Evaluation of Three Automated Extraction Systems for the Detection of SARS-CoV-2 from Clinical Respiratory Specimens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Specimens and Automated Extraction Systems
2.2. Total Nucleic Acids Extraction
2.3. Multiplex RT-qPCR Analysis
2.4. Efficiency of the Extraction Systems
2.5. Limits of Detection (LOD) of the Extraction Systems
2.6. Statistical Analysis
3. Results
3.1. Comparison of the Three Nucleic Acid Extraction Systems
3.2. Extraction Performance of the Three Systems
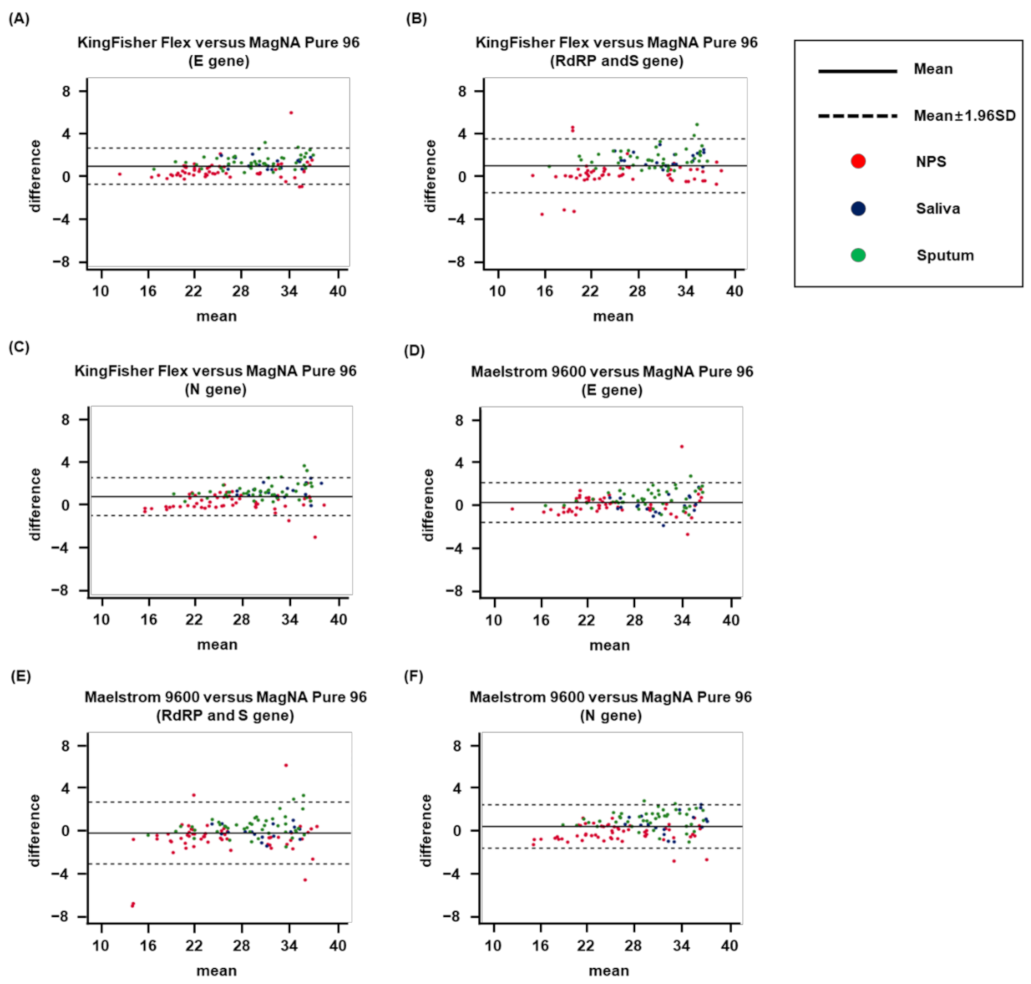
3.3. Intersystem Comparison of Ct Values
3.4. Efficiency of the Extraction Systems for SARS-CoV-2
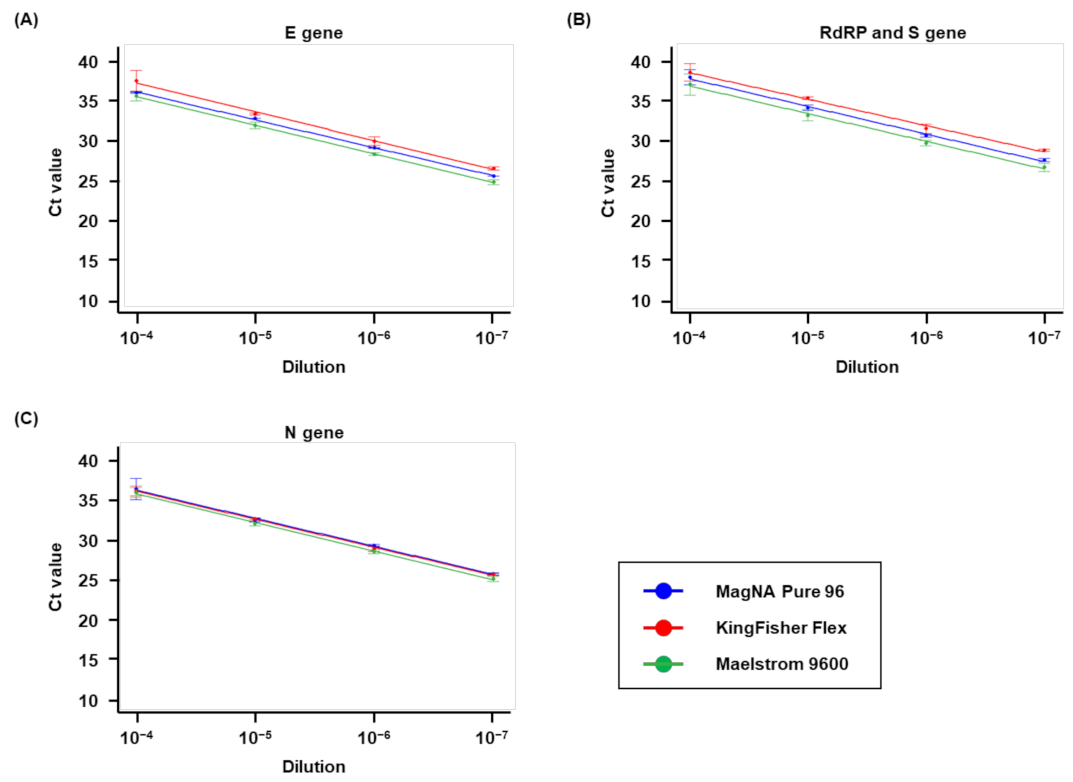
3.5. Analytical Sensitivity and LOD of the Extraction Systems for SARS-CoV-2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hong, K.H.; Lee, S.W.; Kim, T.S.; Huh, H.J.; Lee, J.; Kim, S.Y.; Park, J.S.; Kim, G.J.; Sung, H.; Roh, K.H.; et al. Guidelines for Laboratory Diagnosis of Coronavirus Disease 2019 (COVID-19) in Korea. Ann. Lab. Med. 2020, 40, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Gisondi, P.; S, P.I.; Bordin, C.; Alaibac, M.; Girolomoni, G.; Naldi, L. Cutaneous manifestations of SARS-CoV-2 infection: A clinical update. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 2499–2504. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 21 December 2021).
- Zupin, L.; Licen, S.; Milani, M.; Clemente, L.; Martello, L.; Semeraro, S.; Fontana, F.; Ruscio, M.; Miani, A.; Crovella, S.; et al. Evaluation of Residual Infectivity after SARS-CoV-2 Aerosol Transmission in a Controlled Laboratory Setting. Int. J. Environ. Res. Public Health 2021, 18, 1172. [Google Scholar] [CrossRef]
- Munawar, H.S.; Khan, S.I.; Qadir, Z.; Kiani, Y.S.; Kouzani, A.Z.; Mahmud, M.A.P. Insights into the Mobility Pattern of Australians during COVID-19. Sustainability 2021, 13, 9611. [Google Scholar] [CrossRef]
- Lin, E.-C.; Tu, H.-P.; Hong, C.-H. Halved Incidence of Scrub Typhus after Travel Restrictions to Confine a Surge of COVID-19 in Taiwan. Pathogens 2021, 10, 1386. [Google Scholar] [CrossRef]
- Rajabi, A.; Mantzaris, A.V.; Mutlu, E.C.; Garibay, O.O. Investigating Dynamics of COVID-19 Spread and Containment with Agent-Based Modeling. Appl. Sci. 2021, 11, 5367. [Google Scholar] [CrossRef]
- McFee, D.R.B. COVID-19 Laboratory Testing/CDC Guidelines. Dis. Mon. 2020, 66, 101067. [Google Scholar] [CrossRef] [PubMed]
- Bruce, E.A.; Huang, M.L.; Perchetti, G.A.; Tighe, S.; Laaguiby, P.; Hoffman, J.J.; Gerrard, D.L.; Nalla, A.K.; Wei, Y.; Greninger, A.L.; et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLoS Biol. 2020, 18, e3000896. [Google Scholar] [CrossRef] [PubMed]
- Ravi, N.; Cortade, D.L.; Ng, E.; Wang, S.X. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020, 165, 112454. [Google Scholar] [CrossRef]
- Al-Saud, H.; Al-Romaih, K.; Bakheet, R.; Mahmoud, L.; Al-Harbi, N.; Alshareef, I.; Judia, S.B.; Aharbi, L.; Alzayed, A.; Jabaan, A.; et al. Automated SARS-COV-2 RNA extraction from patient nasopharyngeal samples using a modified DNA extraction kit for high throughput testing. Ann. Saudi Med. 2020, 40, 373–381. [Google Scholar] [CrossRef]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef]
- Lubke, N.; Senff, T.; Scherger, S.; Hauka, S.; Andree, M.; Adams, O.; Timm, J.; Walker, A. Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials. J. Clin. Virol. 2020, 130, 104579. [Google Scholar] [CrossRef] [PubMed]
- Azmi, I.; Faizan, M.I.; Kumar, R.; Raj Yadav, S.; Chaudhary, N.; Kumar Singh, D.; Butola, R.; Ganotra, A.; Datt Joshi, G.; Deep Jhingan, G.; et al. A Saliva-Based RNA Extraction-Free Workflow Integrated with Cas13a for SARS-CoV-2 Detection. Front. Cell. Infect. Microbiol. 2021, 11, 632646. [Google Scholar] [CrossRef]
- Edelmann, A.; Eichenlaub, U.; Lepek, S.; Kruger, D.H.; Hofmann, J. Performance of the MagNA Pure 96 system for cytomegalovirus nucleic acid amplification testing in clinical samples. J. Clin. Microbiol. 2013, 51, 1600–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hindiyeh, M.; Mor, O.; Pando, R.; Mannasse, B.; Kabat, A.; Assraf-Zarfati, H.; Mendelson, E.; Sofer, D.; Mandelboim, M. Comparison of the new fully automated extraction platform eMAG to the MagNA PURE 96 and the well-established easyMAG for detection of common human respiratory viruses. PLoS ONE 2019, 14, e0211079. [Google Scholar] [CrossRef]
- Lau, H.K.; Clotilde, L.M.; Lin, A.P.; Hartman, G.L.; Lauzon, C.R. Comparison of IMS Platforms for detecting and recovering Escherichia coli O157 and Shigella flexneri in foods. J. Lab. Autom. 2013, 18, 178–183. [Google Scholar] [CrossRef]
- Arena, F.; Pollini, S.; Rossolini, G.M.; Margaglione, M. Summary of the Available Molecular Methods for Detection of SARS-CoV-2 during the Ongoing Pandemic. Int. J. Mol. Sci. 2021, 22, 1298. [Google Scholar] [CrossRef] [PubMed]
- Mengelle, C.; Mansuy, J.M.; Sandres-Saune, K.; Barthe, C.; Boineau, J.; Izopet, J. Prospective evaluation of a new automated nucleic acid extraction system using routine clinical respiratory specimens. J. Med. Virol. 2012, 84, 906–911. [Google Scholar] [CrossRef]
- Morecchiato, F.; Coppi, M.; Baccani, I.; Maggini, N.; Ciccone, N.; Antonelli, A.; Rossolini, G.M. Evaluation of extraction-free RT-PCR methods for faster and cheaper detection of SARS-CoV-2 using two commercial systems. Int. J. Infect. Dis. 2021, 112, 264–268. [Google Scholar] [CrossRef]
- So, M.K.; Park, S.; Lee, K.; Kim, S.K.; Chung, H.S.; Lee, M. Variant Prediction by Analyzing RdRp/S Gene Double or Low Amplification Pattern in Allplex SARS-CoV-2 Assay. Diagnostics 2021, 11, 1854. [Google Scholar] [CrossRef]
- Lim, H.J.; Park, J.E.; Park, M.Y.; Baek, J.H.; Jung, S.; Sung, N.; Yang, J.H.; Lee, M.W.; Lee, S.H.; Yang, Y.J. Assay System for Simultaneous Detection of SARS-CoV-2 and Other Respiratory Viruses. Diagnostics 2021, 11, 1084. [Google Scholar] [CrossRef]
- Kubis, P.; Materniak, M.; Kuzmak, J. Comparison of nested PCR and qPCR for the detection and quantitation of BoHV6 DNA. J. Virol. Methods 2013, 194, 94–101. [Google Scholar] [CrossRef]
- van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef] [PubMed]
- Wallis, R.S. PROBIT: A computer program analysis. J. Immunol. Methods 1991, 145, 267–268. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 1, 307–310. [Google Scholar] [CrossRef]
- Cheng, M.P.; Papenburg, J.; Desjardins, M.; Kanjilal, S.; Quach, C.; Libman, M.; Dittrich, S.; Yansouni, C.P. Diagnostic Testing for Severe Acute Respiratory Syndrome-Related Coronavirus 2: A Narrative Review. Ann. Intern. Med. 2020, 172, 726–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Yip, C.C.; To, K.K.; Tang, T.H.; Wong, S.C.; Leung, K.H.; Fung, A.Y.; Ng, A.C.; Zou, Z.; Tsoi, H.W.; et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020, 58, e00310-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossmanith, P.; Wagner, M. A novel poisson distribution-based approach for testing boundaries of real-time PCR assays for food pathogen quantification. J. Food Prot. 2011, 74, 1404–1412. [Google Scholar] [CrossRef]
- Wee, S.K.; Sivalingam, S.P.; Yap, E.P.H. Rapid Direct Nucleic Acid Amplification Test without RNA Extraction for SARS-CoV-2 Using a Portable PCR Thermocycler. Genes 2020, 11, 664. [Google Scholar] [CrossRef] [PubMed]
| Platform | MagNA Pure 96 | KingFisher Flex | Maelstrom 9600 |
|---|---|---|---|
| Platform manufacturer | Roche | Thermo Fisher Scientific | Taiwan Advanced Nanotech Inc. |
| Properties | Fully automated | Fully automated | Fully automated |
| Machine measurements (W × D × H) | 136 cm × 81.5 cm × 100 cm | 68 cm × 60 cm × 38 cm | 87 cm × 57.5 cm × 70 cm |
| Extraction reagent | MagNA Pure 96 DNA and Viral NA Small Volume kit | careGENETM Viral/Pathogen HiFi Nucleic Acid Isolation kit | SGRespiTM Pure kit |
| Reagent manufacturer | Roche | WELLS BIO | Seegene |
| Number of samples | 96 | 96 | 96 |
| Extraction technique | Magnetic beads | Magnetic beads | Magnetic beads |
| Duration of extraction | Approximately 60 min | Approximately 30 min | Approximately 30 min |
| Sample loading | Needed to load the sample into an empty cartridge | Needed to load the sample into a lysis buffer cartridge | Needed to load the sample into a lysis buffer cartridge |
| Nucleic acid collection | Nucleic acid transferred to a 96-well plate | Nucleic acid transferred to a 96-well plate | Nucleic acid transferred to a 96-well plate |
| Identification reader | Barcode system | Not available | Barcode system |
| Extraction Platform | Specimen | Analyte (CV [%] ± SD) | ||
|---|---|---|---|---|
| E Gene | RdRP and S Gene | N Gene | ||
| MagNA Pure 96 | NPS | 1.70 ± 0.49 | 3.28 ± 0.76 | 1.58 ± 0.45 |
| Sputum | 2.80 ± 0.84 | 3.21 ± 0.95 | 2.53 ± 0.78 | |
| Saliva | 3.24 ± 1.00 | 3.87 ± 1.19 | 2.52 ± 0.83 | |
| KingFisher Flex | NPS | 1.32 ± 0.36 | 3.81 ± 0.81 | 1.53 ± 0.43 |
| Sputum | 2.14 ± 0.66 | 2.16 ± 0.67 | 1.78 ± 0.55 | |
| Saliva | 1.45 ± 0.45 | 1.76 ± 0.58 | 1.54 ± 0.52 | |
| Maelstrom 9600 | NPS | 1.18 ± 0.31 | 2.48 ± 0.53 | 1.70 ± 0.44 |
| Sputum | 2.21 ± 0.66 | 1.92 ± 0.59 | 2.16 ± 0.65 | |
| Saliva | 1.10 ± 0.36 | 1.21 ± 0.38 | 1.62 ± 0.55 | |
| Specimens | Target Gene | Platform | No. of Samples | Sen. (%) | Spe. (%) | |||
|---|---|---|---|---|---|---|---|---|
| TP (+/+) | FN (+/–) | FP (–/+) | TN (–/–) | |||||
| NPS | E gene | KingFisher Flex | 57 | 0 | 0 | 25 | 100 | 100 |
| Maelstrom 9600 | 57 | 0 | 0 | 25 | 100 | 100 | ||
| RdRP and S gene | KingFisher Flex | 56 | 1 | 0 | 25 | 98.2 | 100 | |
| Maelstrom 9600 | 57 | 0 | 0 | 25 | 100 | 100 | ||
| N gene | KingFisher Flex | 57 | 0 | 0 | 25 | 100 | 100 | |
| Maelstrom 9600 | 56 | 1 | 0 | 25 | 98.2 | 100 | ||
| Sputum | E gene | KingFisher Flex | 45 | 0 | 0 | 34 | 100 | 100 |
| Maelstrom 9600 | 45 | 0 | 0 | 34 | 100 | 100 | ||
| RdRP and S gene | KingFisher Flex | 45 | 0 | 0 | 34 | 100 | 100 | |
| Maelstrom 9600 | 45 | 0 | 0 | 34 | 100 | 100 | ||
| N gene | KingFisher Flex | 45 | 0 | 0 | 34 | 100 | 100 | |
| Maelstrom 9600 | 44 | 1 | 0 | 34 | 97.8 | 100 | ||
| Saliva | E gene | KingFisher Flex | 17 | 1 | 0 | 66 | 94.4 | 100 |
| Maelstrom 9600 | 18 | 0 | 0 | 66 | 100 | 100 | ||
| RdRP and S gene | KingFisher Flex | 18 | 0 | 0 | 66 | 100 | 100 | |
| Maelstrom 9600 | 18 | 0 | 0 | 66 | 100 | 100 | ||
| N gene | KingFisher Flex | 17 | 1 | 0 | 66 | 94.4 | 100 | |
| Maelstrom 9600 | 18 | 0 | 0 | 66 | 100 | 100 | ||
| Total (NPS + sputum + saliva) | E gene | KingFisher Flex | 119 | 1 | 0 | 125 | 99.2 | 100 |
| Maelstrom 9600 | 120 | 0 | 0 | 125 | 100 | 100 | ||
| RdRP and S gene | KingFisher Flex | 119 | 1 | 0 | 125 | 99.2 | 100 | |
| Maelstrom 9600 | 120 | 0 | 0 | 125 | 100 | 100 | ||
| N gene | KingFisher Flex | 119 | 1 | 0 | 125 | 99.2 | 100 | |
| Maelstrom 9600 | 118 | 2 | 0 | 125 | 98.3 | 100 | ||
| Positive Strain | Target | Conc. (TCID50/mL) | E Gene | RdRp and S Gene | N Gene | |||
|---|---|---|---|---|---|---|---|---|
| Positive Rate (%) | LOD 95% (TCID50/mL) | Positive Rate (%) | LOD 95% (TCID50/mL) | Positive Rate (%) | LOD 95% (TCID50/mL) | |||
| Zeptometrix- 0810589CFHI | MagNA Pure 96 | 7.6 × 104 | 100 | 1.74 × 101 | 100 | 3.15 × 101 | 100 | 1.34 × 101 |
| 7.6 × 103 | 100 | 100 | 100 | |||||
| 7.6 × 102 | 100 | 100 | 100 | |||||
| 7.6 × 101 | 100 | 100 | 100 | |||||
| 7.6 × 100 | 62.5 | 31.3 | 75 | |||||
| 7.6 × 10−1 | 0 | 0 | 0 | |||||
| KingFisher Flex | 7.6 × 104 | 100 | 1.34 × 101 | 100 | 3.62 × 101 | 100 | 9.72 | |
| 7.6 × 103 | 100 | 100 | 100 | |||||
| 7.6 × 102 | 100 | 100 | 100 | |||||
| 7.6 × 101 | 100 | 100 | 100 | |||||
| 7.6 × 100 | 75 | 25 | 87.5 | |||||
| 7.6 × 10−1 | 0 | 0 | 0 | |||||
| Maelstrom 9600 | 7.6 × 104 | 100 | 1.98 × 101 | 100 | 1.98 × 101 | 100 | 1.16 × 101 | |
| 7.6 × 103 | 100 | 100 | 100 | |||||
| 7.6 × 102 | 100 | 100 | 100 | |||||
| 7.6 × 101 | 100 | 100 | 100 | |||||
| 7.6 × 100 | 56.3 | 56.3 | 81.3 | |||||
| 7.6 × 10−1 | 0 | 0 | 0 | |||||
| Zeptometrix- 0810590CFHI | MagNA Pure 96 | 1.4 × 103 | 100 | 3.7 × 10−1 | 100 | 5.3 × 10−1 | 100 | 4.5 × 10−1 |
| 1.4 × 102 | 100 | 100 | 100 | |||||
| 1.4 × 101 | 100 | 100 | 100 | |||||
| 1.4 × 100 | 100 | 100 | 100 | |||||
| 1.4 × 10−1 | 56.3 | 31.3 | 43.8 | |||||
| 1.4 × 10−2 | 0 | 0 | 0 | |||||
| KingFisher Flex | 1.4 × 103 | 100 | 6.5 × 10−1 | 100 | 5.3 × 10−1 | 100 | 4.1 × 10−1 | |
| 1.4 × 102 | 100 | 100 | 100 | |||||
| 1.4 × 101 | 100 | 100 | 100 | |||||
| 1.4 × 100 | 100 | 100 | 100 | |||||
| 1.4 × 10−1 | 18.8 | 31.3 | 50 | |||||
| 1.4 × 10−2 | 0 | 0 | 0 | |||||
| Maelstrom 9600 | 1.4 × 103 | 100 | 3.3 × 10−1 | 100 | 4.5 × 10−1 | 100 | 8.8 × 10−1 | |
| 1.4 × 102 | 100 | 100 | 100 | |||||
| 1.4 × 101 | 100 | 100 | 100 | |||||
| 1.4 × 100 | 100 | 100 | 93.8 | |||||
| 1.4 × 10−1 | 62.5 | 43.8 | 68.8 | |||||
| 1.4 × 10−2 | 0 | 0 | 0 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, H.-J.; Jung, H.-S.; Park, M.-Y.; Baek, Y.-H.; Kannappan, B.; Park, J.-Y.; Yang, J.-H.; Seol, J.-H.; Lee, M.-W.; Jung, S.-K.; et al. Evaluation of Three Automated Extraction Systems for the Detection of SARS-CoV-2 from Clinical Respiratory Specimens. Life 2022, 12, 68. https://doi.org/10.3390/life12010068
Lim H-J, Jung H-S, Park M-Y, Baek Y-H, Kannappan B, Park J-Y, Yang J-H, Seol J-H, Lee M-W, Jung S-K, et al. Evaluation of Three Automated Extraction Systems for the Detection of SARS-CoV-2 from Clinical Respiratory Specimens. Life. 2022; 12(1):68. https://doi.org/10.3390/life12010068
Chicago/Turabian StyleLim, Ho-Jae, Hye-Soo Jung, Min-Young Park, Young-Hyun Baek, Balaji Kannappan, Jin-Young Park, Jae-Hyun Yang, Ja-Hwan Seol, Min-Woo Lee, Sun-Kyung Jung, and et al. 2022. "Evaluation of Three Automated Extraction Systems for the Detection of SARS-CoV-2 from Clinical Respiratory Specimens" Life 12, no. 1: 68. https://doi.org/10.3390/life12010068
APA StyleLim, H.-J., Jung, H.-S., Park, M.-Y., Baek, Y.-H., Kannappan, B., Park, J.-Y., Yang, J.-H., Seol, J.-H., Lee, M.-W., Jung, S.-K., Lee, S.-H., Park, J.-E., & Yang, Y.-J. (2022). Evaluation of Three Automated Extraction Systems for the Detection of SARS-CoV-2 from Clinical Respiratory Specimens. Life, 12(1), 68. https://doi.org/10.3390/life12010068






