Time-Course of Redox Status, Redox-Related, and Mitochondrial-Dynamics-Related Gene Expression after an Acute Bout of Different Physical Exercise Protocols
Abstract
:1. Introduction
2. Methods
2.1. Animals
2.2. Exercise Protocol
2.3. Determination of Total Peroxidase and Glutathione Peroxidase Activities in Muscle Samples
2.4. Determination of Citrate Synthase (CS) Activity
2.5. Quantitation of mRNA by RT-qPCR
2.6. Statistical Analysis
3. Results
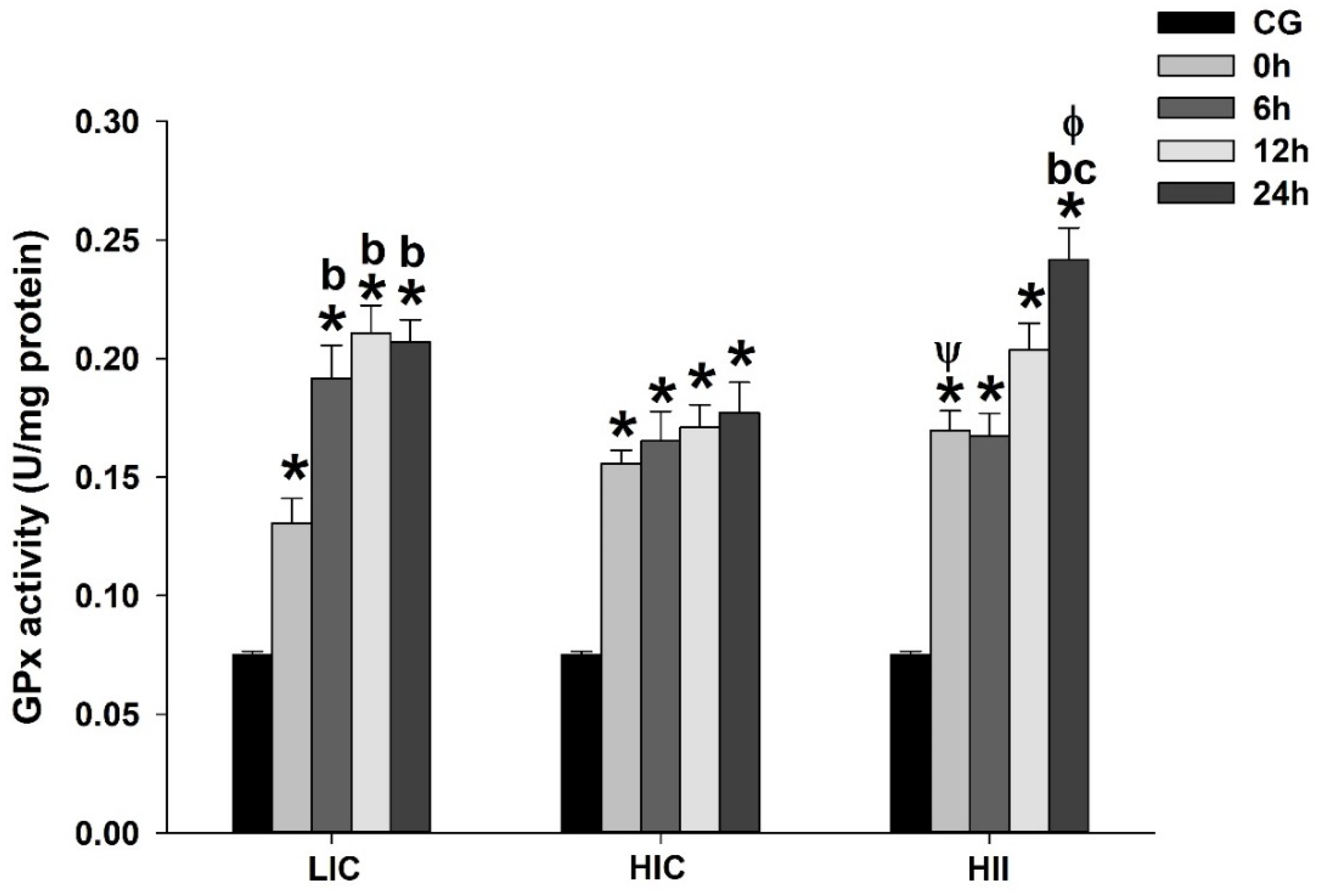
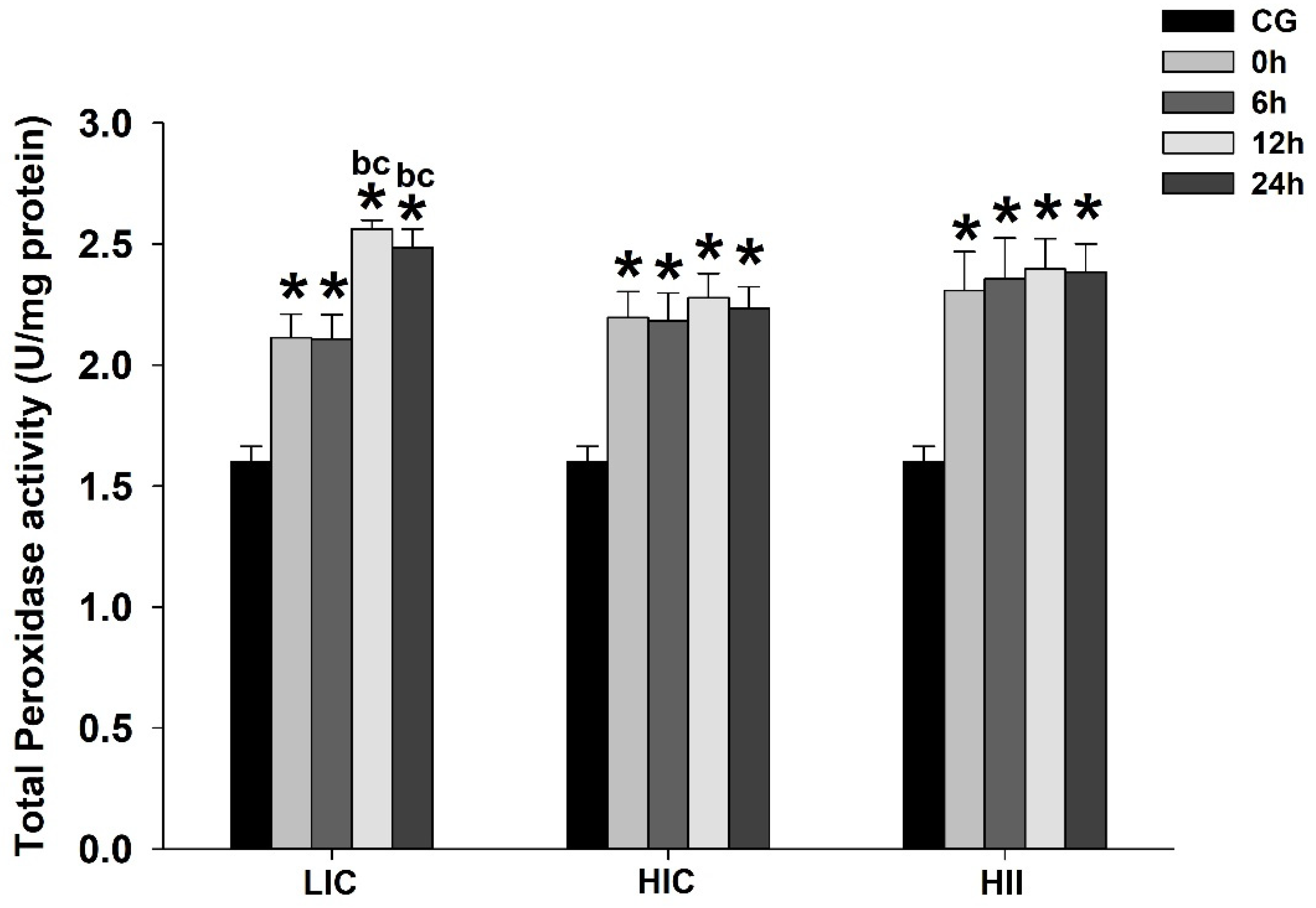
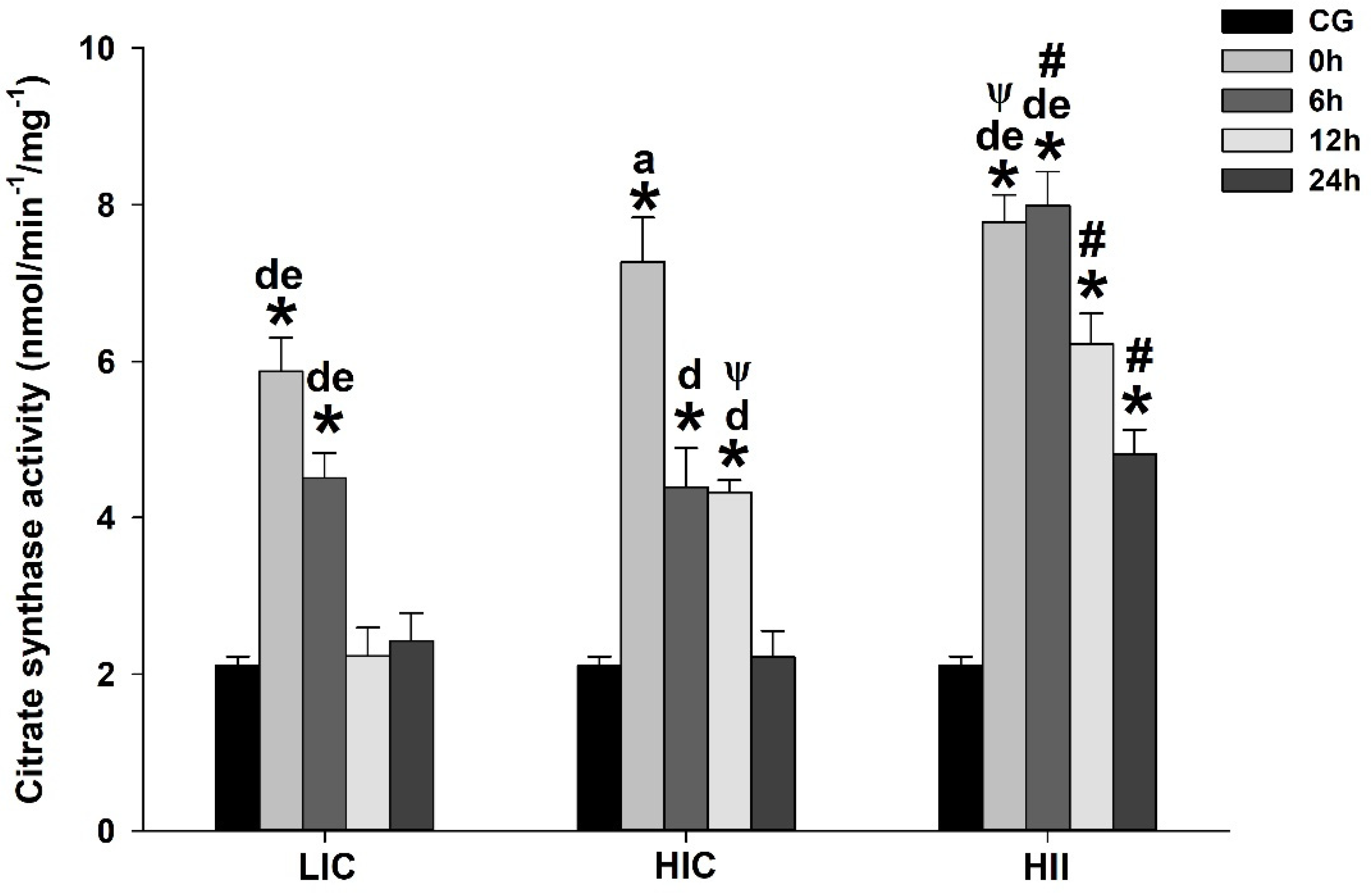
3.1. Serum Lactate, TBARS, and Enzymatic Activity (Total Perosidase, GPx, and CS)
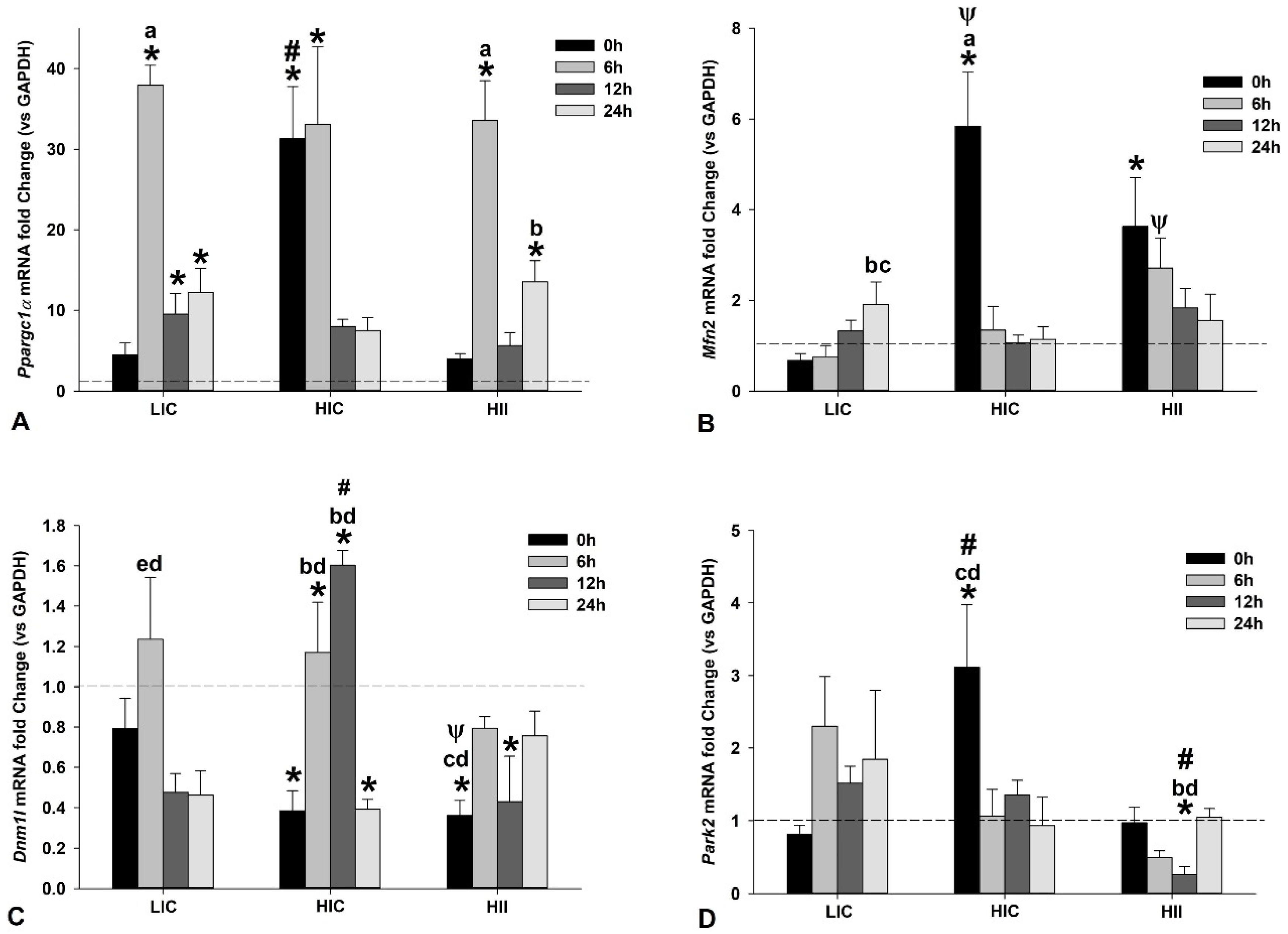
3.2. Mitochondrial Dynamics-Related Gene Expression in Muscle Tissue Samples
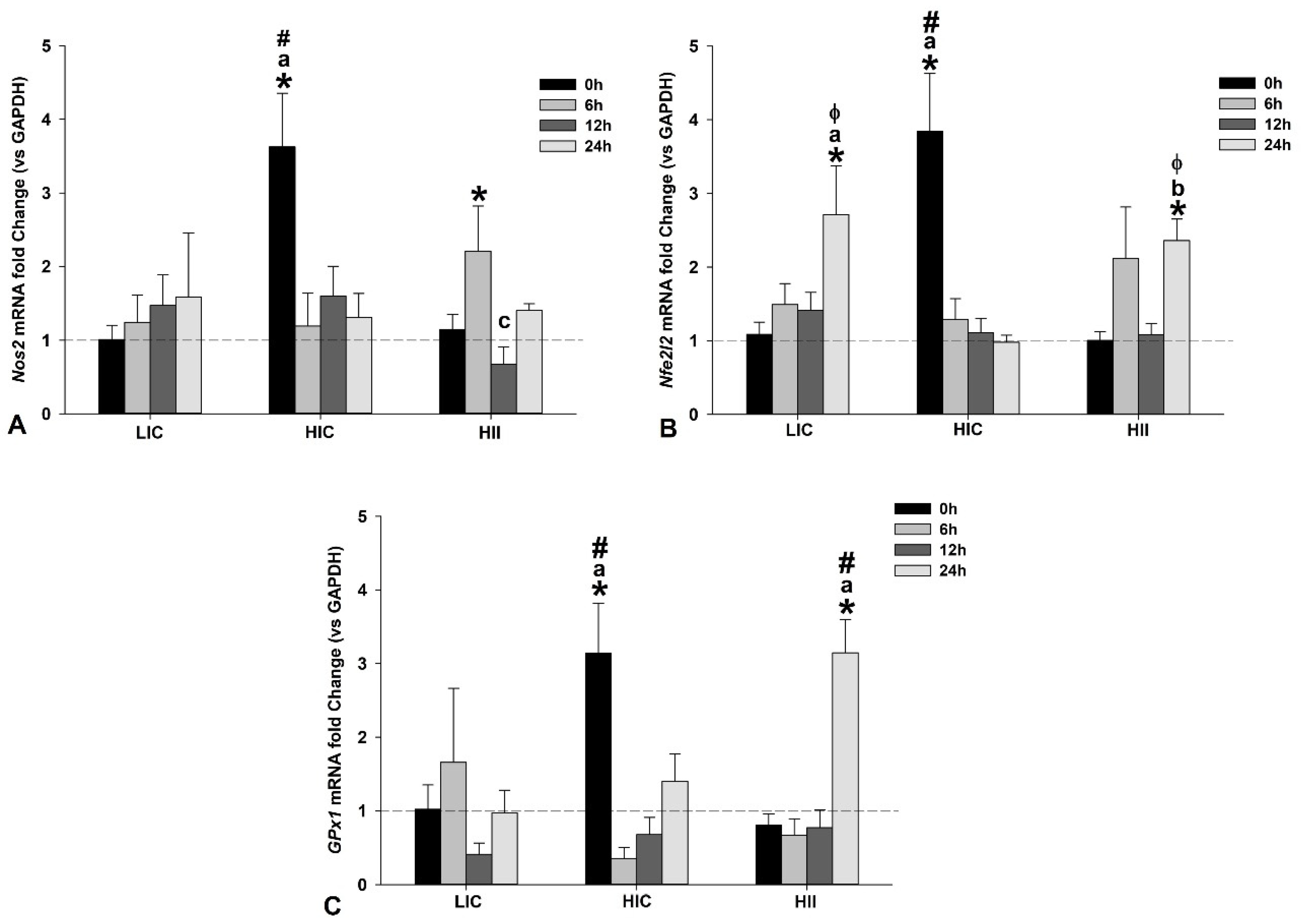
3.3. Redox-Related Gene Expression in Muscle Tissue Samples
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Egan, B.; Zierath, J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, D.J.; Botella, J.; Genders, A.J.; Lee, M.J.; Saner, N.J.; Kuang, J.; Yan, X.; Granata, C. High-intensity exercise and mitochondrial biogenesis: Current controversies and future research directions. Physiology 2019, 34, 56–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorenza, M.; Gunnarsson, T.P.; Hostrup, M.; Iaia, F.M.; Schena, F.; Pilegaard, H.; Bangsbo, J. Metabolic stress-dependent regulation of the mitochondrial biogenic molecular response to high-intensity exercise in human skeletal muscle. J. Physiol. 2018, 596, 2823–2840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, L.L.; Yeo, D.; Kang, C.; Zhang, T. The role of mitochondria in redox signaling of muscle homeostasis. J. Sport Health Sci. 2020, 9, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Cabrera, M.C.; Domenech, E.; Viña, J. Moderate exercise is an antioxidant: Upregulation of antioxidant genes by training. Free Radic. Biol. Med. 2008, 44, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Powers, S.K.; Jackson, M.J. Exercise-induced oxidative stress: Cellular mechanisms and impact on muscle force production. Physiol. Rev. 2008, 88, 1243–1276. [Google Scholar] [CrossRef] [Green Version]
- Pataky, M.W.; Nair, K.S. Too much of a good thing: Excess exercise can harm mitochondria. Cell Metab. 2021, 33, 847–848. [Google Scholar] [CrossRef]
- Cardinale, D.A.; Gejl, K.D.; Petersen, K.G.; Nielsen, J.; Ørtenblad, N.; Larsen, F.J. Short-term intensified training temporarily impairs mitochondrial respiratory capacity in elite endurance athletes. J. Appl. Physiol. 2021, 131, 388–400. [Google Scholar] [CrossRef]
- Evangelista, F.S.; Brum, P.C.; Krieger, J.E. Duration-controlled swimming exercise training induces cardiac hypertrophy in mice. Braz. J. Med. Biol. Res. 2003, 36, 1751–1759. [Google Scholar] [CrossRef] [Green Version]
- Almeida, A.A.; Correia, T.M.L.; Pires, R.A.; da Silva, D.A.; Coqueiro, R.S.; Machado, M.; de Magalhães, A.C.M.; Queiroz, R.F.; Soares, T.J.; Pereira, R. Nephroprotective effect of exercise training in cisplatin-induced renal damage in mice: Influence of training protocol. Braz. J. Med. Biol. Res. 2022, 55, e12116. [Google Scholar]
- Draper, H.H.; Squires, E.J.; Mahmoodi, H.; Wu, J.; Agarwal, S.; Hadley, M. A comparative evaluation of thiobarbituric acid methods for the determination of malondialdehyde in biological materials. Free Radic. Biol. Med. 1993, 15, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Aebi, H. Catalase in vitro. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1984; Volume 105, pp. 121–126. [Google Scholar] [CrossRef]
- Paglia, D.E.; Valentine, W.N. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J. Lab. Clin. Med. 1967, 70, 158–169. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Spinazzi, M.; Casarin, A.; Pertegato, V.; Salviati, L.; Angelini, C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 2012, 7, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Brooks, G.A. Lactate as a fulcrum of metabolism. Redox Biol. 2020, 35, 101454. [Google Scholar] [CrossRef]
- Brooks, G.A.; Arevalo, J.A.; Osmond, A.D.; Leija, R.G.; Curl, C.C.; Tovar, A.P. Lactate in contemporary biology: A phoenix risen. J. Physiol. 2021, 600, 1229–1251. [Google Scholar] [CrossRef]
- Hashimoto, T.; Hussien, R.; Oommen, S.; Gohil, K.; Brooks, G.A. Lactate sensitive transcription factor network in L6 cells: Activation of MCT1 and mitochondrial biogenesis. FASEB J. 2007, 21, 2602–2612. [Google Scholar] [CrossRef]
- Kitaoka, Y.; Takeda, K.; Tamura, Y.; Hatta, H. Lactate administration increases mRNA expression of PGC-1α and UCP3 in mouse skeletal muscle. Appl. Physiol. Nutr. Metab. 2016, 41, 695–698. [Google Scholar] [CrossRef] [Green Version]
- Passarella, S.; de Bari, L.; Valenti, D.; Pizzuto, R.; Paventi, G.; Atlante, A. Mitochondria and L-lactate metabolism. FEBS Lett. 2008, 582, 3569–3576. [Google Scholar] [CrossRef] [Green Version]
- Powers, S.K.; Ji, L.L.; Kavazis, A.N.; Jackson, M.J. Reactive oxygen species: Impact on skeletal muscle. Comp. Physiol. 2011, 1, 941–969. [Google Scholar]
- de Bari, L.; Valenti, D.; Atlante, A.; Passarella, S. L-lactate generates hydrogen peroxide in purified rat liver mitochondria due to the putative L-lactate oxidase localized in the intermembrane space. FEBS Lett. 2010, 584, 2285–2290. [Google Scholar] [CrossRef] [PubMed]
- Veal, E.A.; Day, A.M.; Morgan, B.A. Hydrogen peroxide sensing and signaling. Mol. Cell 2007, 26, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Ristow, M. Mitochondria and metabolic homeostasis. Antioxid. Redox Signal. 2013, 19, 240–242. [Google Scholar] [CrossRef]
- Gaitanos, G.C.; Williams, C.; Boobis, L.H.; Brooks, S. Human muscle metabolism during intermittent maximal exercise. J. Appl. Physiol. 1993, 75, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Bogdanis, G.C.; Nevill, M.E.; Lakomy, H.K.A.; Boobis, L.H. Power output and muscle metabolism during and following recovery from 10 and 20 s of maximal sprint exercise in humans. Acta Physiol. Scand. 1998, 163, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Burgomaster, K.A.; Hughes, S.C.; Heigenhauser, G.J.F.; Bradwell, S.N.; Gibala, M.J. Six sessions of sprint interval training increases muscle oxidative potential and cycle endurance capacity in humans. J. Appl. Physiol. 2005, 98, 1985–1990. [Google Scholar] [CrossRef]
- Yun, J.; Finkel, T. Mitohormesis. Cell Metab. 2014, 19, 757–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merry, T.L.; Ristow, M. Mitohormesis in exercise training. Free Radic. Biol. Med. 2016, 98, 123–130. [Google Scholar] [CrossRef] [Green Version]
- Niess, A.M.; Simon, P. Response and Adaptation of Skeletal Muscle to Exercise--The Role of Reactive Oxygen Species. Front. Biosci.-Landmark 2007, 12, 4826–4838. [Google Scholar] [CrossRef] [Green Version]
- Wai, T.; Langer, T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 2016, 27, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.W.; Erlich, A.T.; Crilly, M.J.; Hood, D.A. Parkin is required for exercise-induced mitophagy in muscle: Impact of aging. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E404–E415. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.W.; Erlich, A.T.; Hood, D.A. Role of Parkin and endurance training on mitochondrial turnover in skeletal muscle. Skelet. Muscle 2018, 8, 10. [Google Scholar] [CrossRef] [Green Version]
- Carter, H.N.; Kim, Y.; Erlich, A.T.; Zarrin-Khat, D.; Hood, D.A. Autophagy and mitophagy flux in young and aged skeletal muscle following chronic contractile activity. J. Physiol. 2018, 596, 3567–3584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Cunha, F.M.; Torelli, N.Q.; Kowaltowski, A.J. Mitochondrial retrograde signaling: Triggers, pathways, and outcomes. Oxid. Med. Cell. Longev. 2015, 2015, 482582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ristow, M.; Zarse, K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis). Exp. Gerontol. 2010, 45, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Quirós, P.M.; Mottis, A.; Auwerx, J. Mitonuclear communication in homeostasis and stress. Nat. Ver. Mol. Cell. Biol. 2016, 17, 213–226. [Google Scholar] [CrossRef]
- Done, A.J.; Traustadóttir, T. Nrf2 mediates redox adaptations to exercise. Redox Biol. 2016, 10, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Margonis, K.; Fatouros, I.G.; Jamurtas, A.Z.; Nikolaidis, M.G.; Douroudos, I.; Chatzinikolaou, A.; Mitrakou, A.; Mastorakos, G.; Papassotiriou, I.; Taxildaris, K.; et al. Oxidative stress biomarkers responses to physical overtraining: Implications for diagnosis. Free Radic. Biol. Med. 2007, 43, 901–910. [Google Scholar] [CrossRef]
- Huber-Abel, F.A.; Gerber, M.; Hoppeler, H.; Baum, O. Exercise-induced angiogenesis correlates with the up-regulated expression of neuronal nitric oxide synthase (nNOS) in human skeletal muscle. Eur. J. Appl. Physiol. 2012, 112, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Dai, L.; Brookes, P.S.; Darley-Usmar, V.M.; Anderson, P.G. Bioenergetics in cardiac hypertrophy: Mitochondrial respiration as a pathological target of NO. Am. J. Physiol. Heart Circ. Physiol. 2001, 281, H2261–H2269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallot, Y.S.; Bohnert, K.R. Confounding Roles of ER stress and the unfolded protein response in skeletal muscle atrophy. Int. J. Mol. Sci. 2021, 22, 2567. [Google Scholar] [CrossRef] [PubMed]
- Ryno, L.M.; Wiseman, R.L.; Kelly, J.W. Targeting unfolded protein response signaling pathways to ameliorate protein misfolding diseases. Curr. Opin. Chem. Biol. 2013, 17, 346–352. [Google Scholar] [CrossRef] [PubMed]



| Gene | Identification | Reference Code | Protein |
|---|---|---|---|
| Gapdh | Mm99999915_g1 | 4453320 | GAPDH |
| Ppargc1α | Mm01208835_m1 | 4453320 | PGC1-α |
| Mfn2 | Mm00500120_m1 | 4453320 | MFN2 |
| Dnm1L | Mm01342903_m1 | 4448892 | Drp1 |
| Park2 | Mm01323528_m1 | 4448892 | Parkin |
| Nfe2l2 | Mm00477784_m1 | 4453320 | NRF-2 |
| Gpx1 | Mm00656767_g1 | 4453320 | GPx1 |
| Nos2 | Mm00440502_m1 | 4453320 | NOS-2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pires, R.A.; Correia, T.M.L.; Almeida, A.A.; Coqueiro, R.d.S.; Machado, M.; Teles, M.F.; Peixoto, Á.S.; Queiroz, R.F.; Pereira, R. Time-Course of Redox Status, Redox-Related, and Mitochondrial-Dynamics-Related Gene Expression after an Acute Bout of Different Physical Exercise Protocols. Life 2022, 12, 2113. https://doi.org/10.3390/life12122113
Pires RA, Correia TML, Almeida AA, Coqueiro RdS, Machado M, Teles MF, Peixoto ÁS, Queiroz RF, Pereira R. Time-Course of Redox Status, Redox-Related, and Mitochondrial-Dynamics-Related Gene Expression after an Acute Bout of Different Physical Exercise Protocols. Life. 2022; 12(12):2113. https://doi.org/10.3390/life12122113
Chicago/Turabian StylePires, Ramon Alves, Thiago Macedo Lopes Correia, Amanda Alves Almeida, Raildo da Silva Coqueiro, Marco Machado, Mauro Fernandes Teles, Álbert Souza Peixoto, Raphael Ferreira Queiroz, and Rafael Pereira. 2022. "Time-Course of Redox Status, Redox-Related, and Mitochondrial-Dynamics-Related Gene Expression after an Acute Bout of Different Physical Exercise Protocols" Life 12, no. 12: 2113. https://doi.org/10.3390/life12122113
APA StylePires, R. A., Correia, T. M. L., Almeida, A. A., Coqueiro, R. d. S., Machado, M., Teles, M. F., Peixoto, Á. S., Queiroz, R. F., & Pereira, R. (2022). Time-Course of Redox Status, Redox-Related, and Mitochondrial-Dynamics-Related Gene Expression after an Acute Bout of Different Physical Exercise Protocols. Life, 12(12), 2113. https://doi.org/10.3390/life12122113






