Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases
Abstract
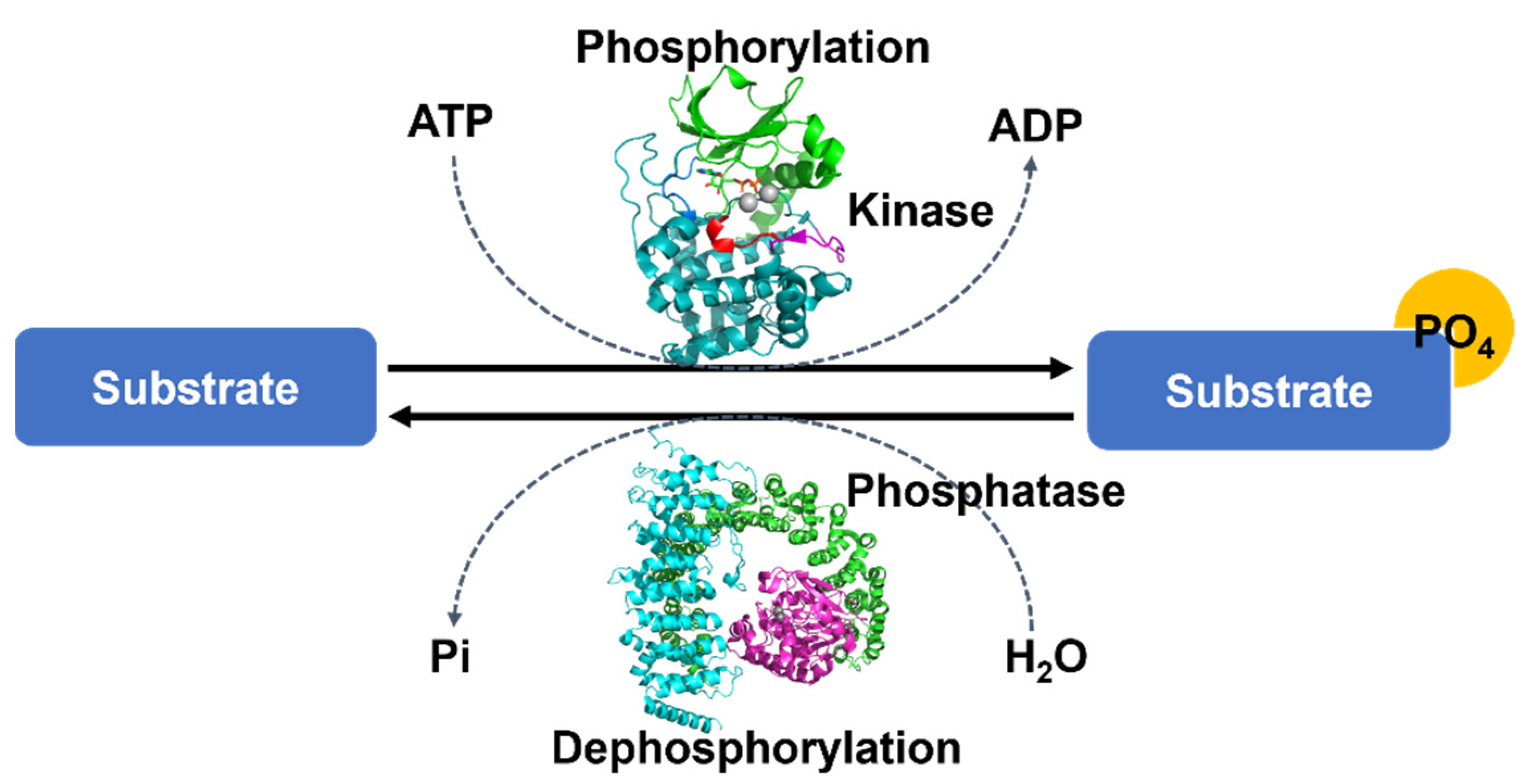
1. Introduction
2. Ser/Thr Protein Kinase Folding
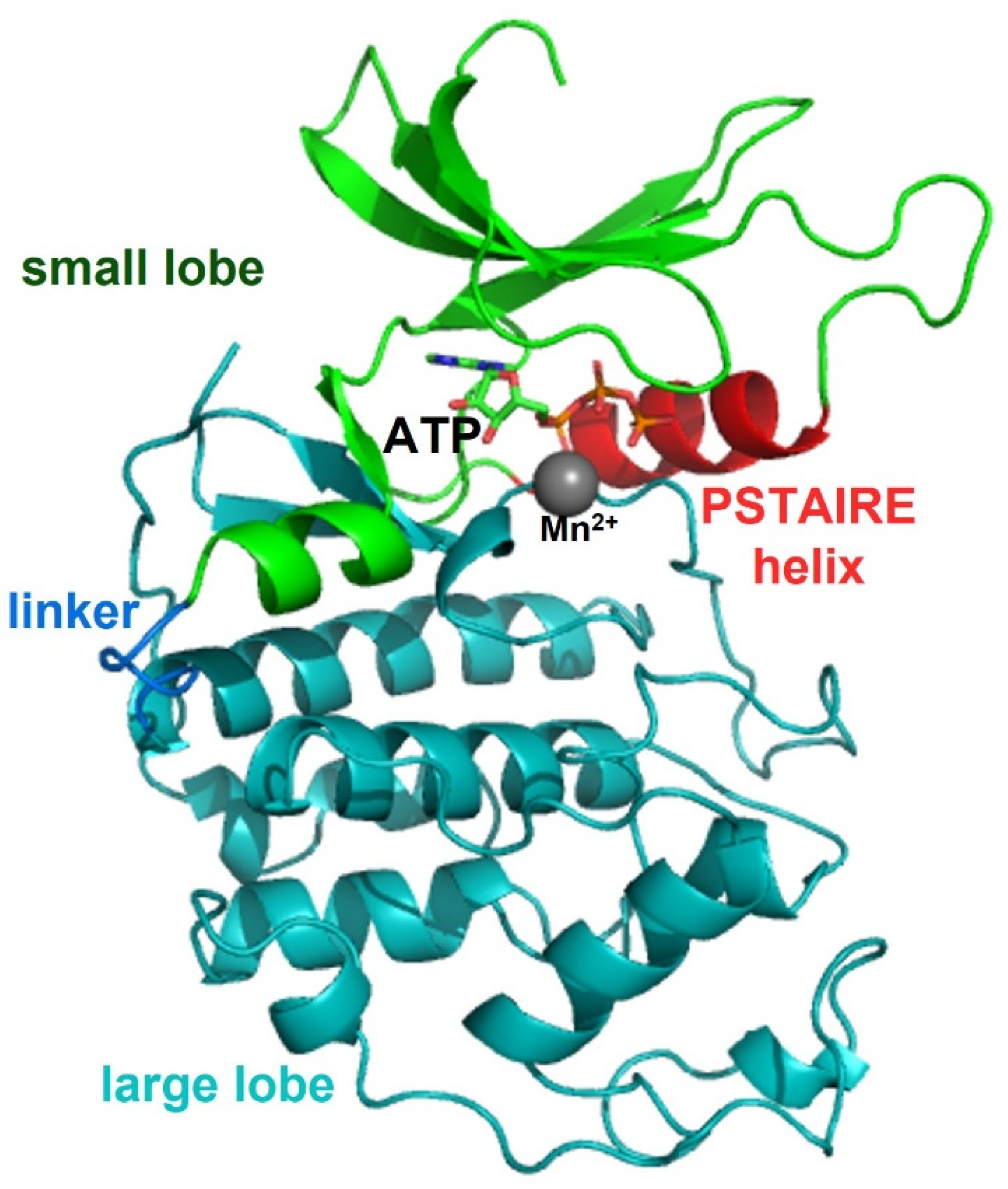
2.1. General Folding of Ser/Thr Protein Kinase
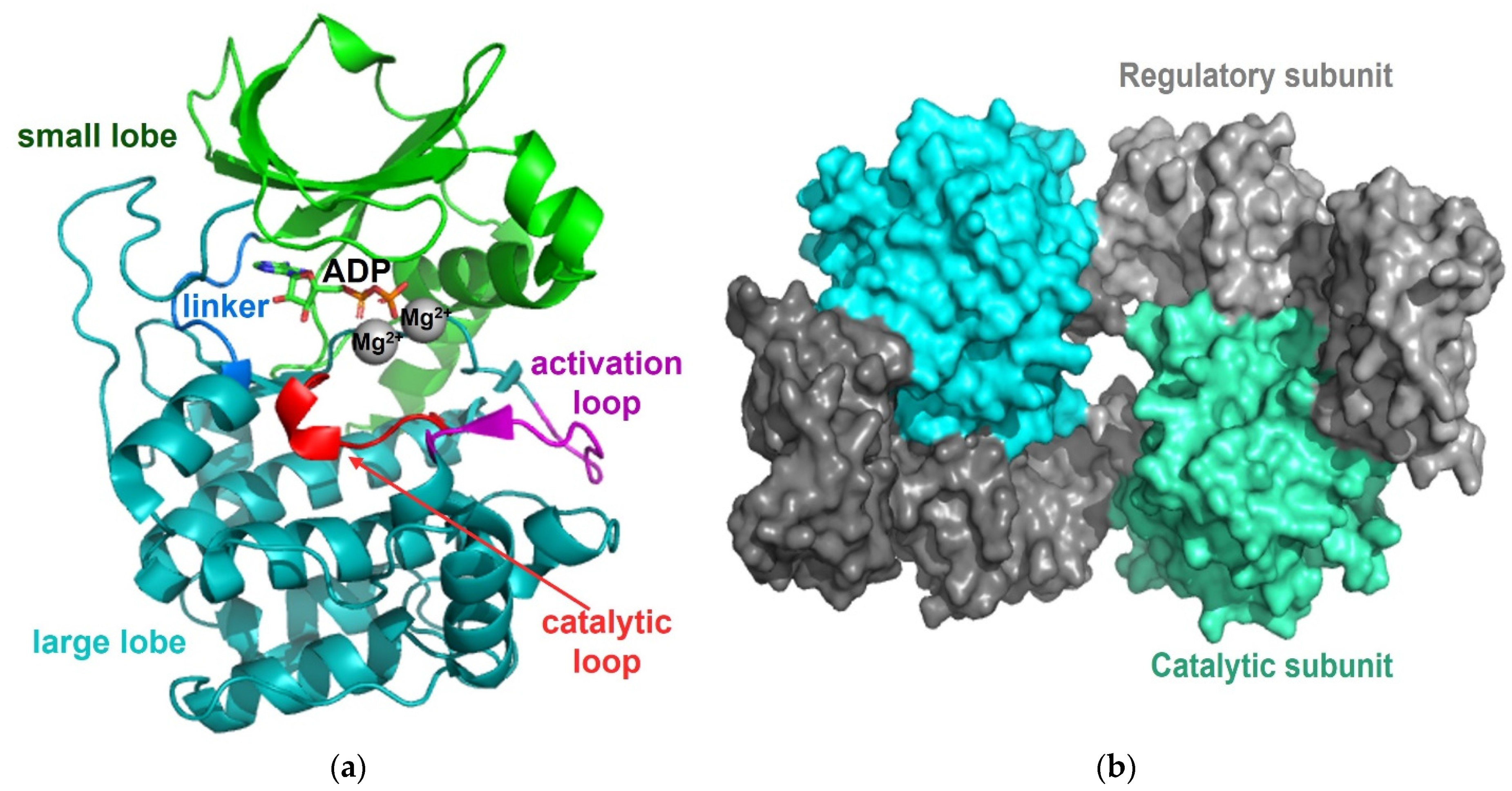
2.2. Protein Kinase A
2.3. Cyclin-Dependent Kinase 2
3. Protein Ser/Thr Phosphatase Folding
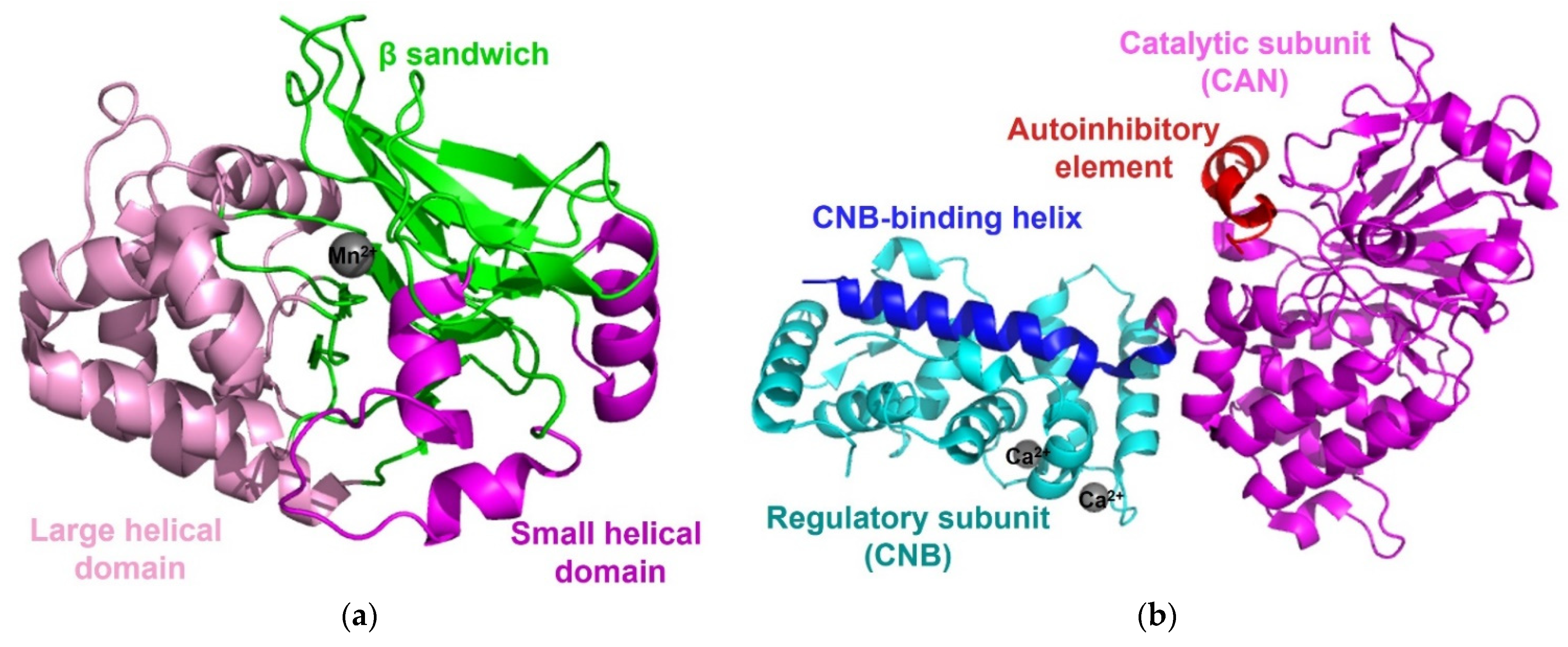
3.1. Protein Phosphatase 1
3.2. Calcineurin/Protein Phosphatase 2B
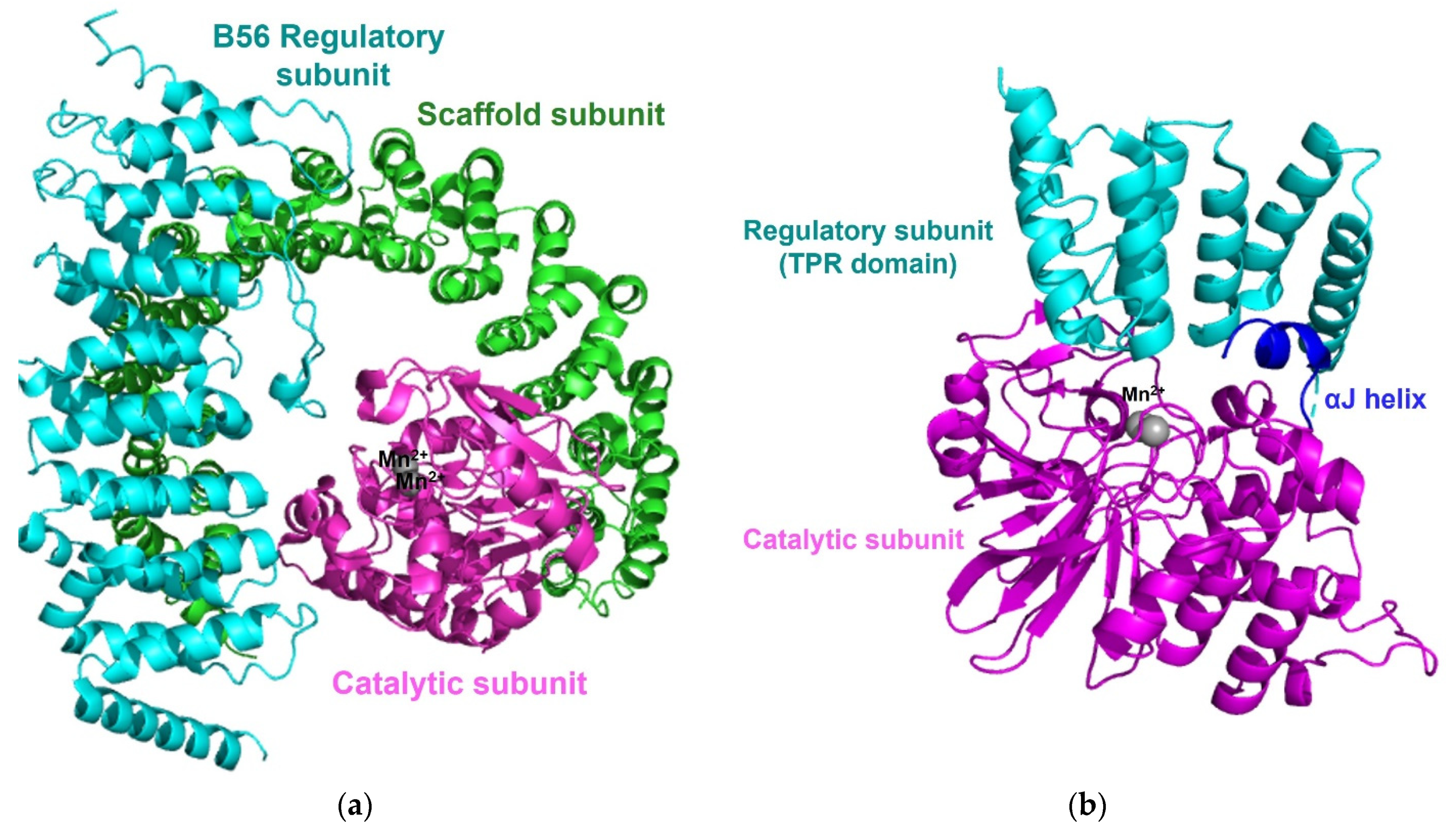
3.3. Protein Phosphatase 2A
3.4. Other Protein Ser/Thr Phosphatases
3.5. Metal-Dependent Protein Phosphatases
4. Substrate Recognition with Short Linear Motifs (SLiMs)
5. Structural Regulation by Phosphorylation
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Venerando, A.; Cesaro, L.; Pinna, L.A. From phosphoproteins to phosphoproteomes: A historical account. FEBS J. 2017, 284, 1936–1951. [Google Scholar] [CrossRef]
- Hunter, T. Signaling—2000 and beyond. Cell 2000, 100, 113–127. [Google Scholar] [CrossRef]
- Brognard, J.; Hunter, T. Protein kinase signaling networks in cancer. Curr. Opin. Genet. Dev. 2011, 21, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Dixon, J.E.; Manning, G. Genomics and evolution of protein phosphatases. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef]
- Nilsson, J. Protein phosphatases in the regulation of mitosis. J. Cell Biol. 2019, 218, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Damle, N.P.; Mohanty, D. Deciphering kinase-substrate relationships by analysis of domain-specific phosphorylation network. Bioinformatics 2014, 30, 1730–1738. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef]
- Turk, B.E. Understanding and exploiting substrate recognition by protein kinases. Curr. Opin. Chem. Biol. 2008, 12, 4–10. [Google Scholar] [CrossRef][Green Version]
- Collins, M.O.; Yu, L.; Campuzano, I.; Grant, S.G.; Choudhary, J.S. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol. Cell Proteom. 2008, 7, 1331–1348. [Google Scholar] [CrossRef]
- Nishi, H.; Hashimoto, K.; Panchenko, A.R. Phosphorylation in protein-protein binding: Effect on stability and function. Structure 2011, 19, 1807–1815. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef]
- Puttick, J.; Baker, E.N.; Delbaere, L.T. Histidine phosphorylation in biological systems. Biochim. Biophys. Acta 2008, 1784, 100–105. [Google Scholar] [CrossRef]
- Ciesla, J.; Fraczyk, T.; Rode, W. Phosphorylation of basic amino acid residues in proteins: Important but easily missed. Acta Biochim. Pol. 2011, 58, 137–148. [Google Scholar] [CrossRef]
- Endicott, J.A.; Noble, M.E.; Johnson, L.N. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 2012, 81, 587–613. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Miranda-Saavedra, D.; Barton, G.J. Kinomer v. 1.0: A database of systematically classified eukaryotic protein kinases. Nucleic Acids Res. 2009, 37, D244–D250. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Huse, M.; Kuriyan, J. The conformational plasticity of protein kinases. Cell 2002, 109, 275–282. [Google Scholar] [CrossRef]
- Nolen, B.; Taylor, S.; Ghosh, G. Regulation of protein kinases: Controlling activity through activation segment conformation. Mol. Cell 2004, 15, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Tarrant, M.K.; Cole, P.A. The chemical biology of protein phosphorylation. Annu. Rev. Biochem. 2009, 78, 797–825. [Google Scholar] [CrossRef] [PubMed]
- Bossemeyer, D. Protein kinases—Structure and function. FEBS Lett. 1995, 369, 57–61. [Google Scholar] [CrossRef]
- London, E.; Bloyd, M.; Stratakis, C.A. PKA functions in metabolism and resistance to obesity: Lessons from mouse and human studies. J. Endocrinol. 2020, 246, R51–R64. [Google Scholar] [CrossRef]
- Juszczak, M.; Krzyminska, A.; Bojanowska, E.; Roszczyk, M. The role of the cAMP/PKA signalling pathway in the inhibitory influence of melatonin on oxytocin and vasopressin secretion from the rat hypothalamo-neurohypophysial system. Endokrynol. Pol. 2018, 69, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.D.; Samelson, B.K.; Scott, J.D. Discovery of cellular substrates for protein kinase A using a peptide array screening protocol. Biochem. J. 2011, 438, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Shabb, J.B. Physiological substrates of cAMP-dependent protein kinase. Chem. Rev. 2001, 101, 2381–2411. [Google Scholar] [CrossRef]
- Imamura, H.; Wagih, O.; Niinae, T.; Sugiyama, N.; Beltrao, P.; Ishihama, Y. Identifications of Putative PKA Substrates with Quantitative Phosphoproteomics and Primary-Sequence-Based Scoring. J. Proteome Res. 2017, 16, 1825–1830. [Google Scholar] [CrossRef]
- Taylor, S.S.; Yang, J.; Wu, J.; Haste, N.M.; Radzio-Andzelm, E.; Anand, G. PKA: A portrait of protein kinase dynamics. Biochim. Biophys. Acta 2004, 1697, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Knighton, D.R.; Zheng, J.H.; Ten Eyck, L.F.; Ashford, V.A.; Xuong, N.H.; Taylor, S.S.; Sowadski, J.M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 1991, 253, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Engh, R.A.; Girod, A.; Kinzel, V.; Huber, R.; Bossemeyer, D. Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity. J. Biol. Chem. 1996, 271, 26157–26164. [Google Scholar] [CrossRef]
- Breitenlechner, C.; Gassel, M.; Hidaka, H.; Kinzel, V.; Huber, R.; Engh, R.A.; Bossemeyer, D. Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: Structural basis of selectivity. Structure 2003, 11, 1595–1607. [Google Scholar] [CrossRef]
- Arencibia, J.M.; Pastor-Flores, D.; Bauer, A.F.; Schulze, J.O.; Biondi, R.M. AGC protein kinases: From structural mechanism of regulation to allosteric drug development for the treatment of human diseases. Biochim. Biophys. Acta 2013, 1834, 1302–1321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Smith-Nguyen, E.V.; Keshwani, M.M.; Deal, M.S.; Kornev, A.P.; Taylor, S.S. Structure and allostery of the PKA RIIbeta tetrameric holoenzyme. Science 2012, 335, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Cheng, C.Y.; Saldanha, S.A.; Taylor, S.S. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell 2007, 130, 1032–1043. [Google Scholar] [CrossRef] [PubMed]
- Bossemeyer, D.; Engh, R.A.; Kinzel, V.; Ponstingl, H.; Huber, R. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24). EMBO J. 1993, 12, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Endicott, J.A.; Noble, M.E.; Tucker, J.A. Cyclin-dependent kinases: Inhibition and substrate recognition. Curr. Opin. Struct. Biol. 1999, 9, 738–744. [Google Scholar] [CrossRef]
- Caruso, J.A.; Duong, M.T.; Carey, J.P.W.; Hunt, K.K.; Keyomarsi, K. Low-Molecular-Weight Cyclin E in Human Cancer: Cellular Consequences and Opportunities for Targeted Therapies. Cancer Res. 2018, 78, 5481–5491. [Google Scholar] [CrossRef]
- Wood, D.J.; Korolchuk, S.; Tatum, N.J.; Wang, L.Z.; Endicott, J.A.; Noble, M.E.M.; Martin, M.P. Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition. Cell Chem. Biol. 2019, 26, 121–130.e5. [Google Scholar] [CrossRef]
- Echalier, A.; Endicott, J.A.; Noble, M.E. Recent developments in cyclin-dependent kinase biochemical and structural studies. Biochim. Biophys. Acta 2010, 1804, 511–519. [Google Scholar] [CrossRef]
- Russo, A.A.; Jeffrey, P.D.; Pavletich, N.P. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat. Struct. Biol. 1996, 3, 696–700. [Google Scholar] [CrossRef]
- Barford, D. Molecular mechanisms of the protein serine/threonine phosphatases. Trends Biochem. Sci. 1996, 21, 407–412. [Google Scholar] [CrossRef]
- Moorhead, G.B.; Trinkle-Mulcahy, L.; Ulke-Lemee, A. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 2007, 8, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Wlodarchak, N.; Xing, Y. PP2A as a master regulator of the cell cycle. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 162–184. [Google Scholar] [CrossRef]
- Shi, Y. Serine/threonine phosphatases: Mechanism through structure. Cell 2009, 139, 468–484. [Google Scholar] [CrossRef]
- Almo, S.C.; Bonanno, J.B.; Sauder, J.M.; Emtage, S.; Dilorenzo, T.P.; Malashkevich, V.; Wasserman, S.R.; Swaminathan, S.; Eswaramoorthy, S.; Agarwal, R.; et al. Structural genomics of protein phosphatases. J. Struct. Funct. Genom. 2007, 8, 121–140. [Google Scholar] [CrossRef]
- Egloff, M.P.; Cohen, P.T.; Reinemer, P.; Barford, D. Crystal structure of the catalytic subunit of human protein phosphatase 1 and its complex with tungstate. J. Mol. Biol. 1995, 254, 942–959. [Google Scholar] [CrossRef]
- Goldberg, J.; Huang, H.B.; Kwon, Y.G.; Greengard, P.; Nairn, A.C.; Kuriyan, J. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 1995, 376, 745–753. [Google Scholar] [CrossRef]
- Rusnak, F.; Mertz, P. Calcineurin: Form and function. Physiol. Rev. 2000, 80, 1483–1521. [Google Scholar] [CrossRef]
- Kissinger, C.R.; Parge, H.E.; Knighton, D.R.; Lewis, C.T.; Pelletier, L.A.; Tempczyk, A.; Kalish, V.J.; Tucker, K.D.; Showalter, R.E.; Moomaw, E.W.; et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature 1995, 378, 641–644. [Google Scholar] [CrossRef]
- Juvvadi, P.R.; Fox, D., 3rd; Bobay, B.G.; Hoy, M.J.; Gobeil, S.M.C.; Venters, R.A.; Chang, Z.; Lin, J.J.; Averette, A.F.; Cole, D.C.; et al. Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents. Nat. Commun. 2019, 10, 4275. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Harrison, S.C. Crystal structure of human calcineurin complexed with cyclosporin A and human cyclophilin. Proc. Natl. Acad. Sci. USA 2002, 99, 13522–13526. [Google Scholar] [CrossRef] [PubMed]
- Ke, H.; Huai, Q. Structures of calcineurin and its complexes with immunophilins-immunosuppressants. Biochem. Biophys. Res. Commun. 2003, 311, 1095–1102. [Google Scholar] [CrossRef]
- Huai, Q.; Kim, H.Y.; Liu, Y.; Zhao, Y.; Mondragon, A.; Liu, J.O.; Ke, H. Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes. Proc. Natl. Acad. Sci. USA 2002, 99, 12037–12042. [Google Scholar] [CrossRef]
- Griffith, J.P.; Kim, J.L.; Kim, E.E.; Sintchak, M.D.; Thomson, J.A.; Fitzgibbon, M.J.; Fleming, M.A.; Caron, P.R.; Hsiao, K.; Navia, M.A. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell 1995, 82, 507–522. [Google Scholar] [CrossRef]
- Ye, Q.; Li, X.; Wong, A.; Wei, Q.; Jia, Z. Structure of calmodulin bound to a calcineurin peptide: A new way of making an old binding mode. Biochemistry 2006, 45, 738–745. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, H.; Zheng, J.; Wei, Q.; Jia, Z. The complex structure of calmodulin bound to a calcineurin peptide. Proteins 2008, 73, 19–27. [Google Scholar] [CrossRef]
- Rumi-Masante, J.; Rusinga, F.I.; Lester, T.E.; Dunlap, T.B.; Williams, T.D.; Dunker, A.K.; Weis, D.D.; Creamer, T.P. Structural basis for activation of calcineurin by calmodulin. J. Mol. Biol. 2012, 415, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Feng, Y.; Yin, Y.; Faucher, F.; Currie, M.A.; Rahman, M.N.; Jin, J.; Li, S.; Wei, Q.; Jia, Z. Structural basis of calcineurin activation by calmodulin. Cell Signal. 2013, 25, 2661–2667. [Google Scholar] [CrossRef] [PubMed]
- Janssens, V.; Goris, J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001, 353, 417–439. [Google Scholar] [CrossRef] [PubMed]
- Lechward, K.; Awotunde, O.S.; Swiatek, W.; Muszynska, G. Protein phosphatase 2A: Variety of forms and diversity of functions. Acta Biochim. Pol. 2001, 48, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Groves, M.R.; Hanlon, N.; Turowski, P.; Hemmings, B.A.; Barford, D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 1999, 96, 99–110. [Google Scholar] [CrossRef]
- Xing, Y.; Xu, Y.; Chen, Y.; Jeffrey, P.D.; Chao, Y.; Lin, Z.; Li, Z.; Strack, S.; Stock, J.B.; Shi, Y. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell 2006, 127, 341–353. [Google Scholar] [CrossRef]
- Xu, Y.; Xing, Y.; Chen, Y.; Chao, Y.; Lin, Z.; Fan, E.; Yu, J.W.; Strack, S.; Jeffrey, P.D.; Shi, Y. Structure of the protein phosphatase 2A holoenzyme. Cell 2006, 127, 1239–1251. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, Y.; Zhang, P.; Jeffrey, P.D.; Shi, Y. Structure of a protein phosphatase 2A holoenzyme: Insights into B55-mediated Tau dephosphorylation. Mol. Cell 2008, 31, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.T.; Philp, A.; Vazquez-Martin, C. Protein phosphatase 4--from obscurity to vital functions. FEBS Lett. 2005, 579, 3278–3286. [Google Scholar] [CrossRef] [PubMed]
- Guergnon, J.; Derewenda, U.; Edelson, J.R.; Brautigan, D.L. Mapping of protein phosphatase-6 association with its SAPS domain regulatory subunit using a model of helical repeats. BMC Biochem. 2009, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Stefansson, B.; Ohama, T.; Daugherty, A.E.; Brautigan, D.L. Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry 2008, 47, 1442–1451. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, J.; Dong, M.; Yu, H.; Dai, X.; Li, K. Inhibition of protein phosphatase 5 (PP5) suppresses survival and growth of colorectal cancer cells. Biotechnol. Appl. Biochem. 2015, 62, 621–627. [Google Scholar] [CrossRef]
- Lu, X.; Nguyen, T.A.; Moon, S.H.; Darlington, Y.; Sommer, M.; Donehower, L.A. The type 2C phosphatase Wip1: An oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008, 27, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Brautigan, D.L.; Shenolikar, S. Protein Serine/Threonine Phosphatases: Keys to Unlocking Regulators and Substrates. Annu. Rev. Biochem. 2018, 87, 921–964. [Google Scholar] [CrossRef]
- Van Roey, K.; Uyar, B.; Weatheritt, R.J.; Dinkel, H.; Seiler, M.; Budd, A.; Gibson, T.J.; Davey, N.E. Short linear motifs: Ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 2014, 114, 6733–6778. [Google Scholar] [CrossRef]
- Ali, M.; Simonetti, L.; Ivarsson, Y. Screening Intrinsically Disordered Regions for Short Linear Binding Motifs. Methods Mol. Biol. 2020, 2141, 529–552. [Google Scholar] [CrossRef]
- Arsenault, R.; Griebel, P.; Napper, S. Peptide arrays for kinome analysis: New opportunities and remaining challenges. Proteomics 2011, 11, 4595–4609. [Google Scholar] [CrossRef]
- Kettenbach, A.N.; Schweppe, D.K.; Faherty, B.K.; Pechenick, D.; Pletnev, A.A.; Gerber, S.A. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 2011, 4, rs5. [Google Scholar] [CrossRef]
- Wu, C.G.; Chen, H.; Guo, F.; Yadav, V.K.; McIlwain, S.J.; Rowse, M.; Choudhary, A.; Lin, Z.; Li, Y.; Gu, T.; et al. PP2A-B’ holoenzyme substrate recognition, regulation and role in cytokinesis. Cell Discov. 2017, 3, 17027. [Google Scholar] [CrossRef]
- Davey, N.E.; Seo, M.H.; Yadav, V.K.; Jeon, J.; Nim, S.; Krystkowiak, I.; Blikstad, C.; Dong, D.; Markova, N.; Kim, P.M.; et al. Discovery of short linear motif-mediated interactions through phage display of intrinsically disordered regions of the human proteome. FEBS J. 2017, 284, 485–498. [Google Scholar] [CrossRef]
- Perez-Mejias, G.; Velazquez-Cruz, A.; Guerra-Castellano, A.; Banos-Jaime, B.; Diaz-Quintana, A.; Gonzalez-Arzola, K.; De la Rosa, M.A.; Diaz-Moreno, I. Exploring protein phosphorylation by combining computational approaches and biochemical methods. Comput. Struct. Biotechnol. J. 2020, 18, 1852–1863. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.E.; Graves, D.J.; Benjamini, E.; Krebs, E.G. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J. Biol. Chem. 1977, 252, 4888–4894. [Google Scholar] [CrossRef]
- Nishikawa, K.; Toker, A.; Johannes, F.J.; Songyang, Z.; Cantley, L.C. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 1997, 272, 952–960. [Google Scholar] [CrossRef] [PubMed]
- Songyang, Z.; Blechner, S.; Hoagland, N.; Hoekstra, M.F.; Piwnica-Worms, H.; Cantley, L.C. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 1994, 4, 973–982. [Google Scholar] [CrossRef]
- Flotow, H.; Graves, P.R.; Wang, A.Q.; Fiol, C.J.; Roeske, R.W.; Roach, P.J. Phosphate groups as substrate determinants for casein kinase I action. J. Biol. Chem. 1990, 265, 14264–14269. [Google Scholar] [CrossRef]
- Meggio, F.; Pinna, L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003, 17, 349–368. [Google Scholar] [CrossRef]
- Gonzalez, F.A.; Raden, D.L.; Davis, R.J. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J. Biol. Chem. 1991, 266, 22159–22163. [Google Scholar] [CrossRef]
- Songyang, Z.; Lu, K.P.; Kwon, Y.T.; Tsai, L.H.; Filhol, O.; Cochet, C.; Brickey, D.A.; Soderling, T.R.; Bartleson, C.; Graves, D.J.; et al. A structural basis for substrate specificities of protein Ser/Thr kinases: Primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell Biol. 1996, 16, 6486–6493. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Lim, D.; Joughin, B.A.; Hegemann, B.; Hutchins, J.R.; Ehrenberger, T.; Ivins, F.; Sessa, F.; Hudecz, O.; Nigg, E.A.; et al. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci. Signal. 2011, 4, ra42. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Beullens, M.; Huang, J.; De Munter, S.; Lesage, B.; Bollen, M. Cdk1 orders mitotic events through coordination of a chromosome-associated phosphatase switch. Nat. Commun. 2015, 6, 10215. [Google Scholar] [CrossRef]
- Egloff, M.P.; Johnson, D.F.; Moorhead, G.; Cohen, P.T.; Cohen, P.; Barford, D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997, 16, 1876–1887. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.S.; Gokhan, E.; De Munter, S.; Bollen, M.; Vagnarelli, P.; Peti, W.; Page, R. The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism. Elife 2016, 5. [Google Scholar] [CrossRef]
- Grigoriu, S.; Bond, R.; Cossio, P.; Chen, J.A.; Ly, N.; Hummer, G.; Page, R.; Cyert, M.S.; Peti, W. The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin. PLoS Biol. 2013, 11, e1001492. [Google Scholar] [CrossRef]
- Bultynck, G.; Heath, V.L.; Majeed, A.P.; Galan, J.M.; Haguenauer-Tsapis, R.; Cyert, M.S. Slm1 and slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell Biol. 2006, 26, 4729–4745. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, L.; Rao, A.; Harrison, S.C.; Hogan, P.G. Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction. J. Mol. Biol. 2007, 369, 1296–1306. [Google Scholar] [CrossRef]
- Hertz, E.P.T.; Kruse, T.; Davey, N.E.; Lopez-Mendez, B.; Sigurethsson, J.O.; Montoya, G.; Olsen, J.V.; Nilsson, J. A Conserved Motif Provides Binding Specificity to the PP2A-B56 Phosphatase. Mol. Cell 2016, 63, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bajaj, R.; Bollen, M.; Peti, W.; Page, R. Expanding the PP2A Interactome by Defining a B56-Specific SLiM. Structure 2016, 24, 2174–2181. [Google Scholar] [CrossRef]
- Li, H.; Rao, A.; Hogan, P.G. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011, 21, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.N. The regulation of protein phosphorylation. Biochem. Soc. Trans. 2009, 37, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Rago, F.; Hori, T.; Tomii, K.; Cheeseman, I.M.; Fukagawa, T. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 2013, 32, 424–436. [Google Scholar] [CrossRef]
- Gascoigne, K.E.; Takeuchi, K.; Suzuki, A.; Hori, T.; Fukagawa, T.; Cheeseman, I.M. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 2011, 145, 410–422. [Google Scholar] [CrossRef]
- Bomblies, R.; Luitz, M.P.; Zacharias, M. Molecular Dynamics Analysis of 4E-BP2 Protein Fold Stabilization Induced by Phosphorylation. J. Phys. Chem. B 2017, 121, 3387–3393. [Google Scholar] [CrossRef]
- Bah, A.; Vernon, R.M.; Siddiqui, Z.; Krzeminski, M.; Muhandiram, R.; Zhao, C.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 2015, 519, 106–109. [Google Scholar] [CrossRef]
- Thapar, R. Structural basis for regulation of RNA-binding proteins by phosphorylation. ACS Chem. Biol. 2015, 10, 652–666. [Google Scholar] [CrossRef]
- Miranda, F.F.; Thorolfsson, M.; Teigen, K.; Sanchez-Ruiz, J.M.; Martinez, A. Structural and stability effects of phosphorylation: Localized structural changes in phenylalanine hydroxylase. Protein Sci. 2004, 13, 1219–1226. [Google Scholar] [CrossRef]
- Chiang, C.M. Phospho-BRD4: Transcription plasticity and drug targeting. Drug Discov. Today Technol. 2016, 19, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Lee, A.Y.; Lai, H.T.; Zhang, H.; Chiang, C.M. Phospho Switch Triggers Brd4 Chromatin Binding and Activator Recruitment for Gene-Specific Targeting. Mol. Cell 2013, 49, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012, 40, D261–D270. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Kornhauser, J.M.; Latham, V.; Murray, B.; Nandhikonda, V.; Nord, A.; Skrzypek, E.; Wheeler, T.; Zhang, B.; Gnad, F. 15 years of PhosphoSitePlus(R): Integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res. 2019, 47, D433–D441. [Google Scholar] [CrossRef]

| Name | Consensus Sequence 1,2,3 | References |
|---|---|---|
| Ser/Thrprotein kinases | ||
| Cyclic AMP-dependent kinase (PKA) | R-R/K-S/T-ϕ | [77] |
| Protein kinase B (PKB, Akt) | R-x-R-x-x-S/T-ϕ | [15] |
| Cyclic GMP-dependent protein kinase (PKG) | R/K-R/K-R/K-x-S/T-x | [77] |
| Protein kinase C (PKC) | x-S/T-x-R/K | [78] |
| Cyclin-dependent kinases (CDKs) | S/T-P-x-K/R | [79] |
| Casein kinase 1 (CK1) | D/E-D/E-D/E-x-x-S/T-ϕ pS/pT-x-x-S/T-ϕ | [80] |
| Casein kinase 2 (CK2) | S/T-D/E-x-D/E | [81] |
| Calcium/Calmodulin-dependent Protein Kinase II (CaM II) | R-x-x-S/T-x | [77] |
| AMP-activated protein kinase (AMPK) | ϕ-x-R-x-x-S-x-x-x-I/L | [82] |
| Phosphorylase kinase (PhK) | R-x-x-S/T-x-ϕ-R | [15] |
| Mitogen-activated protein kinases (MAPKs) | P/ϕ-x-S/T-P | [83] |
| NimA-related kinase (NEK) | ϕ-x-x-S/T | [82] |
| Polo-kike kinase 1 (Plk) | D/E/N-x-S/T-ϕ | [84] |
| Protein Ser/Thrphosphatase | ||
| Protein phosphatase 1 (PP1) | R-V-x-F | [85,86] |
| Protein phosphatase 2A-B56 (PP2A-B56) | L-x-x-I-x-E | [74,87] |
| Calcineurin (PP2B) | P-x-I-x-I-T LxVP | [88,89,90] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seok, S.-H. Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases. Life 2021, 11, 957. https://doi.org/10.3390/life11090957
Seok S-H. Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases. Life. 2021; 11(9):957. https://doi.org/10.3390/life11090957
Chicago/Turabian StyleSeok, Seung-Hyeon. 2021. "Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases" Life 11, no. 9: 957. https://doi.org/10.3390/life11090957
APA StyleSeok, S.-H. (2021). Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases. Life, 11(9), 957. https://doi.org/10.3390/life11090957






