Screening for Volatile α-Unsaturated Ester-Producing Yeasts from the Feces of Wild Animals in South Africa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Materials
2.2. Isolation and Selection of ILV+ Yeast Strains
2.3. Qualitative and Semi-Quantitative Analysis of Volatile Organic Compounds (VOCs)
2.4. Colony PCR and Yeast Identification Using rDNA Sequence Analysis
2.5. Statistical Analysis
3. Results
3.1. Mammalian Feces: An Original Yeast Species Resource
3.2. Selection of ILV+ Strains Producing Volatile Organic Compounds (VOCs)
3.3. VOCs Produced by ILV+ Aroma-Producing Isolates
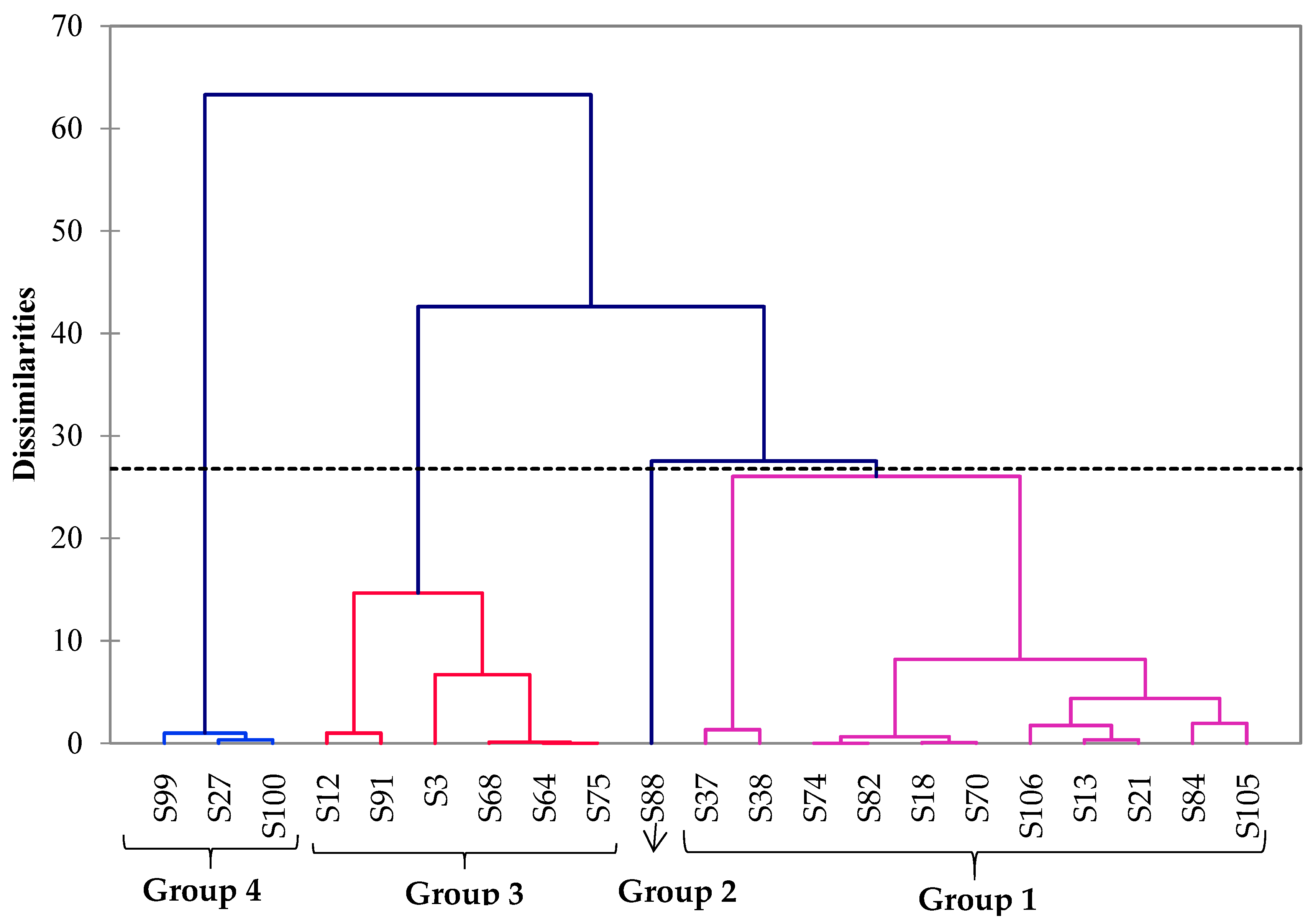
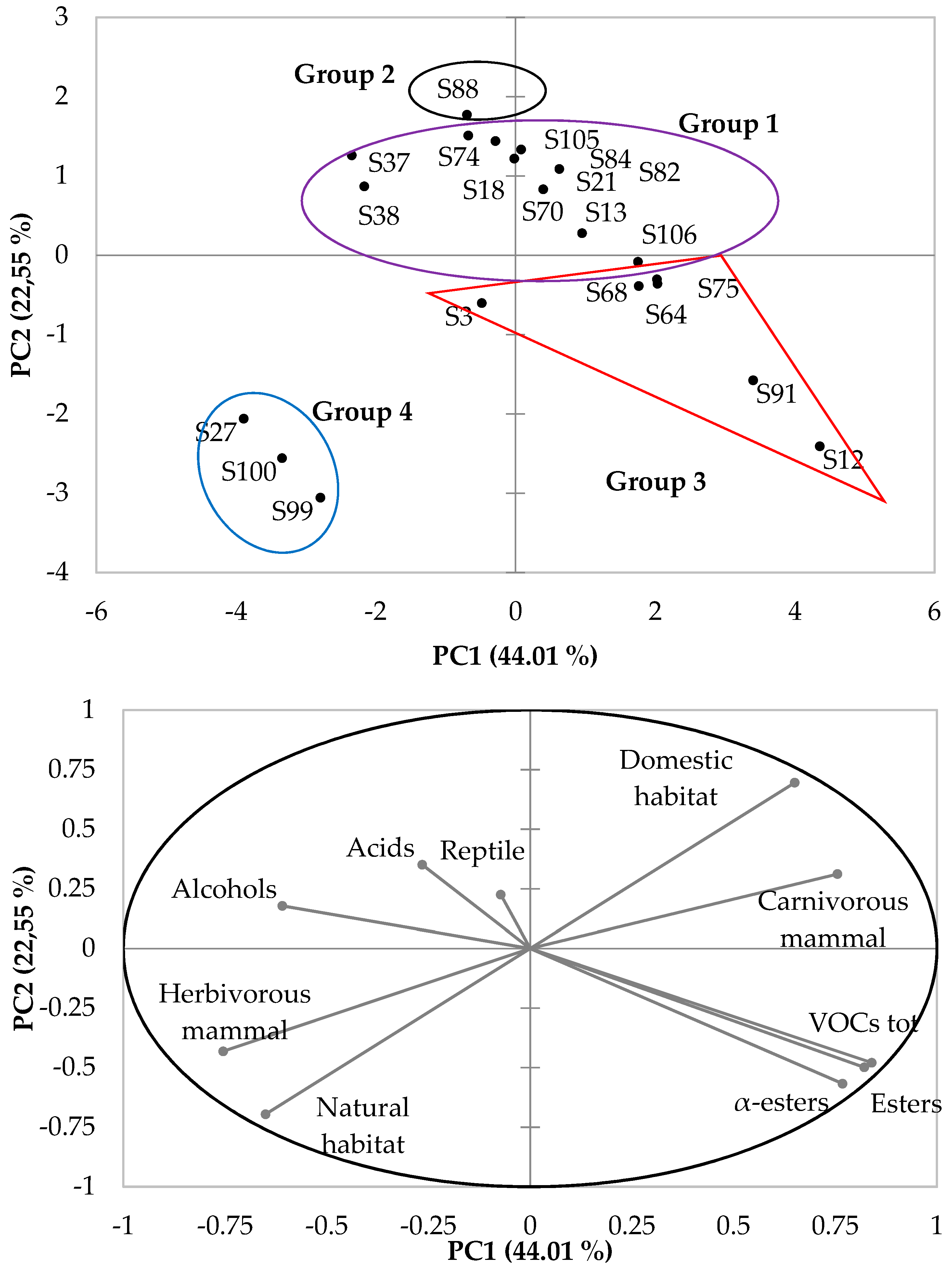
3.4. Multivariate Analysis of ILV+ Aroma-Producing Yeasts, Isolated Based on Their Origins and Their Volatile Organic Compound Production
3.5. Identification of Strains S12 and S91
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bogacz-Radomska, L.; Pietkiewicz, J.J.; Górska, K. Aroma production and application in food products. In Proceedings of the 35th International Conference of Slovak Society of Chemical Engineering, Vysoké Tatry, Slovakia, 26–30 May 2008. [Google Scholar]
- Bhari, R.; Singh, R.S. Microbial production of natural flavours. In Technology of Handling, Packaging, Processing, Preservation of Fruits and Vegetables; Theory and Practicals: New Dehli, India, 2019; pp. 767–813. [Google Scholar]
- Roy, P.; Kumar, V. Production of bioflavour from microbial sources and its health benefits. Indian J. Biochem. Biophys. 2019, 56, 352–357. [Google Scholar]
- Sales, A.; Paulino, B.N.; Pastore, G.M.; Bicas, J.L. Biogeneration of aroma compounds. Curr. Opin. Food Sci. 2018, 19, 77–84. [Google Scholar] [CrossRef]
- Felipe, L.D.O.; de Oliveira, A.M.; Bicas, J.L. Bioaromas—Perspectives for sustainable development. Trends Food Sci. Technol. 2017, 62, 141–153. [Google Scholar] [CrossRef]
- Ebert, B.E.; Blank, L.; Halbfeld, C. Exploration and Exploitation of the Yeast Volatilome. Curr. Metabol. 2017, 5, 102–118. [Google Scholar] [CrossRef]
- Hazelwood, L.A.; Daran, J.-M.; van Maris, A.J.A.; Pronk, J.T.; Dickinson, J.R. The Ehrlich Pathway for Fusel Alcohol Production: A Century of Research on Saccharomyces cerevisiae Metabolism. Appl. Environ. Microbiol. 2008, 74, 3920. [Google Scholar] [CrossRef] [Green Version]
- Budroni, M.; Zara, G.; Ciani, M.; Comitini, F. Saccharomyces and Non-Saccharomyces Starter Yeasts; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Caro, Y.; Shum-Cheong-Sing, A.; Robert, L.; François, J.-M.; Petit, T. Evaluation of mixed-fermentation of Saccharomyces cerevisiae with Saprochaete suaveolens to produce natural fruity beer from industrial wort. Food Chem. 2021, 346, 128804. [Google Scholar] [CrossRef] [PubMed]
- Schifferdecker, A.J.; Dashko, S.; Ishchuk, O.P.; Piškur, J. The wine and beer yeast Dekkera bruxellensis. Yeast 2014, 31, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Tataridis, P.; Kanellis, A.; Logothetis, S.; Nerantzis, E. Use of non-Saccharomyces Torulaspora delbrueckii yeast strains in winemaking and brewing. J. Nat. Sci. 2013, 124, 415–426. [Google Scholar] [CrossRef]
- Ravasio, D.; Carlin, S.; Boekhout, T.; Groenewald, M.; Vrhovsek, U.; Walther, A.; Wendland, J. Adding Flavor to Beverages with Non-Conventional Yeasts. Fermentation 2018, 4, 15. [Google Scholar] [CrossRef] [Green Version]
- Tufariello, M.; Fragasso, M.; Pico, J.; Panighel, A.; Castellarin, S.D.; Flamini, R.; Grieco, F. Influence of Non-Saccharomyces on Wine Chemistry: A Focus on Aroma-Related Compounds. Molecules 2021, 26, 644. [Google Scholar] [CrossRef]
- Buzzini, P.; Gasparetti, C.; Turchetti, B.; Cramarossa, M.R.; Vaughan-Martini, A.; Martini, A.; Pagnoni, U.M.; Forti, L. Production of volatile organic compounds (VOCs) by yeasts isolated from the ascocarps of black (Tuber melanosporum Vitt.) and white (Tuber magnatum Pico) truffles. Arch. Microbiol. 2005, 184, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Buzzini, P.; Martini, A.; Cappelli, F.; Pagnoni, U.M.; Davoli, P. A study on volatile organic compounds (VOCs) produced by tropical ascomycetous yeasts. Antonie Leeuwenhoek 2003, 84, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Grondin, E.; Sing, A.S.C.; Caro, Y.; Raherimandimby, M.; Randrianierenana, A.L.; James, S.; Nueno-Palop, C.; François, J.M.; Petit, T. A comparative study on the potential of epiphytic yeasts isolated from tropical fruits to produce flavoring compounds. Int. J. Food Microbiol. 2015, 203, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grondin, E.; Sing, A.S.C.; James, S.; Nueno-Palop, C.; François, J.M.; Petit, T. Flavour production by Saprochaete and Geotrichum yeasts and their close relatives. Food Chem. 2017, 237, 677–684. [Google Scholar] [CrossRef]
- Contarino, R.; Brighina, S.; Fallico, B.; Cirvilleri, G.; Parafati, L.; Restuccia, C. Volatile organic compounds (VOCs) produced by biocontrol yeasts. Food Microbiol. 2019, 82, 70–74. [Google Scholar] [CrossRef]
- Amoikon, T.L.S.; Marcotte, S.; Djeni, T.N.; N’Sa, K.M.C.; Grondin, C.; Tinsley, C.; Casaregola, S.; Dje, M.K. A study on the potential of yeasts isolated from palm wines to produce flavouring compounds. LWT 2020, 128, 109506. [Google Scholar] [CrossRef]
- Gethins, L.; Guneser, O.; Demirkol, A.; Rea, M.C.; Stanton, C.; Ross, R.P.; Karagul Yuceer, Y.; Morrissey, J.P. Influence of Carbon and Nitrogen source on production of volatile fragrance and flavour metabolites by the yeast Kluyveromyces marxianus: Nutrient effects on volatiles in K. marxianus. Yeast 2014, 32, 67–76. [Google Scholar] [CrossRef]
- Basso, R.F.; Alcarde, A.R.; Portugal, C.B. Could non-Saccharomyces yeasts contribute on innovative brewing fermentations? Food Res. Int. 2016, 86, 112–120. [Google Scholar] [CrossRef]
- Saerens, S.; Swiegers, J.H. Production of Low-Alcohol or Alcohol Free Beer with Pichia Kluyveri Yeast Strains. 2014. Available online: https://www.freepatentsonline.com/EP2964742.html (accessed on 14 November 2022).
- Saerens, S.; Swiegers, J.H. Enhancement of Beer Flavor by a Combination of Pichia Yeast and Different Hop Varieties. 2013. Available online: https://www.semanticscholar.org/paper/ENHANCEMENT-OF-BEER-FLAVOR-BY-A-COMBINATION-OF-AND/b3c0c648bd467fa22e010863b74c0ea9bfe0df25 (accessed on 14 November 2022).
- Michel, M.; Kopecká, J.; Meier-Dörnberg, T.; Zarnkow, M.; Jacob, F.; Hutzler, M. Screening for new brewing yeasts in the non-Saccharomyces sector with Torulaspora delbrueckii as model. Yeast 2016, 33, 129–144. [Google Scholar] [CrossRef] [Green Version]
- Canonico, L.; Agarbati, A.; Comitini, F.; Ciani, M. Torulaspora delbrueckii in the brewing process: A new approach to enhance bioflavour and to reduce ethanol content. Food Microbiol. 2016, 56, 45–51. [Google Scholar] [CrossRef]
- Canonico, L.; Galli, E.; Ciani, E.; Comitini, F.; Ciani, M. Exploitation of Three Non-Conventional Yeast Species in the Brewing Process. Microorganisms 2019, 7, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callejo, M.J.; Navas, J.J.G.; Alba, R.; Escott, C.; Loira, I.; González, M.C.; Morata, A. Wort fermentation and beer conditioning with selected non-Saccharomyces yeasts in craft beers. Eur. Food Res. Technol. 2019, 245, 1229–1238. [Google Scholar] [CrossRef]
- Jayanthi, P.D.K.; Woodcock, C.M.; Caulfield, J.; Birkett, M.; Bruce, T.J.A. Isolation and Identification of Host Cues from Mango, Mangifera indica, That Attract Gravid Female Oriental Fruit fly, Bactrocera dorsalis. J. Chem. Ecol. 2012, 38, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Ceva-Antunes, P.M.; Bizzo, H.R.; Silva, A.S.; Carvalho, C.; Antunes, O. Analysis of volatile composition of siriguela (Spondias purpurea L.) by solid phase microextraction (SPME). LWT 2006, 39, 437–443. [Google Scholar] [CrossRef]
- Quijano, C.E.; Pino, J.A. Analysis of Volatile Compounds of Cacao Maraco (Theobroma bicolorHumb. et Bonpl.) Fruit. J. Essent. Oil Res. 2009, 21, 211–215. [Google Scholar] [CrossRef]
- Tateo, F.; Bononi, M. Headspace-SPME Analysis of Volatiles from Quince Whole Fruits. J. Essent. Oil Res. 2010, 22, 416–418. [Google Scholar] [CrossRef]
- Pino, J.; Marbot, R.; Rosado, A. Volatile constituents of star apple (Chrysophyllum cainito L.) from Cuba. Flavour Fragr. J. 2002, 17, 401–403. [Google Scholar] [CrossRef]
- Grondin, E.; Shum Cheong Sing, A.; Caro, Y.; Billerbeck, G.M.; François, J.M.; Petit, T. Physiological and biochemical char-acteristics of the ethyl tiglate production pathway in the yeast Saprochaete suaveolens. Yeast 2015, 32, 57–66. [Google Scholar]
- Tan, M.; Caro, Y.; Sing, A.S.C.; Reiss, H.; Francois, J.-M.; Petit, T. Selection by UV Mutagenesis and Physiological Characterization of Mutant Strains of the Yeast Saprochaete suaveolens (Former Geotrichum fragrans) with Higher Capacity to Produce Flavor Compounds. J. Fungi 2021, 7, 1031. [Google Scholar] [CrossRef]
- Lorliam, W.; Akaracharanya, A.; Jindamorakot, S.; Suwannarangsee, S.; Tanasupawat, S. Characterization of xylose-utilizing yeasts isolated from herbivore faeces in Thailand. ScienceAsia 2013, 39, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Kumar, C.G.; Mongolla, P.; Joseph, J.; Nageswar, Y.; Kamal, A. Antimicrobial activity from the extracts of fungal isolates of soil and dung samples from Kaziranga National Park, Assam, India. J. Mycol. Méd. 2010, 20, 283–289. [Google Scholar] [CrossRef]
- Van Dijken, J.P.; Bauer, J.; Brambilla, L.; Duboc, P.; Francois, J.M.; Gancedo, C.; Giuseppin, M.L.F.; Heijnen, J.J.; Hoare, M.; Lange, H.C.; et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 2000, 26, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C. Recognition of yeast species gene sequence comparisons. Open Appl. Inf. J. 2011, 5, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Delsuc, F.; Metcalf, J.L.; Parfrey, L.W.; Song, S.J.; González, A.; Knight, R. Convergence of gut microbiomes in myrmecophagous mammals. Mol. Ecol. 2013, 23, 1301–1317. [Google Scholar] [CrossRef] [PubMed]
- Hammer, T.J.; Sanders, J.G.; Fierer, N. Not all animals need a microbiome. FEMS Microbiol. Lett. 2019, 366, fnz117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budd, K.; Gunn, J.C.; Finch, T.; Klymus, K.; Sitati, N.; Eggert, L.S. Effects of diet, habitat, and phylogeny on the fecal microbiome of wild African savanna (Loxodonta africana) and forest elephants (L. cyclotis). Ecol. Evol. 2020, 10, 5637–5650. [Google Scholar] [CrossRef]
- Desselberger, U. The Mammalian Intestinal Microbiome: Composition, Interaction with the Immune System, Significance for Vaccine Efficacy, and Potential for Disease Therapy. Pathogens 2018, 7, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beasley, D.; Koltz, A.M.; Lambert, J.E.; Fierer, N.; Dunn, R. The Evolution of Stomach Acidity and Its Relevance to the Human Microbiome. PLoS ONE 2015, 10, e0134116. [Google Scholar] [CrossRef]
- Oliveira, B.C.M.; Murray, M.; Tseng, F.; Widmer, G. The fecal microbiota of wild and captive raptors. Anim. Microbiome 2020, 2, 15. [Google Scholar] [CrossRef]
- Nicholson, M.J.; McSweeney, C.S.; Mackie, R.I.; Brookman, J.L.; Theodorou, M.K. Diversity of anaerobic gut fungal populations analysed using ribosomal ITS1 sequences in faeces of wild and domesticated herbivores. Anaerobe 2010, 16, 66–73. [Google Scholar] [CrossRef]
- Grenet, E.; Fonty, G.; Jamot, J.; Bonnemoy, F. Influence of diet and monensin on development of anaerobic fungi in the rumen, duodenum, cecum, and feces of cows. Appl. Environ. Microbiol. 1989, 55, 2360–2364. [Google Scholar] [CrossRef] [PubMed]
- Maggio-Hall, L.A.; Lyne, P.; Wolff, J.A.; Keller, N.P. A single acyl-CoA dehydrogenase is required for catabolism of isoleucine, valine and short-chain fatty acids in Aspergillus nidulans. Fungal Genet. Biol. 2008, 45, 180–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maggio-Hall, L.A.; Keller, N.P. Mitochondrial β-oxidation in Aspergillus nidulans. Mol. Microbiol. 2004, 54, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
- Kováts, E. Gas-chromatographische Charakterisierung organischer Verbindungen. Teil 1: Retentionsindices aliphatischer Halogenide, Alkohole, Aldehyde und Ketone. Helvetica Chim. Acta 1958, 41, 1915–1932. [Google Scholar] [CrossRef]
- Nagata, Y. Measurement of odor threshold by triangle odor bag method. Odor Meas. Rev. 2003, 10, 118–127. [Google Scholar]
- Chen, J.L.; Wu, J.H.; Wang, Q.; Deng, A.H.; Hu, X.S. Changes in the Volatile Compounds and Chemical and Physical Properties of Kuerle Fragrant Pear (Pyrus serotina Reld) during Storage. J. Agric. Food Chem. 2006, 54, 8842–8847. [Google Scholar] [CrossRef]
- Czerny, M.; Christlbauer, M.; Christlbauer, M.; Fischer, A.; Granvogl, M.; Hammer, M.; Hartl, C.; Hernandez, N.M.; Schieberle, P. Re-investigation on odour thresholds of key food aroma compounds and development of an aroma language based on odour qualities of defined aqueous odorant solutions. Eur. Food Res. Technol. 2008, 228, 265–273. [Google Scholar] [CrossRef]
- Leffingwell & Associates. Odor & Flavor Detection Thresholds in Water (in Parts Per Billion); Leffingwell & Associates: Canton, GA, USA, 2020; Available online: http://www.leffingwell.com/odorthre.htm (accessed on 13 May 2020).
- Suomalainen, H.; Lehtonen, M. The Production of Aroma Compounds by Yeast. J. Inst. Brew. 1979, 85, 149–156. [Google Scholar] [CrossRef]
- Damasceno, S.; Cereda, M.P.; Pastore, G.; Oliveira, J. Production of volatile compounds by Geotrichum fragrans using cassava wastewater as substrate. Process Biochem. 2003, 39, 411–414. [Google Scholar] [CrossRef]
- İşcan, G.; Demirci, B.; Demirci, F.; Başer, K.H.C. Headspace solid phase microextraction (HS-SPME) and analysis of Geotrichum fragrans volatiles. Nat. Volatiles Essent. Oils 2015, 2, 45–51. [Google Scholar]
| Site Name | Type of Site and Address |
|---|---|
| Moreleta Kloof Nature Reserve | Game reserve (Helios Street, Moreleta park, Pretoria, Gauteng) |
| Kruger National Park | National Park (Kruger National Park, Mpumalanga) |
| Lory Park Zoo | Zoo (180/1 Kruger Road, President Park, Midrand, Gauteng) |
| McCrindle Farm | Game reserve (Pretoria, Gauteng) |
| Emoya big cat sanctuary | Zoo (Farm Milumbe, D1939, Lephalale, Limpopo) |
| Elephant Sanctuary | Zoo (Hartbeespoort dam) |
| Moholoholo Animal Rehabilitation center | Zoo (Limpopo) |
| Animals | Type of Animal | Sampling | Origin | No. of Different Strains |
|---|---|---|---|---|
| Aardwolf (Proteles cristata) | Carnivorous mammal | Freeze-dried | Not communicated | 2 (S1/S2) |
| Abyssinian hornbill (Bucorvus abyssinicus) | Bird | Swab | Lory Park Zoo | 0 |
| Bateleur eagle (Terathopius ecaudatus) | Raptor | Swab | Moholoholo Animal Rehabilitation Center | 0 |
| Black-and-white-casqued hornbill (Bycanistes subcylindricus) | Bird | Swab | Lory Park Zoo | 1 (S31) |
| Black-crested gibbon (Nomascus concolor) | Herbivorous mammal | Swab | Lory Park Zoo | 2 (S3 */S4 *) |
| Black jaguar (Panthera pardus) | Carnivorous mammal | Swab | Lory Park Zoo | 3 (S6/S7/S8) |
| Black-footed cat (Felis nigripes) | Carnivorous mammal | Tubes | Lory Park Zoo | 4 (S12 */S14/S15/S16)) |
| Swab | Lory Park Zoo | 3 (S13 */S17 */S18 *) | ||
| Blesbok (Damaliscus pygarus) | Herbivorous mammal | Tube | Moreleta Kloof Nature Reserve | 2 (S9/S10) |
| Brown lemur (Eulemur fulvus) | Herbivorous mammal | Swab | Lory Park Zoo | 3 (S58 */S59/S60 *) |
| Brown jaguar (Panthera onca) | Carnivorous mammal | Swab | Lory Park Zoo | 5 (S20/S21 */S22 */S23/S24) |
| Buffalo (Syncerus caffer) | Herbivorous mammal | Swab | Kruger National Park | 2 (S28 */S29 *) |
| Freeze-dried | Not communicated | 1 (S30) | ||
| Bushbuck (Tragelaphus sylvaticus) | Herbivorous mammal | Tube | Moreleta Kloof Nature Reserve | 2 (S26/S27 *) |
| Swab | Moreleta Kloof Nature Reserve | 0 | ||
| Cape vulture (Gyps coprotheres) | Raptor | Swab | Moholoholo Animal Rehabilitation Center | 0 |
| Cheetah (Acinonyx jubatus) | Carnivorous mammal | Tube | Lory Park Zoo | 2 (S32/S33 *) |
| Swab | Lory Park Zoo | 2 (S34/S35) | ||
| Freeze-dried | Not communicated | 0 | ||
| Duiker (Sylvicapra grimmia) | Herbivorous mammal | Tube | Moholoholo Animal Rehabilitation Center | 2 (S37 */S38 *) |
| Elephant (Loxodonta africana) | Herbivorous mammal | Swab | Kruger National Park | 0 |
| Freeze-dried | Not communicated | 1 (S42) | ||
| Swab | Elephant Sanctuary | 3 (S39/S40 */S41) | ||
| Gibbon (Hylobates lar) | Herbivorous mammal | Swab | Lory Park Zoo | 3 (S43/S44 */S45) |
| Giraffe (Giraffa cameopardalis) | Herbivorous mammal | Freeze-dried | Not communicated | 1 (S46) |
| Hyena (Crocuta crocuta) | Carnivorous mammal | Swab | Lory Park Zoo | 1 (S47) |
| Freeze-dried | Not communicated | 1 (S48) | ||
| Impala (Aepyceros melampus) | Herbivorous mammal | Tube | Kruger National Park | 2 (S49/S50) |
| Tube | McCrindle Farm | 2 (S51/S52) | ||
| King vulture (Sarcoramphus papa) | Raptor | Swab | Lory Park Zoo | 0 |
| Kudu (Tragelaphus strepsiceros) | Herbivorous mammal | Tube | McCrindle Farm | 2 (S53/S54) |
| Lappet-faced vulture (Torgos tracheliotos) | Raptor | Swab | Moholoholo Animal Rehabilitation Center | 0 |
| Lemur (Varecia variegate) | Herbivorous mammal | Tube | Lory Park Zoo | 3 (S55/S56/S57) |
| Swab | Lory Park Zoo | 0 | ||
| Leopard (Panthera pardus) | Carnivorous mammal | Swab | Lory Park Zoo | 7 (S62 */S63 */S64 */S65 */S68 */S69/S70 *) |
| Tube | Lory Park Zoo | 2 (S66/S67) | ||
| Freeze-dried | Not communicated | 0 | ||
| Lion (Panthera leo) | Carnivorous mammal | Freeze-dried | Not communicated | 1 (S80) |
| Tube | Emoya big cat sanctuary | 10 (S71/S72 */S78 */S73/S74 */S75 */S76/S77/S79/S80) | ||
| Swab | Emoya big cat sanctuary | 1 (S78) | ||
| Long-tailed hornbill (Tropicranus albocristatus) | Raptor | Swab | Lory Park Zoo | 0 |
| Meerkat (Suricata suricatta) | Carnivorous mammal | Tube | Lory Park Zoo | 3 (S82 */S83/S84 *) |
| Nile crocodile (Crocodylus niloticus) | Reptile | Swab | Lory Park Zoo | 4 (S85/S86/S87/S88 *) |
| Ostrich (Struthio camelus) | Bird | Swab | Moreleta Kloof Nature Reserve | 2 (S89/S90) |
| Palm nut vulture (Gypohierax angolensis) | Raptor | Swab | Lory Park Zoo | 0 |
| Serval (Leptailruus serval) | Carnivorous mammal | Tube | Lory Park Zoo | 3 (S91 */S92 * /S93) |
| Swab | Lory Park Zoo | 2 (S94/S95) | ||
| Southern hornbill (Bucorvus leadbeateri) | Raptor | Swab | Lory Park Zoo | 0 |
| Springbok (Antidorcas marsupialis) | Herbivorous mammal | Tube | Moreleta Kloof Nature Reserve | 6 (S96/S97 */S98/S99 */S100 */S101) |
| Tawney eagle (Aquila rapax) | Raptor | Swab | Moholoholo Animal Rehabilitation Center | 2 (S102/S103) |
| Tiger (Panthera tigris) | Carnivorous mammal | Tube | Lory Park Zoo | 4 (S104/S105 */S106 */S108/ S109) |
| Swab | Lory Park Zoo | 1 (S107) | ||
| White-headed vulture (Trigonoceps occipitalis) | Raptor | Swab | Lory Park Zoo | 3 (S114 /S115/S116) |
| Swab | Moholoholo Animal Rehabilitation Center | 0 | ||
| White lion (Panthero leo) | Carnivorous mammal | Swab | Lory Park Zoo | 2 (S117 */S118) |
| White rhino (Ceratotherium simum) | Herbivorous mammal | Freeze-dried | Not communicated | 2 (S119/S120) |
| Wild dog (Lycaon pictus) | Carnivorous mammal | Freeze-dried | Not communicated | 1 (S113) |
| Wildebeest (Connochaetes gnou) | Herbivorous mammal | Tube | Moreleta Kloof Nature Reserve | 2 (S111/S112) |
| Freeze-dried | Not communicated | 1 (S110) | ||
| Zebra (Equus zebra) | Herbivorous mammal | Swab | Moreleta Kloof Nature Reserve | 2 (S121 */S122) |
| Tube | Moreleta Kloof Nature Reserve | 2 (S123 */S124 *) | ||
| Swab | McCrindle Farm | 1 (S125 *) | ||
| TOTAL | 119 |
| Volatile Compounds | Odor Type (1) | IRRexp (2) | IRRth (3) | Producing Strains |
|---|---|---|---|---|
| ACIDS | ||||
| 2-methylbutanoic acid | Over-ripe fruit, cashew, sweet | 856 | 846 | S38 |
| 3-methylbutanoic acid | Sweet | 837 | 828 | S37; S84; S105 |
| Butanoic acid | Rancid | 792 | 780 | S37 |
| ALCOHOL | ||||
| 2-phenylethanol | Flower, honey, rose | 1101 | 1114 | S0; S13; S18; S21; S27; S37; S38; S70; S74; S82; S99; S100; S105; S106 |
| ESTERS | ||||
| 2-methylbutyl 2-methylbutanoate | Fruit, apple, rum, berry | 1110 | 1104 | S0 |
| 2-methylbutyl 3-methylbutanoate | Apple, cheese, earth | 1112 | 1107 | S0 |
| 2-methylbutyl butanoate | Fruit, spice, butter | 1057 | 1056 | S0 |
| 2-methylbutyl propanoate | Sweet, banana, fruit, apple, melon, | 975 | 975 | S0 |
| 2-methylpropyl 2-methylbutanoate | Sweet, fruit | 1002 | 1004 | S0 |
| 2-methylpropyl 2-methylpropanoate | Pineapple, grape skin, tropical | 955 (4) | 909 | S0; S91; S106 |
| 2-methylpropyl 3-methylbutanoate | Fruit, apple, raspberry | 892 | 904 | S0; S12; S13; S18; S68; S75; S99 |
| 2-methylpropyl butanoate | Sweet, fruit | 959 | 955 | S0 |
| 2-methylpropyl propanoate | Fruit, green ether, sweet, banana | 869 | 863 | S12 |
| 2-phenylethyl 2-methylbutanoate | Sweet, fruit, herb, floral | 1494 | 1493 | S0 |
| 2-phenylethyl 3-methylbutanoate | Apricot, sweet, bitter | 1486 | 1488 | S0; S12; S13; S27; S75 |
| 2-phenylethyl acetate | Rose, floral, fruit, sweet | 1261 | 1265 | S0 |
| 3-methylbutyl 2-methylbutanoate | Fruit | 1093 | 1102 | S0; S12; S13; S99 |
| 3-methylbutyl 2-methylpropanoate | Mixed fruit | 1011 | 1013 | S0; S12; S75 |
| 3-methylbutyl 3-methylbutanoate | Sweet, apple, fruit | 1099 | 1103 | S12; S13; S18; S75; S99; S105; |
| 3-methylbutyl acetate | Pear, banana | 872 | 871 | S0; S3; S12; S18; S21; S64; S68; S70; S75; S88; S91; S106 |
| 3-methylbutyl pentanoate | Ripe apple | 1097 | 1090 | S0; S3; S64 |
| 3-methylbutyl propanoate | Apricot, pineapple | 968 | 964 | S0; S12; S68; S75; S91; S106 |
| Butyl 2-methylbutanoate | Fresh, sweet, fruit | 1039 | 1043 | S0; S12; S91; |
| Butyl 2-methylpropanoate | Fruit | 951 | 955 | S0; S12; S64; S68; S91; S106 |
| Butyl 3-methylbutanoate | Apple, pear, sweet, pineapple, green | 1043 | 1047 | S0; S12; S106 |
| Butyl butanoate | Fresh, sweet, fruit | 995 | 993 | S12; S21; S64; S91; S100; S106 |
| Butyl propanoate | Sweet, fruit, rum | 907 | 910 | S12; S64; S75; S91; S106 |
| Ethyl 2-methylbutanoate | Fruit, green, apple, flower | 842 | 846 | S0; S3; S12; S18; S21; S68; S75; S91; S99; S105; S106 |
| Ethyl 3-methylbutanoate | Fruit, blueberry | 847 | 849 | S0; S3; S12; S13; S18; S21; S64; S68; S75; S91; S99; S105; S106 |
| Ethyl butanoate | Fruit, pear, pineapple | 799 | 800 | S0; S3; S12; S21; S37; S38; S64; S68; S70; S75; S84; S88; S91; S100; S105; S106 |
| Ethyl hexanoate | Fruit, strawberry, anise | 999 | 996 | S12; S64; S68; S84; S91 |
| Ethyl octanoate | Flower, fruit, menthol, anise, sweet | 1196 | 1198 | S0; S91 |
| Ethyl pentanoate | Fruit, orange, green | 890 | 898 | S12; S13; S64; S68; S75; S84; S91; S106 |
| Octyl 2-methylpropanoate (4) | Cream, wax, fruit, earth, fat | 1349 | 1394 | S0 |
| Octyl 2-methylbutanoate | Green, must, fruit | 1432 | 1438 | S0 |
| Octyl 3-methylbutanoate | Rose, honey, apple, pineapple | 1436 | 1440 | S0 |
| Octyl acetate | Floral, fruit, sweet | 1115 | 1149 | S0 |
| Octyl butanoate | Herb, fruit, green | 1391 | 1434 | S0 |
| Pentyl butanoate | Apricot, pineapple | 1052 | 1093 | S12; S91 |
| Propyl 3-methylbutanoate | Sweet, fruit | 947 | 943 | S0; S12; S91 |
| α-UNSATURATED ESTERS | ||||
| 2-methylpropyl (E)-2-methylbut-2-enoate | Herb, pungent | 1039 | 1034 | S0; S12; S13; S91 |
| 3-methylbutyl (E)-2-methylbut-2-enoate | Floral | 1190 | 1168 | S0; S12; S91 |
| 3-methylbutyl (E)-3-methylbut-2-enoate | 1178 | 1184 | S12 | |
| Butyl (E)-2-methylbut-2-enoate | Floral, herb, warm, fruit | 1091 | 1068 | S0 |
| Ethyl (E)-2-methylbut-2-enoate | Fruit | 938 | 936 | S0; S3; S12; S13; S64; S68; S75; S84 S91; S99; S100; S105; S106 |
| Ethyl (E)-3-methylbut-2-enoate | - | 922 | 911 | S0; S3; S12; S13; S64; S68; S75; S84; S91; S99; S105; S106 |
| Ethyl (E)-but-2-enoate | Fruit, caramel, pungent | 835 | 833 | S0; S21; S64; S106 |
| Ethyl (E)-hex-2-enoate | Fruit, slight, pungent | 1041 | 1025 | S12; S91 |
| Propyl (E)-2-methylbut-2-enoate | - | 1039 | 1034 | S0 |
| Strains | VOC Tot. | Acids | Alcohols | Esters | α-Esters |
|---|---|---|---|---|---|
| S0 | 38 | - | 1 | 30 | 7 |
| S3 | 7 | - | - | 5 | 2 |
| S12 | 27 | - | - | 21 | 6 |
| S13 | 10 | - | 1 | 6 | 3 |
| S18 | 6 | - | 1 | 5 | - |
| S21 | 8 | - | 1 | 6 | 1 |
| S27 | 2 | - | 1 | 1 | - |
| S37 | 5 | 2 | 1 | 2 | - |
| S38 | 4 | 1 | 1 | 2 | - |
| S64 | 13 | - | - | 10 | 3 |
| S68 | 12 | - | - | 10 | 2 |
| S70 | 4 | - | 1 | 3 | - |
| S74 | 1 | - | 1 | - | - |
| S75 | 14 | - | - | 12 | 2 |
| S82 | 1 | - | 1 | - | - |
| S84 | 6 | 1 | - | 3 | 2 |
| S88 | 3 | - | - | 3 | - |
| S91 | 21 | - | - | 16 | 5 |
| S99 | 8 | - | 1 | 5 | 2 |
| S100 | 5 | - | 1 | 3 | 1 |
| S105 | 8 | 1 | 1 | 5 | 1 |
| S106 | 16 | - | 1 | 12 | 3 |
| Variable | VOC | Ac. | Alc. | Est. | α-Est. | Herb. | Carn. | Rept. | Nat. | Dom. |
|---|---|---|---|---|---|---|---|---|---|---|
| VOCs | 1 | |||||||||
| Ac. | −0.211 | 1 | ||||||||
| Alc. | −0.514 | 0.169 | 1 | |||||||
| Est. | 0.990 | −0.288 | −0.560 | 1 | ||||||
| α-est. | 0.938 | −0.262 | −0.551 | 0.903 | 1 | |||||
| Herb. | −0.336 | 0.315 | 0.279 | −0.378 | −0.278 | 1 | ||||
| Carn. | 0.410 | −0.256 | −0.139 | 0.423 | 0.361 | −0.894 | 1 | |||
| Rept. | −0.194 | −0.101 | −0.285 | −0.134 | −0.209 | −0.141 | −0.316 | 1 | ||
| Nat. | −0.228 | −0.185 | 0.320 | −0.244 | −0.139 | 0.645 | −0.577 | −0.091 | 1 | |
| Dom. | 0.228 | 0.185 | −0.320 | 0.244 | 0.139 | −0.645 | 0.577 | 0.091 | −1.000 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, M.; Caro, Y.; Lebeau, J.; Shum-Cheong-Sing, A.; François, J.M.; Regnier, T.; Petit, T. Screening for Volatile α-Unsaturated Ester-Producing Yeasts from the Feces of Wild Animals in South Africa. Life 2022, 12, 1999. https://doi.org/10.3390/life12121999
Tan M, Caro Y, Lebeau J, Shum-Cheong-Sing A, François JM, Regnier T, Petit T. Screening for Volatile α-Unsaturated Ester-Producing Yeasts from the Feces of Wild Animals in South Africa. Life. 2022; 12(12):1999. https://doi.org/10.3390/life12121999
Chicago/Turabian StyleTan, Mélissa, Yanis Caro, Juliana Lebeau, Alain Shum-Cheong-Sing, Jean Marie François, Thierry Regnier, and Thomas Petit. 2022. "Screening for Volatile α-Unsaturated Ester-Producing Yeasts from the Feces of Wild Animals in South Africa" Life 12, no. 12: 1999. https://doi.org/10.3390/life12121999
APA StyleTan, M., Caro, Y., Lebeau, J., Shum-Cheong-Sing, A., François, J. M., Regnier, T., & Petit, T. (2022). Screening for Volatile α-Unsaturated Ester-Producing Yeasts from the Feces of Wild Animals in South Africa. Life, 12(12), 1999. https://doi.org/10.3390/life12121999









