Metagenomic Analysis of Garden Soil-Derived Microbial Consortia and Unveiling Their Metabolic Potential in Mitigating Toxic Hexavalent Chromium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Elemental Characterization
2.2. DNA Extraction and Metagenomic Sequencing
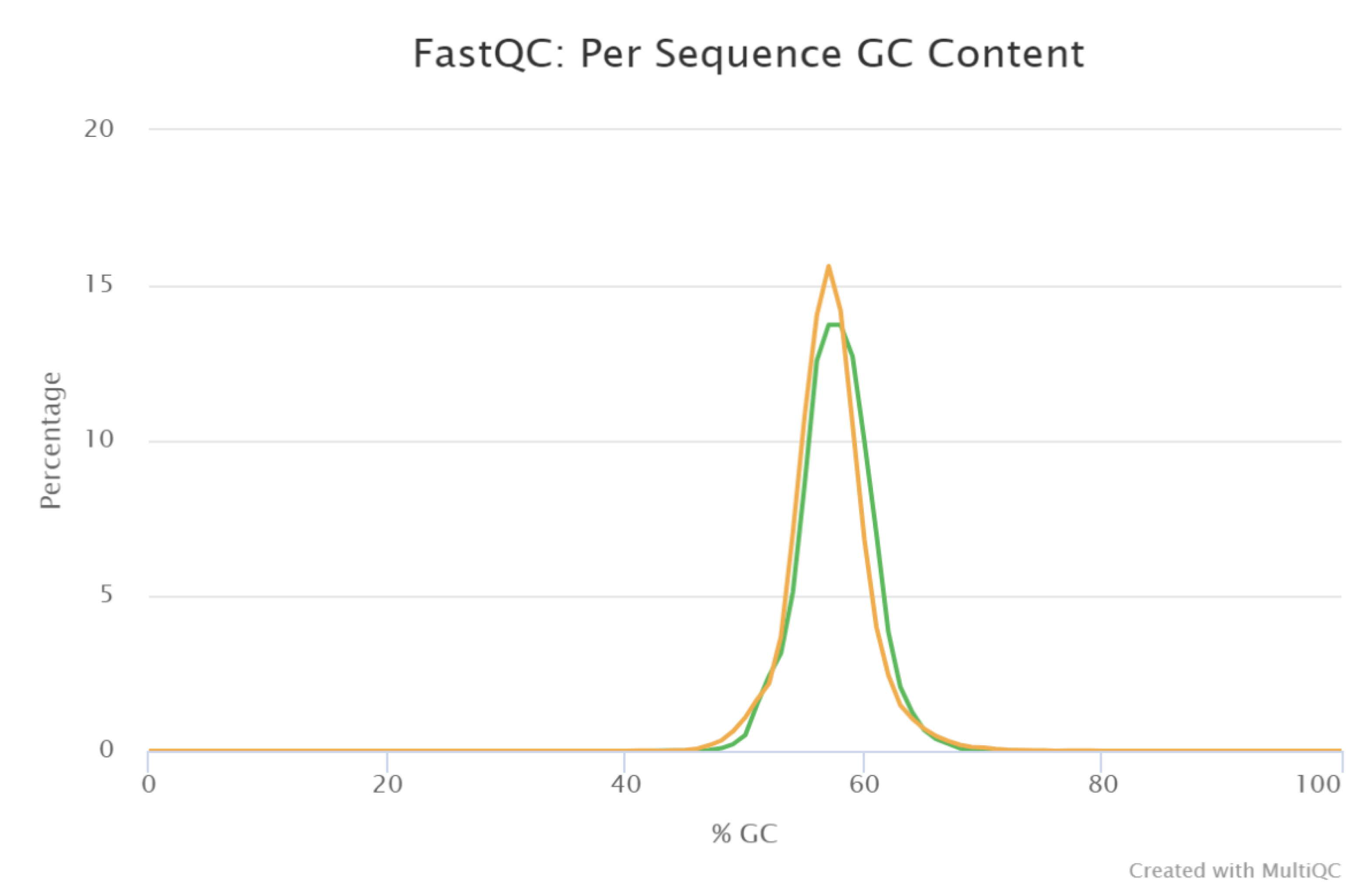
2.3. Bioinformatics Analysis
2.4. Isolation of Soil-Derived Microbial Consortia and Cr (VI) Removal Using Microbial Consortia
2.5. Statistical Analysis
3. Result
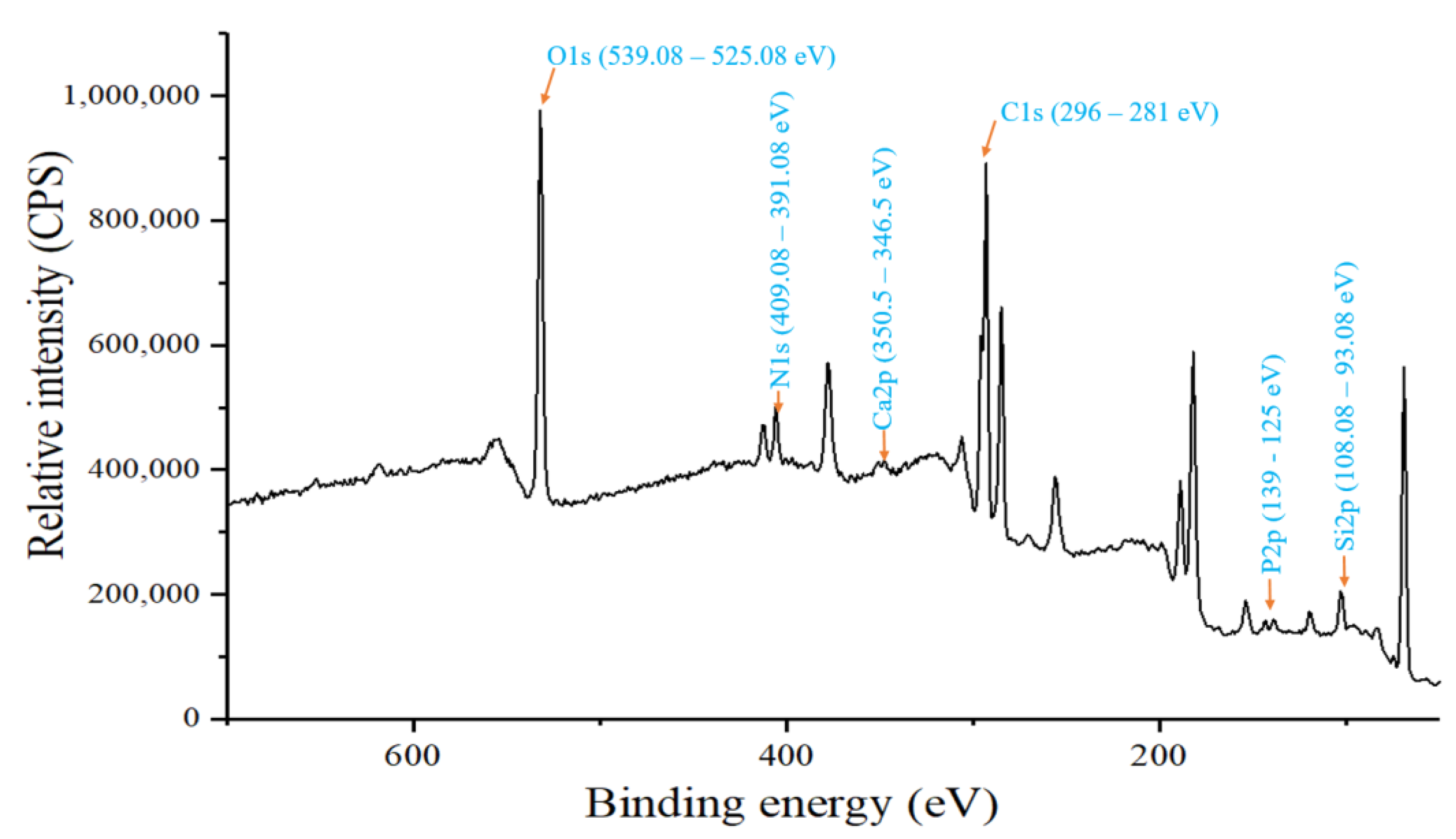
3.1. Elemental Composition of Garden Soil
3.2. Cr (VI) Removal by Using Soil-Derived Microbial Consortia
3.3. High-Throughput Data Analysis
3.4. Taxonomic Composition Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rawat, N.; Joshi, G.K. Bacterial community structure analysis of a hot spring soil by next generation sequencing of ribosomal RNA. Genomics 2019, 111, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Whitman, W.; Coleman, D.; Wiebe, W. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef] [Green Version]
- Girvan, M.S.; Bullimore, J.; Pretty, J.N. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 2003, 69, 1800–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franz, E.; Schijven, J.; de Roda Husman, A.M.; Blaal, H. Meta-regression analysis of commensal and pathogenic Escherichia coli survival in soil and water. Environ. Sci. Technol. 2014, 48, 6763–6771. [Google Scholar] [CrossRef] [PubMed]
- van Veen, J.A.; van Overbeek, L.S.; van Elsas, J.D. Fate and activity of microorganisms introduced into soil Microbiol. Mol. Biol. Rev. 1997, 61, 121–135. [Google Scholar]
- Wu, X.; Yang, J.; Ruan, H.; Wang, S.; Yang, Y.; Naeem, I.; Wang, L.; Liu, L.; Wang, D. The diversity and co-occurrence network of soil bacterial and fungal communities andtheir implications for a new indicator of grassland degradation. Ecol. Indic. 2021, 129, 107989. [Google Scholar] [CrossRef]
- Stenuit, B.; Agathos, S.N. Deciphering microbial community robustness through synthetic ecology and molecular systems synecology. Curr. Opin Biotechnol. 2015, 33, 305–317. [Google Scholar] [CrossRef]
- da C Jesus, E.; Marsh, T.; Tiedje, J.M.; de S Moreira, F.M. Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME J. 2009, 3, 1004–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakelin, S.; Macdonald, L.; Rogers, S.; Gregg, A.L.; Bolger, T.P.; Baldock, J.A. Habitat selective factors influencing the structural composition and functional capacity of microbial communities in agricultural soils. Soil Biol. Biochem. 2008, 40, 803–813. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Mocali, S.; Benedetti, A. Exploring research frontiers in microbiology: The challenge of metagenomics in soil microbiology. Res. Microbiol. 2010, 161, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Rajendhran, J.; Gunasekaran, P. Strategies for accessing soil metagenome for desired applications. Biotechnol. Adv. 2008, 26, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, S.A.; Chen, W.Y.; Wong, T.; Kanakamedala, B.S.; Deveson, I.W.; Ongley, S.E.; Santini, N.S.; Marcellin, E.; Smith, M.A.; Nielsen, L.K.; et al. Synthetic microbe communities provide internal reference standards for metagenome sequencing and analysis. Nat. Commun. 2018, 9, 3096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevenson, B.S.; Eichorst, S.A.; Wertz, J.T.; Schmidt, T.M.; Breznak, J.A. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 2004, 70, 4748–4755. [Google Scholar] [CrossRef] [Green Version]
- Rivera, M.C.; Izard, J. Promises and Prospects of Microbiome Studies, 1st ed.; Izard, J., Rivera, M.C., Eds.; Elsevier: London, UK, 2015; pp. 144–159. [Google Scholar]
- Gupta, S.; Kumar, M.; Kumar, J.; Ahmad, V.; Pandey, R.; Chauhan, N.S. Systemic analysis of soil microbiome deciphers anthropogenic influence on soil ecology and ecosystem functioning. Int. J. Environ. Sci. Technol. 2017, 14, 2229–2238. [Google Scholar] [CrossRef]
- Hemmat-Jou, M.H.; Safari-Sinegani, A.A.; Mirzaie-Asl, A.; Tahmourespour, A. Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach. Ecotoxicology 2018, 27, 1281–1291. [Google Scholar] [CrossRef]
- Almeida, O.G.G.; De Martinis, E.C.P. Bioinformatics tools to assess metagenomic data for applied microbiology. Appl. Microbiol. Biotechnol. 2019, 103, 69–82. [Google Scholar] [CrossRef]
- Chiang, A.D.; Dekker, J.P. From the Pipeline to the Bedside: Advances and Challenges in Clinical Metagenomics. J. Infect. Dis. 2020, 221, S331–S340. [Google Scholar] [CrossRef]
- Zethof, J.H.T.; Bettermann, A.; Vogel, C.; Babin, D.; Cammeraat, E.L.H.; Solé-Benet, A.; Lázaro, R.; Luna, L.; Nesme, J.; Woche, S.K.; et al. Prokaryotic Community Composition and Extracellular Polymeric Substances Affect Soil Microaggregation in Carbonate Containing Semiarid Grasslands. Front. Environ. Sci. 2020, 8, 51. [Google Scholar] [CrossRef]
- de Lima Brossi, M.J.; Jiménez, D.J.; Cortes-Tolalpa, L.; van Elsas, J.D. Soil-Derived Microbial Consortia Enriched with Different Plant Biomass Reveal Distinct Players Acting in Lignocellulose Degradation. Microb. Ecol. 2016, 71, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wei, L.; Yang, J.; Ahmed, W.; Wang, Y.; Fu, L.; Ji, G. Probiotic Consortia: Reshaping the Rhizospheric Microbiome and Its Role in Suppressing Root-Rot Disease of Panax notoginseng. Front. Microbiol. 2020, 11, 701. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Yang, J.; Ahmed, W.; Xiong, X.; Liu, Q.; Huang, Q.; Ji, G. Unraveling the Association between Metabolic Changes in Inter-Genus and Intra-Genus Bacteria to Mitigate Clubroot Disease of Chinese Cabbage. Agronomy 2021, 11, 2424. [Google Scholar] [CrossRef]
- Zhang, J.; Ahmed, W.; Dai, Z.; Zhou, X.; He, Z.; Wei, L.; Ji, G. Microbial Consortia: An Engineering Tool to Suppress Clubroot of Chinese Cabbage by Changing the Rhizosphere Bacterial Community Composition. Biology 2022, 11, 918. [Google Scholar] [CrossRef]
- Phieler, R.; Voit, A.; Kothe, E. Microbially supported phytoremediation of heavy metal contaminated soils: Strategies and applications. Adv. Biochem. Eng. Biotechnol. 2014, 141, 211–235. [Google Scholar]
- Singh, V.; Singh, M.P.; Mishra, V. Bioremediation of toxic metal ions from coal washery effluent. Desalin. Water Treat. 2020, 197, 300–318. [Google Scholar] [CrossRef]
- Singh, V.; Singh, J.; Mishra, V. Development of a cost-effective, recyclable and viable metal ion doped adsorbent for simultaneous adsorption and reduction of toxic Cr (VI) ions. J. Environ. Chem. Eng. 2021, 9, 105124. [Google Scholar] [CrossRef]
- Singh, V.; Mishra, V. Microbial Removal of Cr (VI) by a New Bacterial Strain Isolated from the Site Contaminated with Coal Mine Effluents. J. Environ. Chem. Eng. 2021, 9, 106279. [Google Scholar] [CrossRef]
- Singh, V.; Singh, J.; Mishra, V. Sorption kinetics of an eco-friendly and sustainable Cr (VI) ion scavenger in a batch reactor. J. Environ. Chem. Eng. 2021, 9, 105125. [Google Scholar] [CrossRef]
- Singh, V.; Mishra, V. Sustainable reduction of Cr (VI) and its elemental mapping on chitosan coated citrus limetta peels biomass in synthetic wastewater. Sep. Sci. Technol. 2021, 57, 1609–1626. [Google Scholar] [CrossRef]
- Qasem, N.A.A.; Mohammed, R.H.; Lawal, D.U. Removal of heavy metal ions from wastewater: A comprehensive and critical review. npj Clean Water 2021, 4, 36. [Google Scholar] [CrossRef]
- Bazrafshan, E.; Mohammadi, L.; Ansari-Moghaddam, A.; Mahvi, A.H. Heavy metals removal from aqueous environments by electrocoagulation process—A systematic review. J. Environ. Health Sci. Eng. 2015, 13, 74. [Google Scholar] [CrossRef] [PubMed]
- Li, M.H.; Gao, X.Y.; Li, C.; Yang, C.L.; Fu, C.A.; Liu, J.; Wang, R.; Chen, L.X.; Lin, J.Q.; Liu, X.M.; et al. Isolation and Identification of Chromium Reducing Bacillus Cereus Species from Chromium-Contaminated Soil for the Biological Detoxification of Chromium. Int. J. Environ. Res. Public Health 2020, 17, 2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; You, S.; Liu, L. Characterization of Microbial Communities, Identification of Cr(VI) Reducing Bacteria in Constructed Wetland and Cr(VI) Removal Ability of Bacillus cereus. Sci. Rep. 2019, 9, 12873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Singh, M.P. Computational analysis of microbial community using amplicon sequencing of 16S rRNA gene. Res. J. Biotechnol. 2022, 17, 143–150. [Google Scholar] [CrossRef]
- Singh, N.; Singh, V.; Singh, M.P. Microbial degradation of lignocellulosic biomass for bioenergy production: A metagenomic-based approach. Biocatal. Biotransform. 2022. [Google Scholar] [CrossRef]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Devi, T.; Verma, V.; Rasool, S. Comparative studies on the extraction of metagenomic DNA from various soil and sediment samples of Jammu and Kashmir region in prospect for novel biocatalysts. IOSR J. Environ. Sci. Toxicol. Food Technol. 2014, 8, 46–56. [Google Scholar]
- Ahmed, W.; Dai, Z.; Zhang, J.; Li, S.; Ahmed, A.; Munir, S.; Liu, Q.; Tan, Y.; Ji, G.; Zhao, Z. Plant-Microbe Interaction: Mining the Impact of Native Bacillus amyloliquefaciens WS-10 on Tobacco Bacterial Wilt Disease and Rhizosphere Microbial Communities. Microbiol. Spectr. 2022, 10, e0147122. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2020. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 3 May 2020).
- Babraham Bioinformatics—TrimGalore! 2020. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 3 May 2020).
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N. Greengenes, a Chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, W.A.; Jin, Z.; Youngblut, N.; Wallace, J.G.; Sutter, J.; Zhang, W.; González-Peña, A.; Peiffer, J.; Koren, O.; Shi, Q. Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Natl. Acad. Sci. USA 2018, 115, 7368–7373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Jenkins, C.; Webb, R.I. Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl. Environ. Microbiol. 2002, 68, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.L.; Zhang, C.F.; Jin, R.; Zhang, Y.Q. Steroidobacter flavus sp. nov., a microcystin-degrading Gammaproteobacterium isolated from soil. Antonie Van Leeuwenhoek 2016, 109, 1073–1079. [Google Scholar] [CrossRef]
- Holmes, N.A.; Innocent, T.M.; Heine, D.; Bassam, M.A.; Worsley, S.F.; Trottmann, F.; Patrick, E.H.; Yu, D.W.; Murrell, J.C.; Schiøtt, M. Genome Analysis of Two Pseudonocardia Phylotypes Associated with Acromyrmex Leafcutter Ants Reveals Their Biosynthetic Potential. Front. Microbiol. 2016, 7, 2073. [Google Scholar] [CrossRef] [Green Version]
- Srinivas, A.; Sasikala, C.; Ramana, C.V. Rhodoplanes oryzaesp. nov., a phototrophicalphaproteobacterium isolated from the rhizospheresoil of paddy. Int. J. Syst. Evol. Microbiol. 2014, 64, 2198–2203. [Google Scholar] [CrossRef]
- Li, C.; Hu, H.W.; Chen, Q.L.; Chen, D.; He, J.Z. Comammox Nitrospira play an active role in nitrification of agricultural soils amended with nitrogen fertilizers. Soil Biol. Biochem. 2019, 138, 107609. [Google Scholar] [CrossRef]
- Busti, E.; Monciardini, P.; Cavaletti, L.; Bamonte, R.; Lazzarini, A.; Sosio, M.; Donadio, S. Antibiotic-producing ability by representatives of a newly discovered lineage of actinomycetes. Microbiology 2006, 152, 675–683. [Google Scholar] [CrossRef] [Green Version]
- Sit, C.S.; Ruzzini, A.C.; Van Arnam, E.B.; Ramadhar, T.R.; Currie, C.R.; Clardy, J. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc. Natl. Acad. Sci. USA 2015, 112, 13150–13154. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.; Park, W. Acinetobacter species as model microorganisms in environmental microbiology: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 2533–2548. [Google Scholar] [CrossRef]
- Lopez-Nunez, R.; Ajmal-Poley, F.; González-Pérez, J.A.; Bello-López, M.A.; Burgos-Doménech, P. Quick Analysis of Organic Amendments via Portable X-ray Fluorescence Spectrometry. Int. J. Environ. Res. Public Health 2019, 16, 4317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zocche, J.J.; Rohr, P.; Damiani, A.P.; Leffa, D.D.; Martins, M.C.; Zocche, C.M.; Teixeira, K.O.; Borges, G.D.; Jesus, M.M.; Santos, C.E.I.D.; et al. Elemental composition of vegetables cultivated over coal-mining waste. An. Acad. Bras. Cienc. 2017, 89, 2383–2398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krupenikov, I.A.; Boincean, B.P.; Dent, D. Soil Mineralogy and Elemental Composition. In The Black Earth. International Year of Planet Earth; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar] [CrossRef]
- Upadhyay, N.; Vishwakarma, K.; Singh, J. Tolerance and Reduction of Chromium (VI) by Bacillus sp. MNU16 Isolated from Contaminated Coal Mining Soil. Front. Plant Sci. 2017, 8, 778–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masood, F.; Malik, A. Hexavalent chromium reduction by Bacillus sp. strain FM1 isolated from heavy-metal contaminated soil. Bull. Environ. Contam. Toxicol. 2011, 86, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Yilmazer, P.; Saracoglu, N. Bioaccumulation and biosorption of copper(II) and chromium(III) from aqueous solutions by Pichia stipitis yeast. J. Chem. Technol. Biotechnol. 2009, 84, 604–610. [Google Scholar] [CrossRef]
- Feng, G.; Xie, T.; Wang, X.; Bai, J.; Tang, L.; Zhao, H.; Wei, W.; Wang, M.; Zhao, Y. Metagenomic analysis of microbial community and function involved in cd-contaminated soil. BMC Microbiol. 2018, 18, 11. [Google Scholar] [CrossRef] [Green Version]
- Iliev, I.; Marhova, M.; Kostadinova, S.; Gochev, V.; Tsankova, M.; Ivanova, A.; Yahubyan, G.; Baev, V. Metagenomic analysis of the microbial community structure in protected wetlands in the Maritza River Basin. Biotechnol. Biotechnol. Equip. 2019, 33, 1721–1732. [Google Scholar] [CrossRef] [Green Version]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2015, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Bond, J.G.; Marina, C.F.; Williams, T. The naturally derived insecticide spinosad is highly toxic to Aedes and Anopheles mosquito larvae. Med. Vet. Entomol. 2004, 18, 50–56. [Google Scholar] [CrossRef]
- Oliveira, C.; Gunderman, L.; Coles, C.A.; Lochmann, J.; Parks, M.; Ballard, E.; Glazko, G.; Rahmatallah, Y.; Tackett, A.J.; Thomas, D.J. 16S rRNA Gene-Based Metagenomic Analysis of Ozark Cave Bacteria. Diversity 2017, 9, 31. [Google Scholar] [CrossRef] [Green Version]
- Szymańska, S.; Sikora, M.; Hrynkiewicz, K.; Tyburski, J.; Tretyn, A.; Golebiewski, M. Choosing source of microorganisms and processing technology for next generation beet bioinoculant. Sci. Rep. 2021, 11, 2829. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Bhattacherjee, A.K.; Shukla, P.K.; Singh, B. Influence of imidacloprid on bacterial community diversity of mango orchard soil assessed through 16S rRNA sequencing-based metagenomic analysis. Environ. Monit. Assess. 2021, 193, 102. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, H.; Sun, L.; Qi, G.; Chen, S.; Zhao, X. Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 2017, 7, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucker, S.; Wagner, M.; Maixner, F.; Pelletier, E.; Koch, H.; Vacherie, B.; Rattei, T.; Damsté, J.S.; Spieck, E.; Le Paslier, D. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 13479–13484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Kessel, M.A.H.J.; Speth, D.R.; Albertsen, M.; Nielsen, P.H.; Op den Camp, H.J.; Kartal, B.; Jetten, M.S.; Lücker, S. Complete nitrification by a single microorganism. Nature 2015, 528, 555–559. [Google Scholar] [CrossRef] [Green Version]
- Hruska, K.; Kaevska, M. Mycobacteria in water, soil, plants and air: A review. Vet. Med. 2012, 57, 623–679. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Perez, M.; Morales-Manzo, I.I.; Fita, A.; Rodriguez-Burruezo, A. Mitigation of drought stress in solanaceae vegetables through symbiosis with plant growth—Promoting bacteria and arbuscular mycorrhizal fungi. A review. AgroLife Sci. J. 2022, 11, 86–98. [Google Scholar]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Suciu, L.; Tarau, A.; Urda, C.; Chetan, F.; Muresanu, F.; Sopterean, L.; Boiu-Sicuia, O.A.; Barbu, L.D.N. The influence of bacterial inoculats on pathogens, yield and quality in soybean crop. AgroLife Sci. J. 2021, 10, 221–226. [Google Scholar]



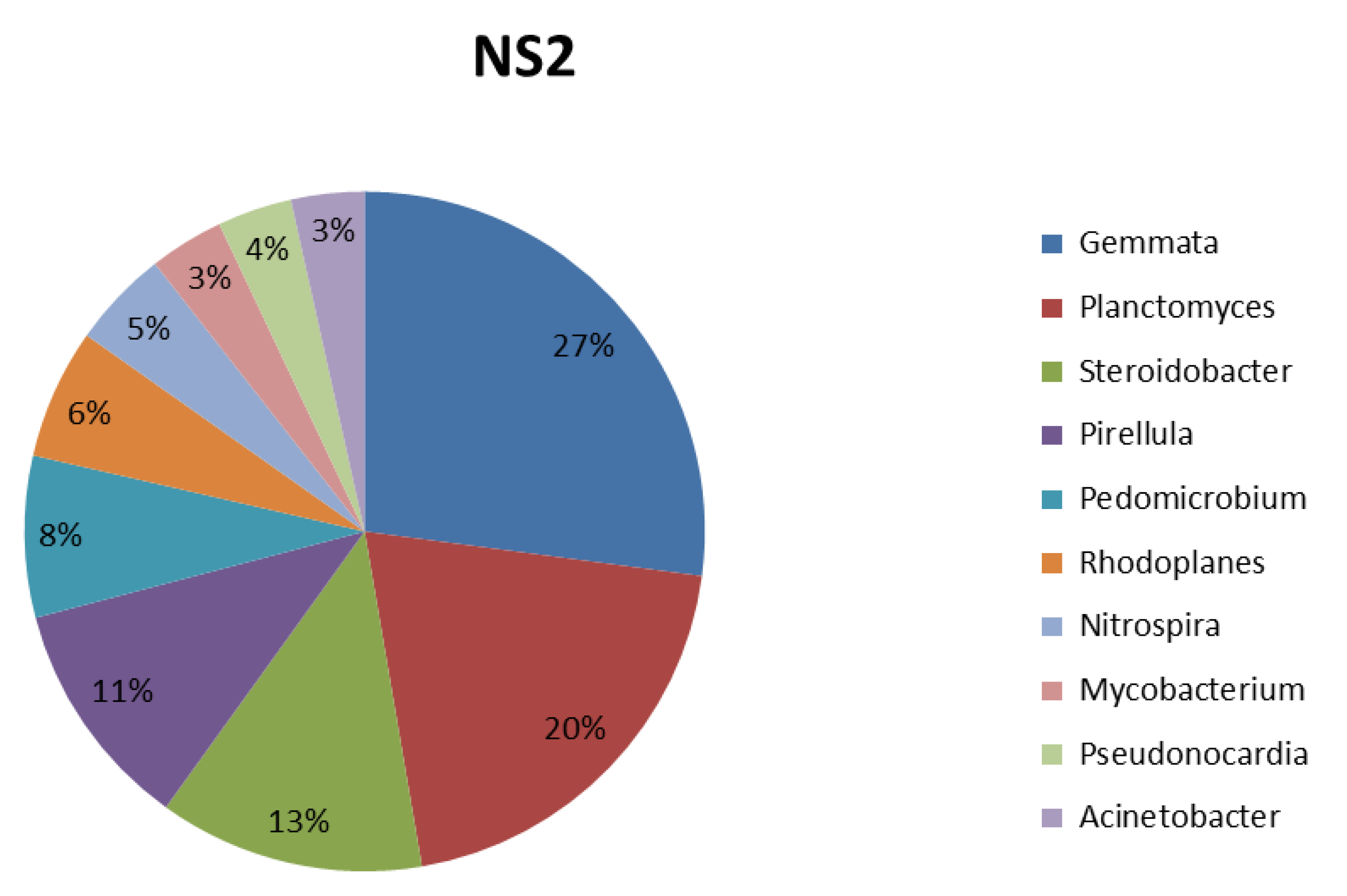
| Bacteria (Genus) | Function (Characteristics) | References |
|---|---|---|
| Gemmata | Chemoheterotrophic aerobes | [46] |
| Planctomyces | It is found in microbial fuel cell systems; it plays a role in bioconversion and energy-transfer processes. | [46] |
| Steroidobacter | Agar-degrading bacteria, | [47] |
| Pirellula | Chemoheterotrophic aerobes play a role in the degradation of sulfated glycopolymers. | [46] |
| Pedomicrobium | More dominant in the crop field rhizosphere. Primarily found in sugar beet. It shows beneficial interaction with plants and comprises numerous bacteria with N2-fixing capability. | [48] |
| Rhodoplanes | Phototrophic bacteria are present in the rhizosphere soil of paddy. | [49] |
| Nitrospira | They are ubiquitous bacteria that play a role in the nitrification of fertilized soil. | [50] |
| Mycobacterium | It is significantly enriched in the rhizosphere soil. | [51] |
| Pseudonocardia | It is a plant-associated microbial community. It improves soil nutrients, promotes plant growth, and controls soil-borne disease. It also plays a vital role in the degradation of xylan through the production of xylanase. | [52] |
| Acinetobacter | It implies active participation in the nutrient cycle in the ecosystem. It involves the degradation of various long-chain dicarboxylic acids and aromatic and hydroxylated aromatic compounds associated with plant degradation products. | [53] |
| OTU Number | Kingdom | Phylum | Class | Order | Family | Genus |
|---|---|---|---|---|---|---|
| 910 | Bacteria | Planctomycetes | Planctomycetia | Gemmatales | Gemmataceae | Gemmata |
| 679 | Bacteria | Planctomycetes | Planctomycetia | Planctomycetales | Planctomycetaceae | Planctomyces |
| 424 | Bacteria | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Sinobacteraceae | Steroidobacter |
| Bacteria | Planctomycetes | Planctomycetia | Pirellulales | Pirellulaceae | Pirellula | |
| 257 | Bacteria | Proteobacteria | Alphaproteobacteria | Rhizobiales | Hyphomicrobiaceae | Pedomicrobium |
| 208 | Bacteria | Proteobacteria | Alphaproteobacteria | Rhizobiales | Hyphomicrobiaceae | Rhodoplanes |
| 156 | Bacteria | Nitrospirae | Nitrospira | Nitrospirales | Nitrospiraceae | Nitrospira |
| 119 | Bacteria | Actinobacteria | Actinobacteria | Actinomycetales | Mycobacteriaceae | Mycobacterium |
| 119 | Bacteria | Actinobacteria | Actinobacteria | Actinomycetales | Pseudonocardiaceae | Pseudonocardia |
| 115 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Acinetobacter |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, N.; Singh, V.; Rai, S.N.; Vamanu, E.; Singh, M.P. Metagenomic Analysis of Garden Soil-Derived Microbial Consortia and Unveiling Their Metabolic Potential in Mitigating Toxic Hexavalent Chromium. Life 2022, 12, 2094. https://doi.org/10.3390/life12122094
Singh N, Singh V, Rai SN, Vamanu E, Singh MP. Metagenomic Analysis of Garden Soil-Derived Microbial Consortia and Unveiling Their Metabolic Potential in Mitigating Toxic Hexavalent Chromium. Life. 2022; 12(12):2094. https://doi.org/10.3390/life12122094
Chicago/Turabian StyleSingh, Nidhi, Veer Singh, Sachchida Nand Rai, Emanuel Vamanu, and Mohan P. Singh. 2022. "Metagenomic Analysis of Garden Soil-Derived Microbial Consortia and Unveiling Their Metabolic Potential in Mitigating Toxic Hexavalent Chromium" Life 12, no. 12: 2094. https://doi.org/10.3390/life12122094
APA StyleSingh, N., Singh, V., Rai, S. N., Vamanu, E., & Singh, M. P. (2022). Metagenomic Analysis of Garden Soil-Derived Microbial Consortia and Unveiling Their Metabolic Potential in Mitigating Toxic Hexavalent Chromium. Life, 12(12), 2094. https://doi.org/10.3390/life12122094







