Discovery of New Microbial Collagenase Inhibitors
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collagenase Activity Assay
2.2. Molecular Docking
2.3. Statistical Analysis
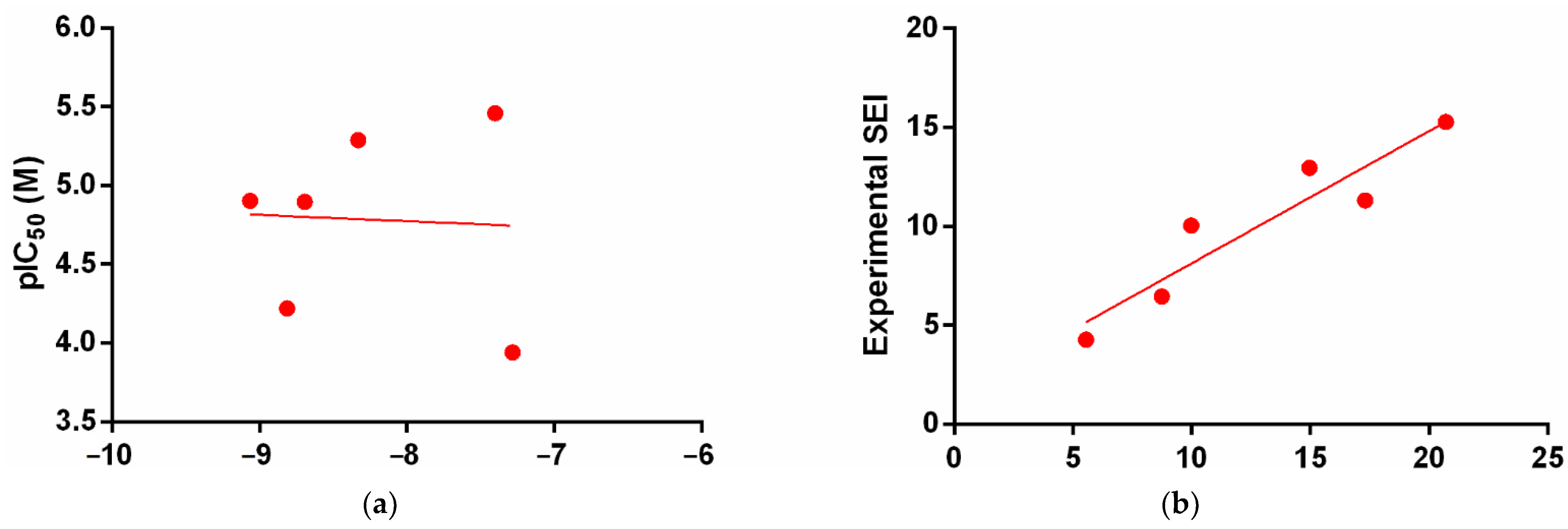
3. Results
3.1. Collagenase Activity Assay
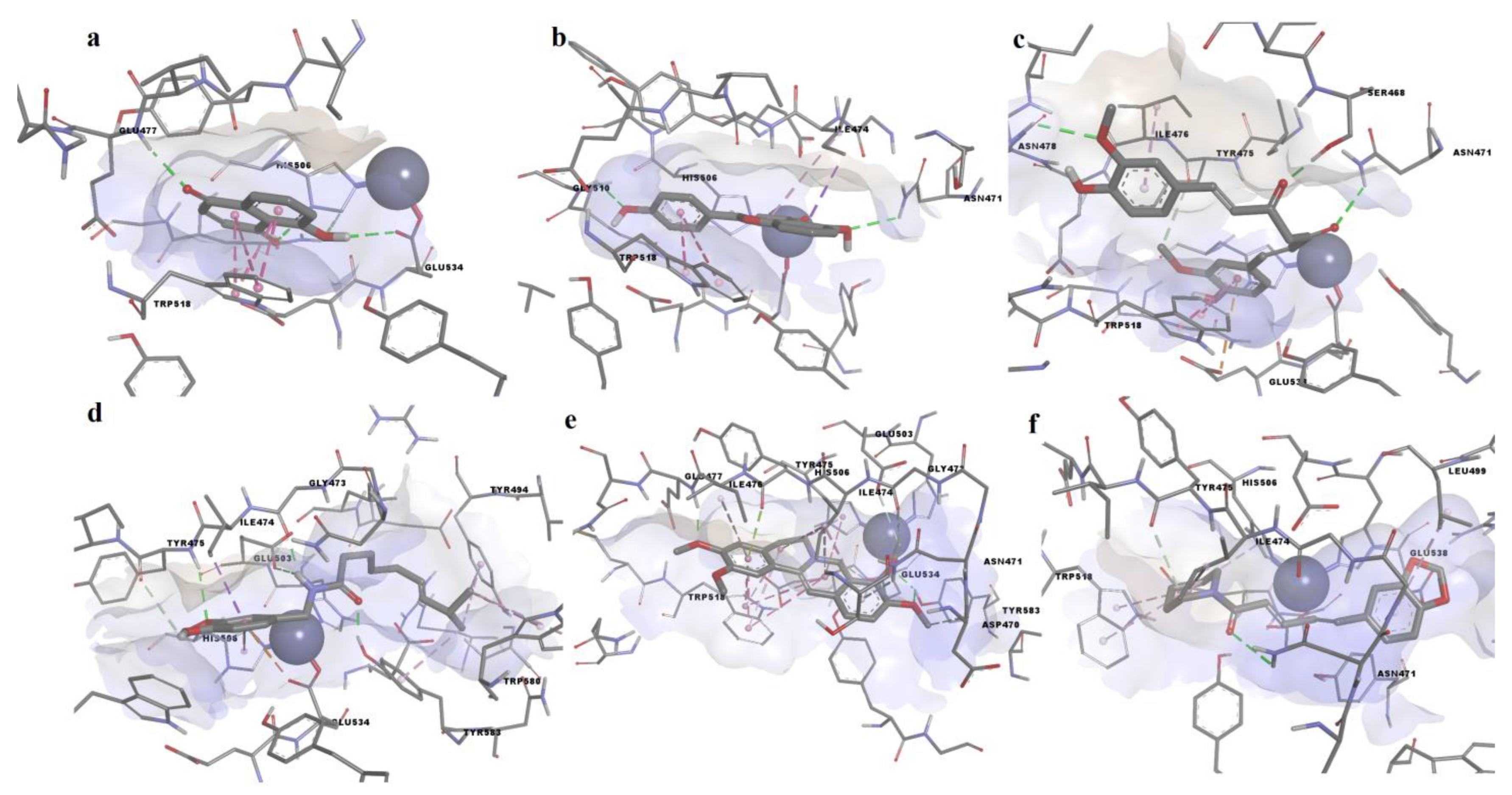
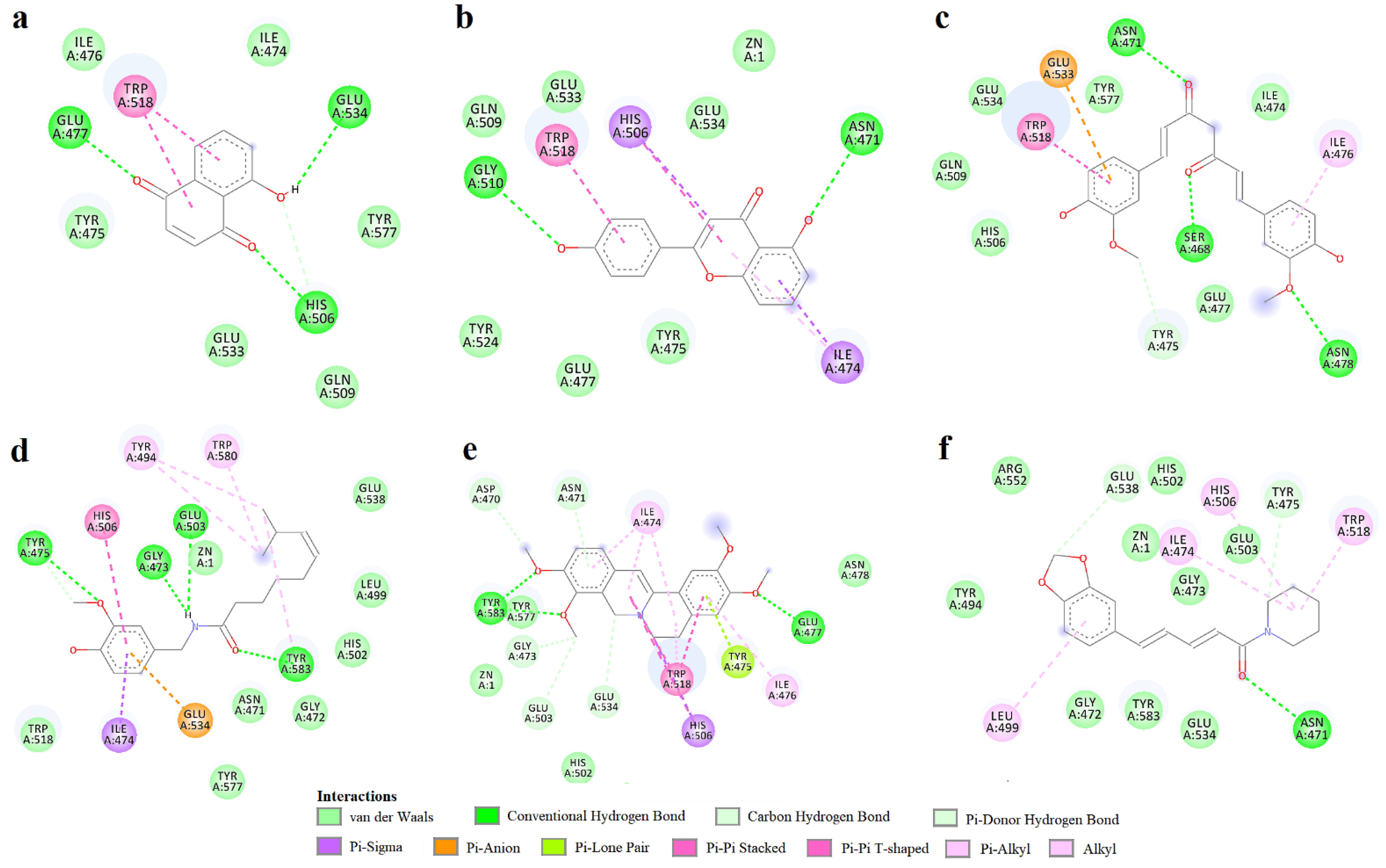
3.2. Molecular Docking Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Windels, E.M.; Van den Bergh, B.; Michiels, J. Bacteria under antibiotic attack: Different strategies for evolutionary adaptation. PLoS Pathog. 2020, 16, e1008431. [Google Scholar] [CrossRef] [PubMed]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 15. [Google Scholar] [CrossRef] [Green Version]
- Totsika, M. Benefits and Challenges of Antivirulence Antimicrobials at the Dawn of the Post-Antibiotic Era. Drug Deliv. Lett. 2016, 6, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Iskandar, K.; Murugaiyan, J.; Hammoudi Halat, D.; Hage, S.E.; Chibabhai, V.; Adukkadukkam, S.; Roques, C.; Molinier, L.; Salameh, P.; Van Dongen, M. Antibiotic Discovery and Resistance: The Chase and the Race. Antibiotics 2022, 11, 182. [Google Scholar] [CrossRef]
- McCarthy, M.; Goncalves, M.; Powell, H.; Morey, B.; Turner, M.; Merrill, A.R. A structural approach to anti-virulence: A discovery pipeline. Microorganisms 2021, 9, 2514. [Google Scholar] [CrossRef]
- Escajadillo, T.; Nizet, V. Pharmacological targeting of pore-forming toxins as adjunctive therapy for invasive bacterial infection. Toxins 2018, 10, 542. [Google Scholar] [CrossRef] [Green Version]
- Psonis, J.J.; Thanassi, D.G. Therapeutic Approaches Targeting the Assembly and Function of Chaperone-Usher Pili. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef]
- Nitulescu, G.; Margina, D.; Zanfirescu, A.; Olaru, O.T.; Nitulescu, G.M. Targeting bacterial sortases in search of anti-virulence therapies with low risk of resistance development. Pharmaceuticals 2021, 14, 415. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Muciño, E.; Arenas-Hernández, M.M.P.; Luna-Guevara, M.L. Mechanisms of Inhibition of Quorum Sensing as an Alternative for the Control of E. coli and Salmonella. Microorganisms 2022, 10, 884. [Google Scholar] [CrossRef] [PubMed]
- Martínez, O.F.; Rigueiras, P.O.; Pires, Á.d.S.; Porto, W.F.; Silva, O.N.; de la Fuente-Nunez, C.; Franco, O.L. Interference With Quorum-Sensing Signal Biosynthesis as a Promising Therapeutic Strategy Against Multidrug-Resistant Pathogens. Front. Cell. Infect. Microbiol. 2019, 8, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, O.F.; Cardoso, M.H.; Ribeiro, S.M.; Franco, O.L. Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition. Front. Cell. Infect. Microbiol. 2019, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Krukiewicz, K.; Kazek-Kęsik, A.; Brzychczy-Włoch, M.; Łos, M.J.; Ateba, C.N.; Mehrbod, P.; Ghavami, S.; Shyntum, D.Y. Recent Advances in the Control of Clinically Important Biofilms. Int. J. Mol. Sci. 2022, 23, 9526. [Google Scholar] [CrossRef]
- Duarte, A.S.; Correia, A.; Esteves, A.C. Bacterial collagenases—A review. Crit. Rev. Microbiol. 2016, 42, 106–126. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef]
- Graef, F.; Vukosavljevic, B.; Michel, J.P.; Wirth, M.; Ries, O.; De Rossi, C.; Windbergs, M.; Rosilio, V.; Ducho, C.; Gordon, S.; et al. The bacterial cell envelope as delimiter of anti-infective bioavailability—An in vitro permeation model of the Gram-negative bacterial inner membrane. J. Control. Release 2016, 243, 214–224. [Google Scholar] [CrossRef]
- Eckhard, U.; Schönauer, E.; Nüss, D.; Brandstetter, H. Structure of collagenase G reveals a chew-and-digest mechanism of bacterial collagenolysis. Nat. Struct. Mol. Biol. 2010, 18, 1109–1114. [Google Scholar] [CrossRef] [Green Version]
- Eckhard, U.; Schönauer, E.; Brandstetter, H. Structural basis for activity regulation and substrate preference of clostridial collagenases G, H, and T. J. Biol. Chem. 2013, 288, 20184–20194. [Google Scholar] [CrossRef] [Green Version]
- Supuran, C.T.; Briganti, F.; Mincione, G.; Scozzafava, A. Protease inhibitors: Synthesis of L-alanine hydroxamate sulfonylated derivatives as inhibitors of Clostridium histolyticum collagenase. J. Enzyme Inhib. 2000, 15, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Scozzafava, A. Protease inhibitors. Part 7: Inhibition of Clostridium histolyticum collagenase with sulfonylated derivatives of L-valine hydroxamate. Eur. J. Pharm. Sci. 2000, 10, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Scozzafava, A.; Supuran, C.T. Protease inhibitors: Synthesis of matrix metalloproteinase and bacterial collagenase inhibitors incorporating 5-amino-2-mercapto-1,3,4-thiadiazole zinc binding functions. Bioorganic Med. Chem. Lett. 2002, 12, 2667–2672. [Google Scholar] [CrossRef]
- Ilies, M.; Banciu, M.D.; Scozzafava, A.; Ilies, M.A.; Caproiu, M.T.; Supuran, C.T. Protease inhibitors: Synthesis of bacterial collagenase and matrix metalloproteinase inhibitors incorporating arylsulfonylureido and 5-dibenzo-suberenyl/suberyl moieties. Bioorganic Med. Chem. 2003, 11, 2227–2239. [Google Scholar] [CrossRef]
- Schönauer, E.; Kany, A.M.; Haupenthal, J.; Hüsecken, K.; Hoppe, I.J.; Voos, K.; Yahiaoui, S.; Elsässer, B.; Ducho, C.; Brandstetter, H.; et al. Discovery of a Potent Inhibitor Class with High Selectivity toward Clostridial Collagenases. J. Am. Chem. Soc. 2017, 139, 12696–12703. [Google Scholar] [CrossRef] [Green Version]
- Ducho, C.; Görbig, U.; Jessel, S.; Gisch, N.; Balzarini, J.; Meier, C. Bis-cycloSal-d4T-monophosphates: Drugs that deliver two molecules of bioactive nucleotides. J. Med. Chem. 2007, 50, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Gostner, J.; Fuchs, J.E.; Chaita, E.; Aligiannis, N.; Skaltsounis, L.; Ganzera, M. Inhibition of Collagenase by Mycosporine-like Amino Acids from Marine Sources. Planta Med. 2015, 81, 813–820. [Google Scholar] [CrossRef] [Green Version]
- Pallela, R.; Na-Young, Y.; Kim, S.K. Anti-photoaging and photoprotective compounds derived from marine organisms. Mar. Drugs 2010, 8, 1189–1202. [Google Scholar] [CrossRef] [Green Version]
- Nitulescu, G.; Nitulescu, G.M.; Zanfirescu, A.; Paul Mihai, D.; Gradinaru, D. Candidates for Repurposing as Anti-Virulence Agents Based on the Structural Profile Analysis of Microbial Collagenase Inhibitors. Pharmaceutics 2022, 14, 62. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Land, H.; Humble, M.S. YASARA: A Tool to Obtain Structural Guidance in Biocatalytic Investigations; Humana Press: New York, NY, USA, 2018; pp. 43–67. [Google Scholar]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Sin, B.Y.; Kim, H.P. Inhibition of collagenase by naturally-occurring flavonoids. Arch. Pharm. Res. 2005, 28, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zhang, P.; Lou, L.; Wang, Y. Perspectives Regarding the Role of Biochanin A in Humans. Front. Pharmacol. 2019, 10, 793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uivarosi, V.; Munteanu, A.C.; Sharma, A.; Singh Tuli, H. Metal Complexation and Patent Studies of Flavonoid—Current Aspects of Flavonoids: Their Role in Cancer Treatment; Tuli, H.S., Ed.; Springer Singapore: Singapore, 2019; pp. 39–89. [Google Scholar]
- Islam, A.K.M.M.; Widhalm, J.R. Agricultural uses of juglone: Opportunities and challenges. Agronomy 2020, 10, 1500. [Google Scholar] [CrossRef]
- Nitulescu, G.; Mihai, D.P.; Nicorescu, I.M.; Olaru, O.T.; Ungurianu, A.; Zanfirescu, A.; Nitulescu, G.M.; Margina, D. Discovery of natural naphthoquinones as sortase A inhibitors and potential anti-infective solutions against Staphylococcus aureus. Drug Dev. Res. 2019, 80, 1136–1145. [Google Scholar] [CrossRef]
- Soto-Maldonado, C.; Vergara-Castro, M.; Jara-Quezada, J.; Caballero-Valdés, E.; Müller-Pavez, A.; Zúñiga-Hansen, M.E.; Altamirano, C. Polyphenolic extracts of walnut (Juglans regia) green husk containing juglone inhibit the growth of HL-60 cells and induce apoptosis. Electron. J. Biotechnol. 2019, 39, 1–7. [Google Scholar] [CrossRef]
- Long, J.; Song, J.; Zhong, L.; Liao, Y.; Liu, L.; Li, X. Palmatine: A review of its pharmacology, toxicity and pharmacokinetics. Biochimie 2019, 162, 176–184. [Google Scholar] [CrossRef]
- Kim, J.H.; Ryu, Y.B.; Lee, W.S.; Kim, Y.H. Neuraminidase inhibitory activities of quaternary isoquinoline alkaloids from Corydalis turtschaninovii rhizome. Bioorganic Med. Chem. 2014, 22, 6047–6052. [Google Scholar] [CrossRef] [Green Version]
- Grabarska, A.; Wróblewska-Łuczka, P.; Kukula-Koch, W.; Łuszczki, J.J.; Kalpoutzakis, E.; Adamczuk, G.; Skaltsounis, A.L.; Stepulak, A. Palmatine, a Bioactive Protoberberine Alkaloid Isolated from Berberis cretica, Inhibits the Growth of Human Estrogen Receptor-Positive Breast Cancer Cells and Acts Synergistically and Additively with Doxorubicin. Molecules 2021, 26, 6253. [Google Scholar] [CrossRef]
- Gong, F.; Cui, L.; Zhang, X.; Zhan, X.; Gong, X.; Wen, Y. Piperine ameliorates collagenase-induced Achilles tendon injury in the rat. Connect. Tissue Res. 2018, 59, 21–29. [Google Scholar] [CrossRef]
- Shao, B.; Cui, C.; Ji, H.; Tang, J.; Wang, Z.; Liu, H.; Qin, M.; Li, X.; Wu, L. Enhanced oral bioavailability of piperine by self-emulsifying drug delivery systems: In vitro, in vivo and in situ intestinal permeability studies. Drug Deliv. 2015, 22, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Marini, E.; Magi, G.; Mingoia, M.; Pugnaloni, A.; Facinelli, B. Antimicrobial and anti-virulence activity of capsaicin against erythromycin-resistant, cell-invasive group A streptococci. Front. Microbiol. 2015, 6, 1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menezes, R.d.P.; Bessa, M.A.d.S.; Siqueira, C.d.P.; Teixeira, S.C.; Ferro, E.A.V.; Martins, M.M.; Cunha, L.C.S.; Martins, C.H.G. Antimicrobial, Antivirulence, and Antiparasitic Potential of Capsicum chinense Jacq. Extracts and Their Isolated Compound Capsaicin. Antibiotics 2022, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Karami-Zarandi, M.; Ghale, H.E.; Ranjbar, R. Characterization of virulence factors and antibacterial activity of curcumin in hypervirulent Klebsiella pneumoniae. Future Microbiol. 2022, 17, 529–540. [Google Scholar] [CrossRef]
- Chainani-Wu, N. Safety and anti-inflammatory activity of curcumin: A component of tumeric (Curcuma longa). J. Altern. Complement. Med. 2003, 9, 161–168. [Google Scholar] [CrossRef] [Green Version]
- Surh, Y.J.; Lee, S.S. Capsaicin, a double-edged sword: Toxicity, metabolism, and chemopreventive potential. Life Sci. 1995, 56, 1845–1855. [Google Scholar] [CrossRef]

| Compound | Inhibition (%) | ||||
|---|---|---|---|---|---|
| 1 µM | 5 µM | 10 µM | 50 µM | 100 µM | |
| Palmatine chloride | 15.5 ± 3.9 | 35.4 ± 1.8 | 84.9 ± 1.5 | 97.9 ± 1.6 | 98.7 ± 0.4 |
| Piperine | 84.5 ± 0.4 | 82.4 ± 0.4 | 72.5 ± 2.7 | NT * | NT * |
| 2′-Hydroxychalcone | −12.1 ± 0.2 | 1.2 ± 0.9 | 6.5 ± 1.3 | 21.8 ± 1.2 | 97.3 ± 1.3 |
| 3-Hydroxyflavone | −13.4 ± 2.7 | −17.0 ± 5.1 | 17.4 ± 4.5 | 25.1 ± 6.8 | 31.0 ± 4.3 |
| Myricetin | −15.4 ± 2.7 | −4.5 ± 1.1 | −1.0 ± 1.1 | 13.6 ± 0.6 | 23.2 ± 2.0 |
| Myricitrin | −37.9 ± 2.3 | −17.4 ± 2.4 | −20.8 ± 1.0 | 16.7 ± 3.1 | 37.2 ± 3.8 |
| Dihydrorobinetin | −16.5 ± 1.6 | −29.7 ± 0.5 | 105.1 ± 0.5 | NT * | NT * |
| Primuletin | −9.3 ± 1.7 | −8.8 ± 2.1 | −0.7 ± 1.3 | 8.5 ± 2.1 | 25.4 ± 1.8 |
| 4′,5-Dihydroxyflavone | −48.0 ± 3.6 | 101.6 ± 0.6 | 107.5 ± 1.0 | 101.0 ± 1.9 | 104.8 ± 2.9 |
| Biochanin A | −24.8 ± 5.3 | 23.0 ± 2.6 | 34.1 ± 2.2 | 99.1 ± 1.0 | 76.1 ± 0.7 |
| Juglone | 13.8 ± 1.5 | 50.6 ± 0.5 | 69.2 ± 0.4 | 80.8 ± 0.7 | 94.8 ± 0.1 |
| Rhein | 2.6 ± 0.6 | 22.9 ± 2.7 | 37.4 ± 3.2 | 77.3 ± 2.2 | 80.6 ± 1.0 |
| Curcumin | −10.0 ± 3.9 | 101.8 ± 0.6 | 102.5 ± 0.8 | 101.5 ± 0.4 | 101.8 ± 0.7 |
| Capsaicin | 94.8 ± 0.8 | 100.5 ± 0.8 | NT * | NT * | NT * |
| Citrulline | −4.3 ± 3.5 | −14.1 ± 1.4 | 27.8 ± 3.0 | 24.3 ± 1.7 | 7.2 ± 0.5 |
| 6-Methoxy-1,2,3,4-tetrahydro-9H-pyrido [3,4-b]indole-1-carboxylic acid | −19.9 ± 2.7 | −1.3 ± 2.1 | −5.2 ± 0.7 | 2.8 ± 1.3 | 19.6 ± 3.9 |
| Compound | IC50 (µM) | 95% CI of IC50 (µM) | r2 |
|---|---|---|---|
| Palmatine chloride | 5.139 | 0.1076 to 245.4 | 0.918 |
| Piperine | <1 | NC * | NC * |
| 2′-Hydroxychalcone | 60.395 | NC * | NC * |
| 3-Hydroxyflavone | >100 | NC * | NC * |
| Myricetin | >100 | NC * | NC * |
| Myricitrin | >100 | NC * | NC * |
| Dihydrorobinetin | <10 | NC * | NC * |
| Primuletin | >100 | NC * | NC * |
| 4′,5-Dihydroxyflavone | <1 | NC * | NC * |
| Biochanin A | 12.474 | 0.2371 to 167.3 | 0.939 |
| Juglone | 3.473 | 0.5838 to 20.66 | 0.984 |
| Rhein | 12.710 | 5.225 to 30.92 | 0.996 |
| Curcumin | <5 | NC * | NC * |
| Capsaicin | <1 | NC * | NC * |
| Citrulline | >100 | NC * | NC * |
| 6-Methoxy-1,2,3,4-tetrahydro-9H-pyrido [3,4-b]indole-1-carboxylic acid | >100 | NC * | NC * |
| 1,10-Phenanthroline | 114.810 | NC * | NC * |
| Compound | LE | ΔG (kcal/mol) | Residue Contacts | Zn Contact |
|---|---|---|---|---|
| Dihydrorobinetin | 0.4313 | −9.489 | Asn471, Gly472, Gly473, Ile474, Tyr475, Ile476, Glu477, His502, Glu503, His506, Gln509, Gly510, Trp518, Tyr524, Glu533, Glu534, Tyr577, Tyr583 | yes |
| Myricitrin | 0.2788 | −9.2 | Asp470, Asn471, Gly472, Gly473, Tyr485, Arg487, Ser492, Ile493, Tyr494, Leu499, His502, Glu503, Glu538, Tyr577, Gly578, Ser579, Trp580, Phe582, Tyr583 | no |
| Curcumin | 0.3358 | −9.185 | Asn471, Ile474, Tyr475, Ile476, Glu477, Asn478, His506, Gln509, Trp518, Glu533, Glu534, Tyr577 | no |
| 4′,5-dihiydroxyflavone | 0.4782 | −9.099 | Asn471, Ile474, Tyr475, Ile476, Glu477, His506, Gln509, Gly510, Trp518, Tyr524, Glu533, Glu534 | yes |
| Biochanin A | 0.4316 | −9.064 | Asp470, Asn471, Gly473, Ile474, Tyr475, Ile476, Glu477, His506, Gln509, Gly510, Trp518, Tyr524, Glu533, Glu534, Tyr577, Tyr583 | yes |
| 5-hydroxyflavone (primuletin) | 0.5011 | −9.02 | Asn471, Gly473, Ile474, Tyr475, Ile476, Glu477, Glu503, His506, Gln509, Gly510, Trp518, Tyr524, Glu533 | yes |
| Myricetin | 0.3847 | −8.847 | Asn471, Gly473, Ile474, Tyr475, Ile476, Glu477, Glu503, His506, Gln509, Trp518, Tyr524, Glu533, Glu534, Tyr577, Tyr583 | yes |
| 2′-hydroxychalcone | 0.5184 | −8.813 | Ile474, Tyr475, Ile476, Glu477, His506, Gln509, Gly510, Trp518, Tyr524, Glu533 | no |
| Rhein | 0.4139 | −8.692 | Ile474, Tyr475, Ile476, Glu477, Asn478, His506, Trp518, Glu534, Tyr577 | no |
| 3-hydroxyflavone | 0.468 | −8.424 | Asn471, Gly473, Ile474, Tyr475, Ile476, Glu477, Glu503, His506, Gln509, Gly510, Trp518, Glu533, Glu534, Tyr577 | no |
| Piperine | 0.4079 | −8.404 | Asn471, Gly472, Gly473, Ile474, Tyr475, Ile476, Tyr494, Leu499, His502, Glu503, His506, Trp518, Glu534, Glu538, Arg552, Tyr577, Trp580, Tyr583 | yes |
| Palmatine | 0.3223 | −8.329 | Asp470, Asn471, Gly473, Ile 474, Tyr475, Ile476, Glu477, Asn478, His506, Met517, Trp518, Gln520, Glu534, Tyr577, Tyr583 | yes |
| Capsaicin | 0.3514 | −8.012 | Asn471, Gly472, Gly473, Ile474, Tyr475, Tyr494, Leu499, His502, Glu503, His506, Trp518, Glu534, Glu538, Tyr577, Trp580, Tyr583, Asn584 | yes |
| 6-Methoxy-1,2,3,4-tetrahydro-9H-pyrido [3,4-b]indole-1-carboxylic acid | 0.4364 | −7.855 | Asn471, Ile474, Tyr475, Ile476, Glu477, His506, Gln509, Gly510, Trp518, Tyr524, Glu533, Glu534, Tyr577, Tyr583 | no |
| Juglone | 0.5688 | −7.402 | Ile474, Tyr475, Ile476, Glu477, His506, Gln509, Gly510, Trp518, Glu533, Glu534, Tyr577 | no |
| 1,10-Phenanthroline | 0.5203 | −7.284 | Asn471, Gly472, Gly473, Tyr494, Leu499, His502, Glu503, Glu538, Trp580, Tyr583 | no |
| Citrulline | 0.4984 | −5.981 | Ile474, Tyr475, Ile476, Glu477, His506, Gln509, Gly510, Val514, Trp518, Tyr524, Glu533, Glu534, Tyr577 | no |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nitulescu, G.; Mihai, D.P.; Zanfirescu, A.; Stan, M.S.; Gradinaru, D.; Nitulescu, G.M. Discovery of New Microbial Collagenase Inhibitors. Life 2022, 12, 2114. https://doi.org/10.3390/life12122114
Nitulescu G, Mihai DP, Zanfirescu A, Stan MS, Gradinaru D, Nitulescu GM. Discovery of New Microbial Collagenase Inhibitors. Life. 2022; 12(12):2114. https://doi.org/10.3390/life12122114
Chicago/Turabian StyleNitulescu, Georgiana, Dragos Paul Mihai, Anca Zanfirescu, Miruna Silvia Stan, Daniela Gradinaru, and George Mihai Nitulescu. 2022. "Discovery of New Microbial Collagenase Inhibitors" Life 12, no. 12: 2114. https://doi.org/10.3390/life12122114
APA StyleNitulescu, G., Mihai, D. P., Zanfirescu, A., Stan, M. S., Gradinaru, D., & Nitulescu, G. M. (2022). Discovery of New Microbial Collagenase Inhibitors. Life, 12(12), 2114. https://doi.org/10.3390/life12122114









