B.J.; Chen, R.; Masaki, K.H.; Allsopp, R.C.; Willcox, D.C.; Tiirikainen, M.; Willcox, B.J. Analysis of polymorphisms in 59 potential candidate genes for association with human longevity. J. Gerontol. Ser. A 2018, 73, 1459–1464. [Google Scholar] [CrossRef]
- Tian, X.; Firsanov, D.; Zhang, Z.; Cheng, Y.; Luo, L.; Tombline, G.; Tan, R.; Simon, M.; Henderson, S.; Steffan, J.; et al. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell 2019, 177, 622–638.e622. [Google Scholar] [CrossRef]
- Kanfi, Y.; Naiman, S.; Amir, G.; Peshti, V.; Zinman, G.; Nahum, L.; Bar-Joseph, Z.; Cohen, H.Y. The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012, 483, 218–221. [Google Scholar] [CrossRef]
- Satoh, A.; Brace, C.S.; Rensing, N.; Cliften, P.; Wozniak, D.F.; Herzog, E.D.; Yamada, K.A.; Imai, S.I. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013, 18, 416–430. [Google Scholar] [CrossRef]
- Mostoslavsky, R.; Chua, K.F.; Lombard, D.B.; Pang, W.W.; Fischer, M.R.; Gellon, L.; Liu, P.; Mostoslavsky, G.; Franco, S.; Murphy, M.M. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006, 124, 315–329. [Google Scholar] [CrossRef]
- Vazquez, B.N.; Thackray, J.K.; Simonet, N.G.; Kane-Goldsmith, N.; Martinez-Redondo, P.; Nguyen, T.; Bunting, S.; Vaquero, A.; Tischfield, J.A.; Serrano, L. SIRT 7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 2016, 35, 1488–1503. [Google Scholar] [CrossRef]
- Haigis, M.C.; Deng, C.-X.; Finley, L.W.; Kim, H.-S.; Gius, D. SIRT3 is a mitochondrial tumor suppressor: A scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res. 2012, 72, 2468–2472. [Google Scholar] [CrossRef]
- Jeong, S.M.; Hwang, S.; Seong, R.H. SIRT4 regulates cancer cell survival and growth after stress. Biochem. Biophys. Res. Commun. 2016, 470, 251–256. [Google Scholar] [CrossRef]
- Serrano, L.; Martínez-Redondo, P.; Marazuela-Duque, A.; Vazquez, B.N.; Dooley, S.J.; Voigt, P.; Beck, D.B.; Kane-Goldsmith, N.; Tong, Q.; Rabanal, R.M.; et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013, 27, 639–653. [Google Scholar] [CrossRef]
- Kratz, E.M.; Sołkiewicz, K.; Kubis-Kubiak, A.; Piwowar, A. Sirtuins as important factors in pathological states and the role of their molecular activity modulators. Int. J. Mol. Sci. 2021, 22, 630. [Google Scholar] [CrossRef]
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 2005, 16, 4623–4635. [Google Scholar] [CrossRef]
- Gomes, P.; Outeiro, T.F.; Cavadas, C. Emerging role of sirtuin 2 in the regulation of mammalian metabolism. Trends Pharmacol. Sci. 2015, 36, 756–768. [Google Scholar] [CrossRef]
- Haigis, M.C.; Guarente, L.P. Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006, 20, 2913–2921. [Google Scholar] [CrossRef]
- Chianese, R.; Viggiano, A.; Urbanek, K.; Cappetta, D.; Troisi, J.; Scafuro, M.; Guida, M.; Esposito, G.; Ciuffreda, L.P.; Rossi, F. Chronic exposure to low dose of bisphenol A impacts on the first round of spermatogenesis via SIRT1 modulation. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Ding, R.-B.; Bao, J.; Deng, C. Emerging roles of SIRT1 in fatty liver diseases. Int. J. Biol. Sci. 2017, 13, 852. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, F.; Ge, X.; Yan, T.; Chen, X.; Shi, X.; Zhai, Q. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007, 6, 307–319. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Liang, Y.; Vanhoutte, P. SIRT1 in metabolic syndrome: Where to target matters. Pharmacol. Ther. 2012, 136, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Xu, Y.; Wan, W.; Shou, X.; Qian, J.; You, Z.; Liu, B.; Chang, C.; Zhou, T.; Lippincott-Schwartz, J. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 2015, 57, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, M.; Yang, X.; Jardine, K.; He, X.; Jacobsen, K.; Staines, W.; Harper, M.; McBurney, M. The Sirt1 deacetylase modulates the insulin-like growth factor signaling pathway in mammals. Mech. Ageing Dev. 2005, 126, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- de Mello, N.P.; Orellana, A.M.; Mazucanti, C.H.; de Morais Lima, G.; Scavone, C.; Kawamoto, E. Insulin and autophagy in neurodegeneration. Front. Neurosci. 2019, 13, 491. [Google Scholar] [CrossRef]
- Henrique Mazucanti, C.; Victor Cabral-Costa, J.; Rodrigues Vasconcelos, A.; Zukas Andreotti, D.; Scavone, C.; Mitiko Kawamoto, E. Longevity pathways (mTOR, SIRT, Insulin/IGF-1) as key modulatory targets on aging and neurodegeneration. Curr. Top. Med. Chem. 2015, 15, 2116–2138. [Google Scholar] [CrossRef]
- Xu, J.; Jackson, C.W.; Khoury, N.; Escobar, I.; Perez-Pinzon, M. Brain SIRT1 mediates metabolic homeostasis and neuroprotection. Front. Endocrinol. 2018, 9, 702. [Google Scholar] [CrossRef]
- Chang, H.-C.; Guarente, L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. 2014, 25, 138–145. [Google Scholar] [CrossRef]
- Chalkiadaki, A.; Guarente, L. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab. 2012, 16, 180–188. [Google Scholar] [CrossRef]
- Gillum, M.P.; Kotas, M.E.; Erion, D.M.; Kursawe, R.; Chatterjee, P.; Nead, K.T.; Muise, E.S.; Hsiao, J.J.; Frederick, D.W.; Yonemitsu, S. SirT1 regulates adipose tissue inflammation. Diabetes 2011, 60, 3235–3245. [Google Scholar] [CrossRef]
- Picard, F.; Kurtev, M.; Chung, N.; Topark-Ngarm, A.; Senawong, T.; De Oliveira, R.M.; Leid, M.; McBurney, M.W.; Guarente, L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 2004, 429, 771–776. [Google Scholar] [CrossRef]
- Purushotham, A.; Schug, T.T.; Xu, Q.; Surapureddi, S.; Guo, X.; Li, X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009, 9, 327–338. [Google Scholar] [CrossRef]
- Rodgers, J.T.; Puigserver, P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. USA 2007, 104, 12861–12866. [Google Scholar] [CrossRef]
- Boutant, M.; Cantó, C. SIRT1 metabolic actions: Integrating recent advances from mouse models. Mol. Metab. 2014, 3, 5–18. [Google Scholar] [CrossRef]
- Moynihan, K.A.; Grimm, A.A.; Plueger, M.M.; Bernal-Mizrachi, E.; Ford, E.; Cras-Méneur, C.; Permutt, M.A.; Imai, S. Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005, 2, 105–117. [Google Scholar] [CrossRef]
- Bell, E.L.; Nagamori, I.; Williams, E.O.; Del Rosario, A.M.; Bryson, B.D.; Watson, N.; White, F.M.; Sassone-Corsi, P.; Guarente, L.J.D. SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 2014, 141, 3495–3504. [Google Scholar] [CrossRef]
- McBurney, M.W.; Yang, X.; Jardine, K.; Hixon, M.; Boekelheide, K.; Webb, J.R.; Lansdorp, P.M.; Lemieux, M.J.M. The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 2003, 23, 38–54. [Google Scholar] [CrossRef]
- Coussens, M.; Maresh, J.G.; Yanagimachi, R.; Maeda, G.; Allsopp, R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS ONE 2008, 3, e1571. [Google Scholar] [CrossRef]
- Seifert, E.L.; Caron, A.Z.; Morin, K.; Coulombe, J.; He, X.H.; Jardine, K.; Dewar-Darch, D.; Boekelheide, K.; Harper, M.E.; McBurney, M. SirT1 catalytic activity is required for male fertility and metabolic homeostasis in mice. FASEB J. 2012, 26, 555–566. [Google Scholar] [CrossRef]
- Allemand, I.; Anglo, A.; Jeantet, A.-Y.; Cerutti, I.; May, E.J.O. Testicular wild-type p53 expression in transgenic mice induces spermiogenesis alterations ranging from differentiation defects to apoptosis. Oncogene 1999, 18, 6521–6530. [Google Scholar] [CrossRef]
- Beumer, T.L.; Roepers-Gajadien, H.L.; Gademan, I.S.; van Buul, P.P.; Gil-Gomez, G.; Rutgers, D.H.; de Rooij, D. The role of the tumor suppressor p53 in spermatogenesis. Cell Death Differ. 1998, 5, 669–677. [Google Scholar] [CrossRef]
- Yin, Y.; Stahl, B.C.; DeWolf, W.C.; Morgentaler, A. p53-mediated germ cell quality control in spermatogenesis. Dev. Biol. 1998, 204, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Kolthur-Seetharam, U.; Teerds, K.; de Rooij, D.G.; Wendling, O.; McBurney, M.; Sassone-Corsi, P.; Davidson, I. The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol. Reprod. 2009, 80, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Bouras, T.; Fu, M.; Sauve, A.A.; Wang, F.; Quong, A.A.; Perkins, N.D.; Hay, R.T.; Gu, W.; Pestell, R. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem. 2005, 280, 10264–10276. [Google Scholar] [CrossRef] [PubMed]
- Stankovic-Valentin, N.; Deltour, S.; Seeler, J.; Pinte, S.; Vergoten, G.; Guérardel, C.; Dejean, A.; Leprince, D.J.M. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved ψKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol. Cell. Biol. 2007, 27, 2661–2675. [Google Scholar] [CrossRef]
- Andreou, A.M.; Tavernarakis, N. SUMOylation and cell signalling. Biotechnol. J. Healthc. Nutr. Technol. 2009, 4, 1740–1752. [Google Scholar] [CrossRef]
- Brown, P.W.; Hwang, K.; Schlegel, P.N.; Morris, P. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum. Reprod. 2008, 23, 2850–2857. [Google Scholar] [CrossRef]
- Metzler-Guillemain, C.; Depetris, D.; Luciani, J.J.; Mignon-Ravix, C.; Mitchell, M.J.; Mattei, M. In human pachytene spermatocytes, SUMO protein is restricted to the constitutive heterochromatin. Chromosome Res. 2008, 16, 761–782. [Google Scholar] [CrossRef]
- Rogers, R.S.; Inselman, A.; Handel, M.A.; Matunis, M. SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma 2004, 113, 233–243. [Google Scholar] [CrossRef]
- Vigodner, M.; Ishikawa, T.; Schlegel, P.N.; Morris, P. Metabolism: SUMO-1, human male germ cell development, and the androgen receptor in the testis of men with normal and abnormal spermatogenesis. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E1022–E1033. [Google Scholar] [CrossRef]
- Vigodner, M.; Morris, P. Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: Silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev. Biol. 2005, 282, 480–492. [Google Scholar] [CrossRef]
- Vigodner, M. Sumoylation precedes accumulation of phosphorylated H2AX on sex chromosomes during their meiotic inactivation. Chromosome Res. 2009, 17, 37–45. [Google Scholar] [CrossRef]
- Kong, X.; Wang, R.; Xue, Y.; Liu, X.; Zhang, H.; Chen, Y.; Fang, F.; Chang, Y. Sirtuin 3, a new target of PGC-1α, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE 2010, 5, e11707. [Google Scholar] [CrossRef]
- Rato, L.; Duarte, A.I.; Tomás, G.D.; Santos, M.S.; Moreira, P.I.; Socorro, S.; Cavaco, J.E.; Alves, M.G.; Oliveira, P. Pre-diabetes alters testicular PGC1-α/SIRT3 axis modulating mitochondrial bioenergetics and oxidative stress. Biochim. Et Biophys. Acta-Bioenerg. 2014, 1837, 335–344. [Google Scholar] [CrossRef]
- Rato, L.; Alves, M.; Dias, T.; Lopes, G.; Cavaco, J.; Socorro, S.; Oliveira, P. High-energy diets may induce a pre-diabetic state altering testicular glycolytic metabolic profile and male reproductive parameters. Andrology 2013, 1, 495–504. [Google Scholar] [CrossRef]
- Ye, X.; Li, M.; Hou, T.; Gao, T.; Zhu, W.-G.; Yang, Y. Sirtuins in glucose and lipid metabolism. Oncotarget 2017, 8, 1845. [Google Scholar] [CrossRef]
- Boussouar, F.; Benahmed, M. Lactate and energy metabolism in male germ cells. Trends Endocrinol. 2004, 15, 345–350. [Google Scholar] [CrossRef]
- Oliveira, P.F.; Martins, A.D.; Moreira, A.C.; Cheng, C.Y.; Alves, M. The Warburg effect revisited—Lesson from the Sertoli cell. Med. Res. Rev. 2015, 35, 126–151. [Google Scholar] [CrossRef]
- Rato, L.; GAlves, M.; MSilva, B.; Sousa, M.; Oliveira, P. Sirtuins: Novel players in male reproductive health. Curr. Med. Chem. 2016, 23, 1084–1099. [Google Scholar] [CrossRef]
- Tremellen, K. Oxidative stress and male infertility—A clinical perspective. Hum. Reprod. Update 2008, 14, 243–258. [Google Scholar] [CrossRef]
- Aitken, R.; Harkiss, D.; Buckingham, D. Analysis of lipid peroxidation mechanisms in human spermatozoa. Mol. Reprod. Dev. 1993, 35, 302–315. [Google Scholar] [CrossRef]
- Barbonetti, A.; Cinque, B.; Vassallo, M.R.C.; Mineo, S.; Francavilla, S.; Cifone, M.G.; Francavilla, F.J.F. Effect of vaginal probiotic lactobacilli on in vitro–induced sperm lipid peroxidation and its impact on sperm motility and viability. Fertil. Steril. 2011, 95, 2485–2488. [Google Scholar] [CrossRef]
- Wagner, B.A.; Buettner, G.R.; Burns, C. Free radical-mediated lipid peroxidation in cells: Oxidizability is a function of cell lipid bis-allylic hydrogen content. Biochemistry 1994, 33, 4449–4453. [Google Scholar] [CrossRef]
- Wu, C.-C.; Bratton, S. Regulation of the intrinsic apoptosis pathway by reactive oxygen species. Antioxid. Redox Signal. 2013, 19, 546–558. [Google Scholar] [CrossRef]
- Cheng, H.-L.; Mostoslavsky, R.; Saito Si Manis, J.P.; Gu, Y.; Patel, P.; Bronson, R.; Appella, E.; Alt, F.W.; Chua, K. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 2003, 100, 10794–10799. [Google Scholar] [CrossRef]
- Liu, C.; Song, Z.; Wang, L.; Yu, H.; Liu, W.; Shang, Y.; Xu, Z.; Zhao, H.; Gao, F.; Wen, J. Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 2017, 144, 441–451. [Google Scholar]
- Mostafa, T.; Nabil, N.; Rashed, L.; Makeen, K.; El-Kasas, M.; Mohamaed, H. Seminal SIRT 1 expression in infertile oligoasthenoteratozoospermic men with varicocoele. Andrology 2018, 6, 301–305. [Google Scholar] [CrossRef]
- Cho, C.; Willis, W.D.; Goulding, E.H.; Jung-Ha, H.; Choi, Y.-C.; Hecht, N.B.; Eddy, E. Haploinsufficiency of protamine-1 or-2 causes infertility in mice. Nat. Genet. 2001, 28, 82–86. [Google Scholar] [CrossRef]
- Montellier, E.; Boussouar, F.; Rousseaux, S.; Zhang, K.; Buchou, T.; Fenaille, F.; Shiota, H.; Debernardi, A.; Héry, P.; Curtet, S.J.G.; et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 2013, 27, 1680–1692. [Google Scholar] [CrossRef]
- Morinière, J.; Rousseaux, S.; Steuerwald, U.; Soler-López, M.; Curtet, S.; Vitte, A.-L.; Govin, J.; Gaucher, J.; Sadoul, K.; Hart, D. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 2009, 461, 664–668. [Google Scholar] [CrossRef]
- Gaucher, J.; Boussouar, F.; Montellier, E.; Curtet, S.; Buchou, T.; Bertrand, S.; Hery, P.; Jounier, S.; Depaux, A.; Vitte, A. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 2012, 31, 3809–3820. [Google Scholar] [CrossRef]
- Zini, A.; Libman, J. Sperm DNA damage: Clinical significance in the era of assisted reproduction. Cmaj 2006, 175, 495–500. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Palmer, N.O.; Fullston, T.; Mitchell, M.; Setchell, B.P.; Lane, M. SIRT6 in mouse spermatogenesis is modulated by diet-induced obesity. Reprod. Fertil. Dev. 2011, 23, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Takahashi, Y. The essential role of SIRT1 in hypothalamic-pituitary axis. Front. Endocrinol. 2018, 9, 605. [Google Scholar] [CrossRef] [PubMed]
- Sam, S.; Frohman, L. Normal physiology of hypothalamic pituitary regulation. Endocrinol. Metab. Clin. N. Am. 2008, 37, 1–22. [Google Scholar] [CrossRef]
- Veldhuis, J.D.; Keenan, D.M.; Liu, P.Y.; Iranmanesh, A.; Takahashi, P.Y.; Nehra, A. The aging male hypothalamic–pituitary–gonadal axis: Pulsatility and feedback. Mol. Cell. Endocrinol. 2009, 299, 14–22. [Google Scholar] [CrossRef]
- Lannes, J.; L’Hôte, D.; Garrel, G.; Laverrière, J.-N.; Cohen-Tannoudji, J. Quérat BJMe: Rapid communication: A microRNA-132/212 pathway mediates GnRH activation of FSH expression. Mol. Endocrinol. 2015, 29, 364–372. [Google Scholar] [CrossRef]
- Vazquez, M.; Toro, C.; Castellano, J.; Ruiz-Pino, F.; Roa, J.; Beiroa, D.; Heras, V.; Velasco, I.; Dieguez, C.; Pinilla, L. SIRT1 mediates obesity-and nutrient-dependent perturbation of pubertal timing by epigenetically controlling Kiss1 expression. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Di Sante, G.; Wang, L.; Wang, C.; Jiao, X.; Casimiro, M.C.; Chen, K.; Pestell, T.G.; Yaman, I.; Di Rocco, A.; Sun, X. Sirt1-deficient mice have hypogonadotropic hypogonadism due to defective GnRH neuronal migration. Mol. Endocrinol. 2015, 29, 200–212. [Google Scholar] [CrossRef][Green Version]
- Ramadori, G.; Fujikawa, T.; Anderson, J.; Berglund, E.D.; Frazao, R.; Michán, S.; Vianna, C.R.; Sinclair, D.A.; Elias, C.F.; Coppari, R. SIRT1 deacetylase in SF1 neurons protects against metabolic imbalance. Cell Metab. 2011, 14, 301–312. [Google Scholar] [CrossRef]
- Ramadori, G.; Fujikawa, T.; Fukuda, M.; Anderson, J.; Morgan, D.A.; Mostoslavsky, R.; Stuart, R.C.; Perello, M.; Vianna, C.R.; Nillni, E. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010, 12, 78–87. [Google Scholar] [CrossRef]
- Sasaki, T.; Kikuchi, O.; Shimpuku, M.; Susanti, V.Y.; Yokota-Hashimoto, H.; Taguchi, R.; Shibusawa, N.; Sato, T.; Tang, L.; Amano, K.J.D. Hypothalamic SIRT1 prevents age-associated weight gain by improving leptin sensitivity in mice. Diabetologia 2014, 57, 819–831. [Google Scholar] [CrossRef]
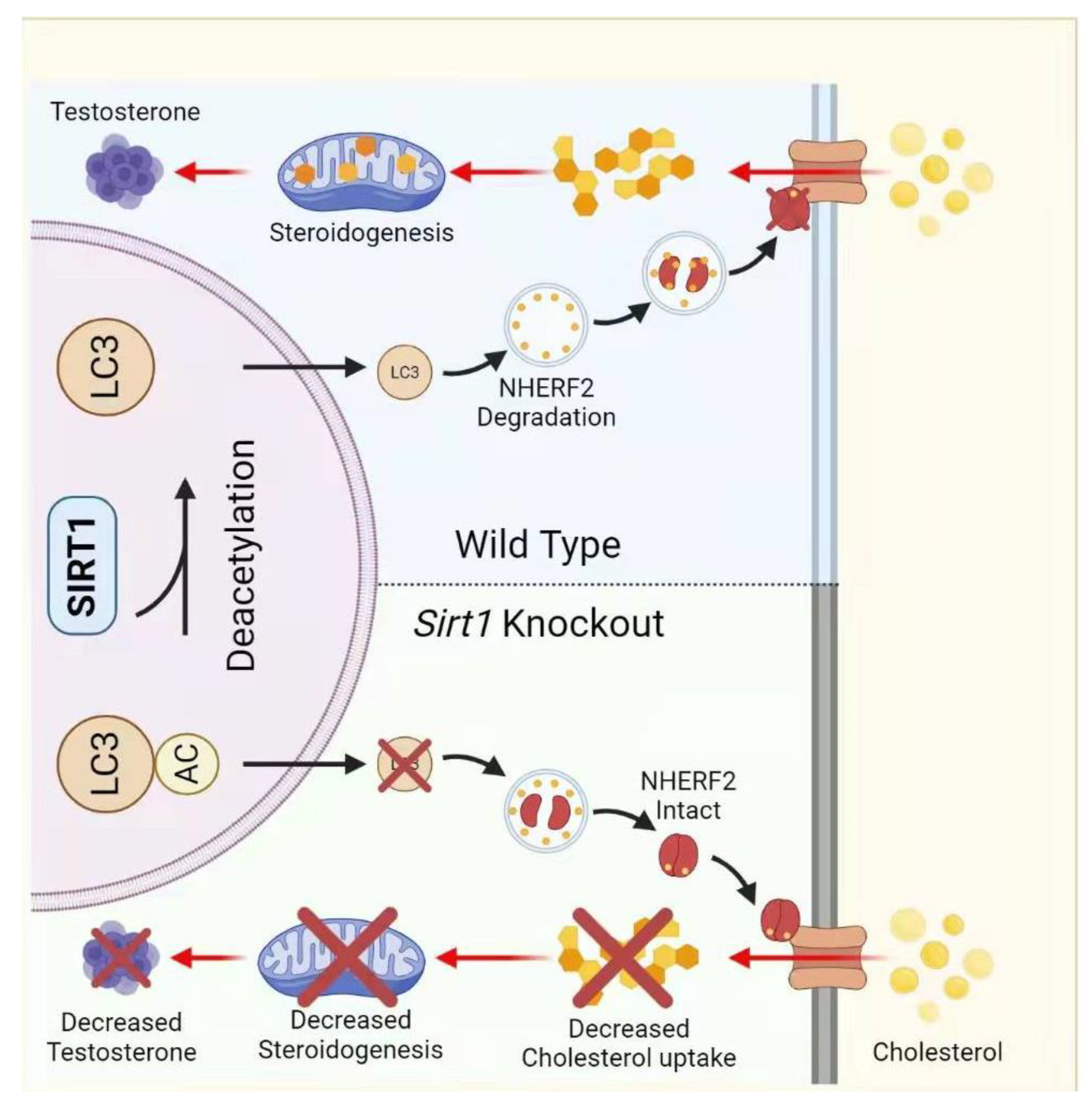
- Khawar, M.B.; Liu, C.; Gao, F.; Gao, H.; Liu, W.; Han, T.; Wang, L.; Li, G.; Jiang, H.; Li, W.J.P.; et al. Sirt1 regulates testosterone biosynthesis in Leydig cells via modulating autophagy. Protein Cell 2021, 12, 67–75. [Google Scholar] [CrossRef]
- Leisegang, K.; Henkel, R. The in vitro modulation of steroidogenesis by inflammatory cytokines and insulin in TM3 Leydig cells. Reprod. Biol. Endocrinol. 2018, 16, 1–11. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, A.; Sun, Y.; Zhu, X.; Fan, W.; Lu, X.; Yang, Q.; Feng, Y. Sirt1 exerts anti-inflammatory effects and promotes steroidogenesis in Leydig cells. Fertil. Steril. 2012, 98, 194–199. [Google Scholar] [CrossRef]
- Wei, H.; Khawar, M.B.; Tang, W.; Wang, L.; Wang, L.; Liu, C.; Jiang, H.; Li, W. Sirt6 is required for spermatogenesis in mice. Aging (Albany NY) 2020, 12, 17099. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khawar, M.B.; Sohail, A.M.; Li, W. SIRT1: A Key Player in Male Reproduction. Life 2022, 12, 318. https://doi.org/10.3390/life12020318
Khawar MB, Sohail AM, Li W. SIRT1: A Key Player in Male Reproduction. Life. 2022; 12(2):318. https://doi.org/10.3390/life12020318
Chicago/Turabian StyleKhawar, Muhammad Babar, Abdullah Muhammad Sohail, and Wei Li. 2022. "SIRT1: A Key Player in Male Reproduction" Life 12, no. 2: 318. https://doi.org/10.3390/life12020318
APA StyleKhawar, M. B., Sohail, A. M., & Li, W. (2022). SIRT1: A Key Player in Male Reproduction. Life, 12(2), 318. https://doi.org/10.3390/life12020318





