Recombinant Globular Domain of TcpA Pilin from Vibrio cholerae El Tor: Recovery from Inclusion Bodies and Structural Characterization
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation Conditions
2.2. Preparation of Cell Lysates
2.3. Purification of the Protein from the Soluble Fraction
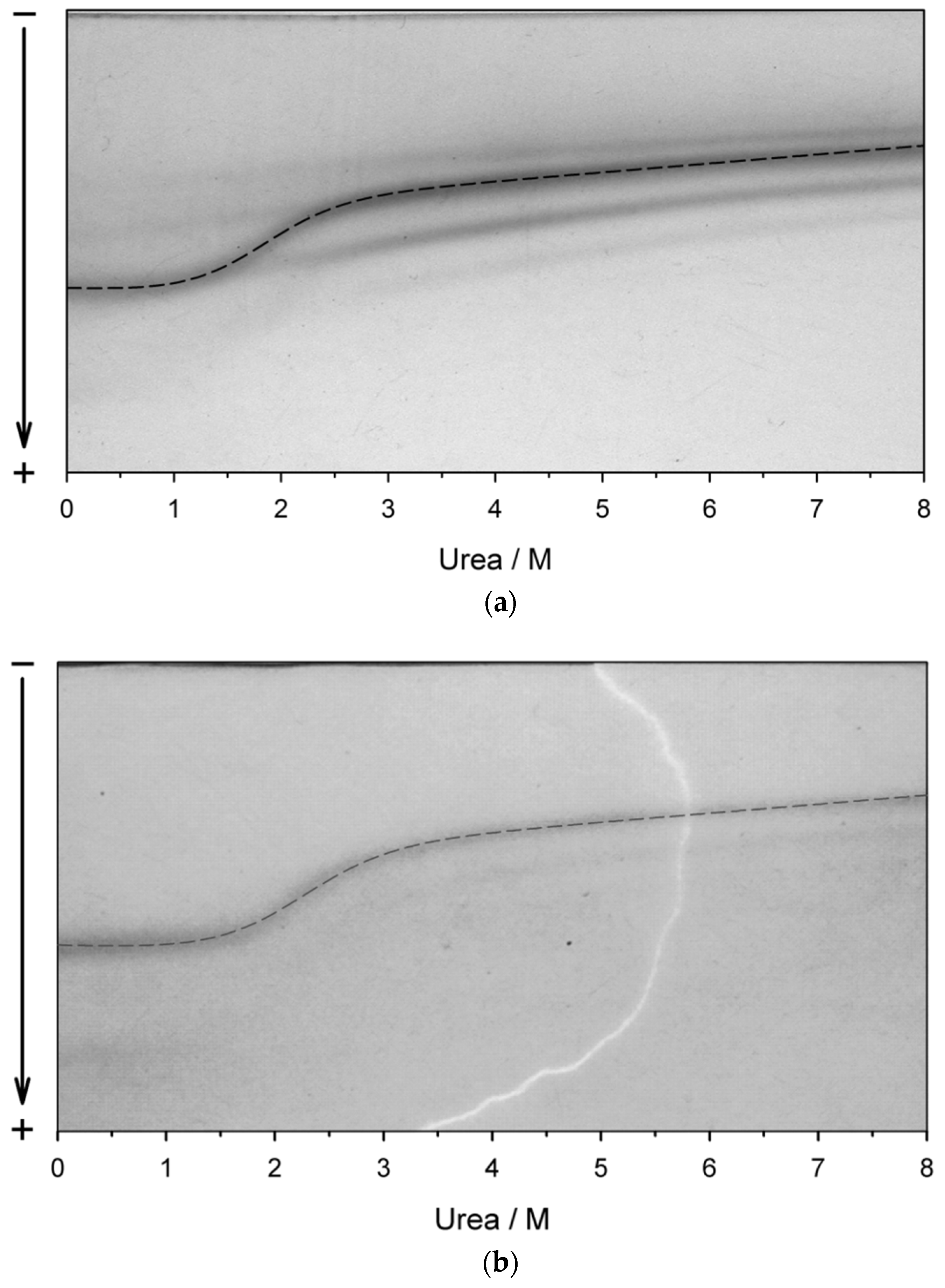
2.4. Recovery of the Target Protein from Inclusion Bodies through the Urea-Unfolded State
2.5. Mass Spectrometry Analysis of the Recombinant Protein TcpA-C
2.6. Structural Characterization of the Recombinant Protein TcpA-C
3. Results
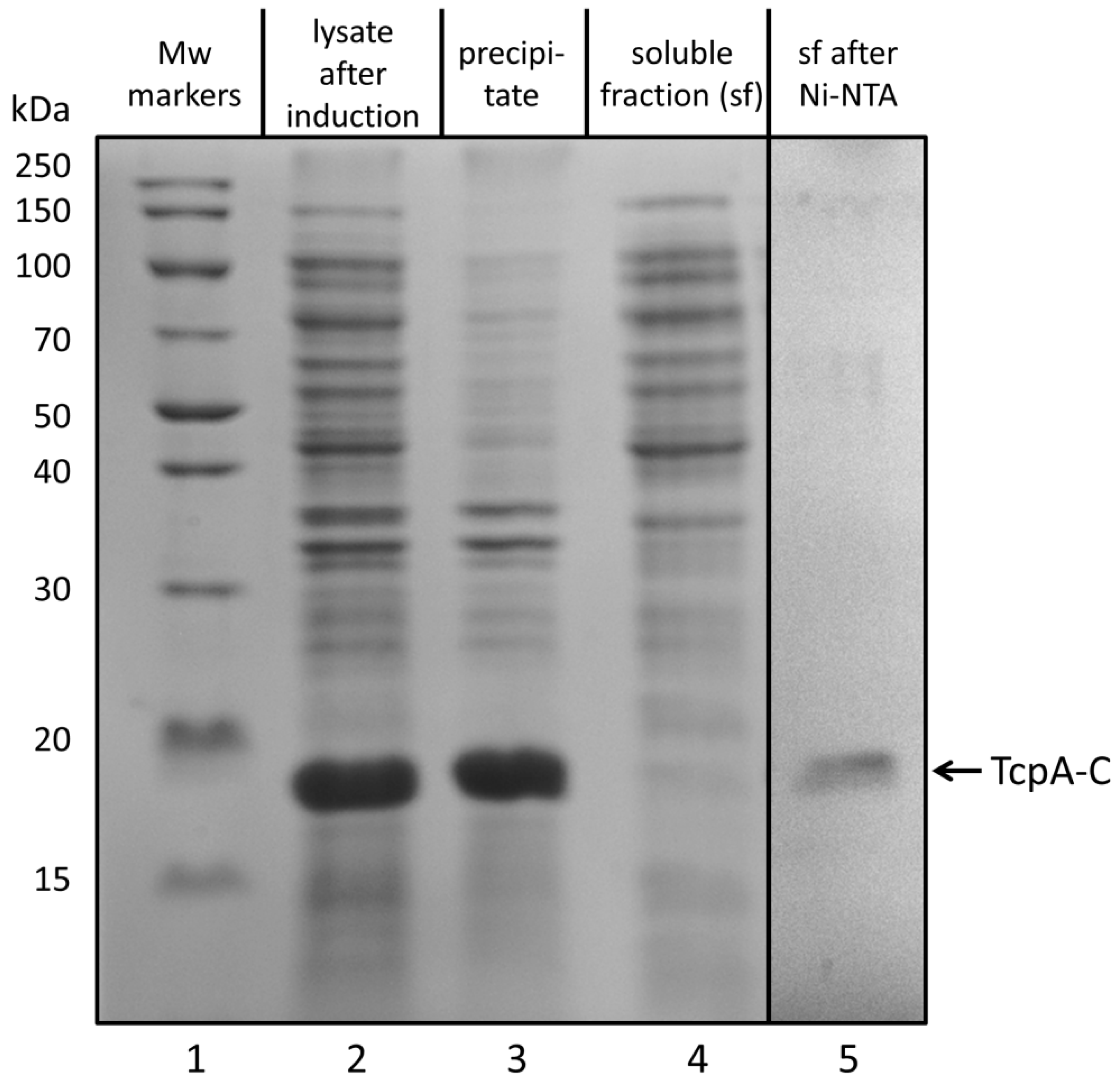
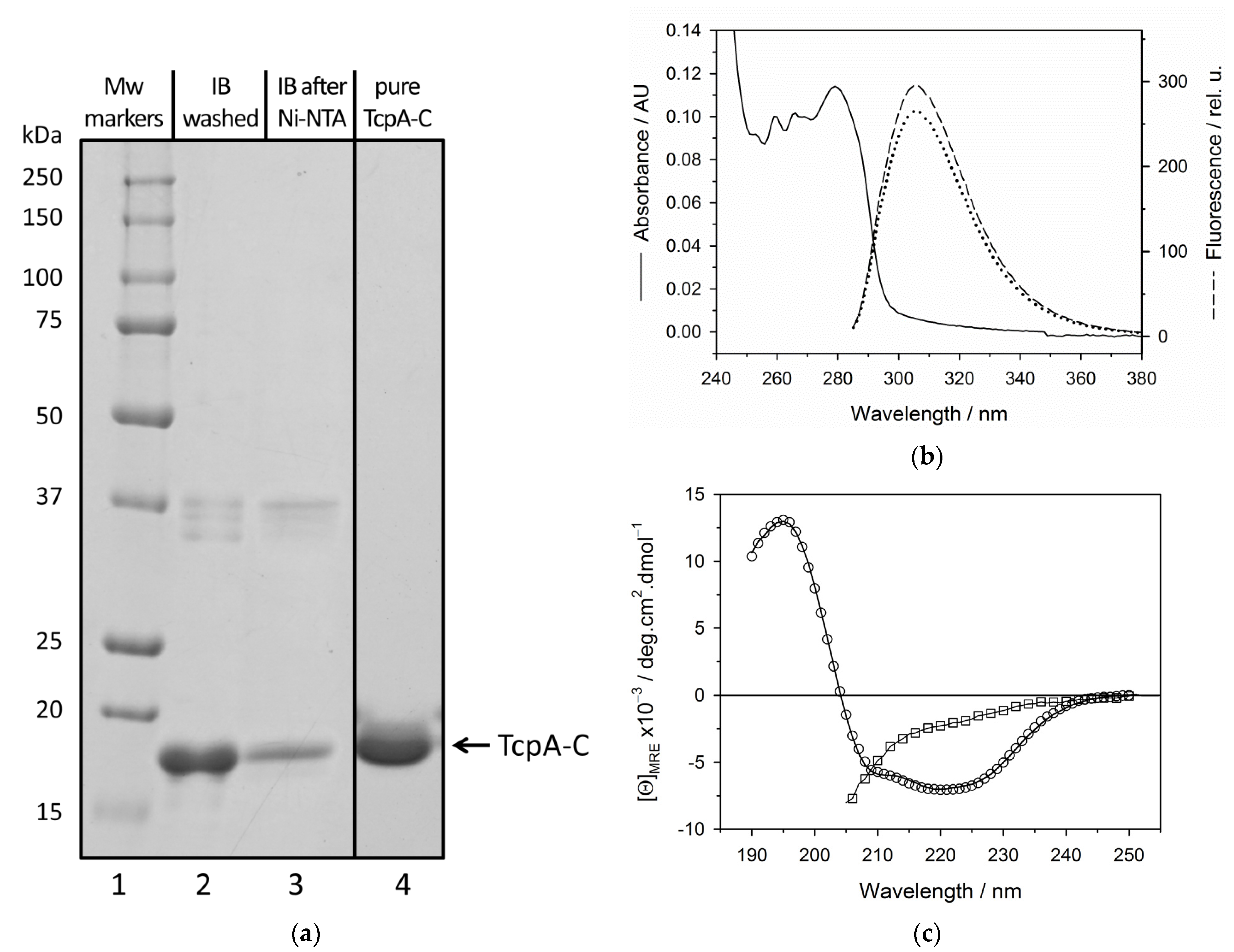
3.1. Biosynthesis of TcpA-C in E. coli Cells and Its Purification
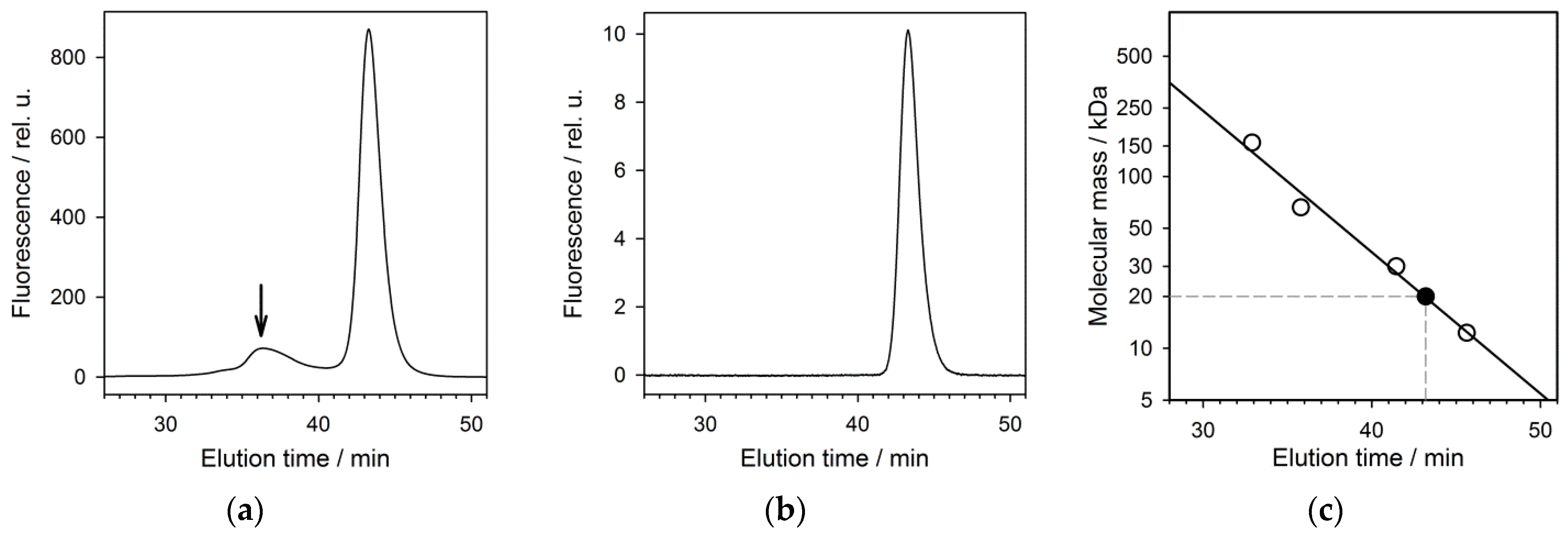
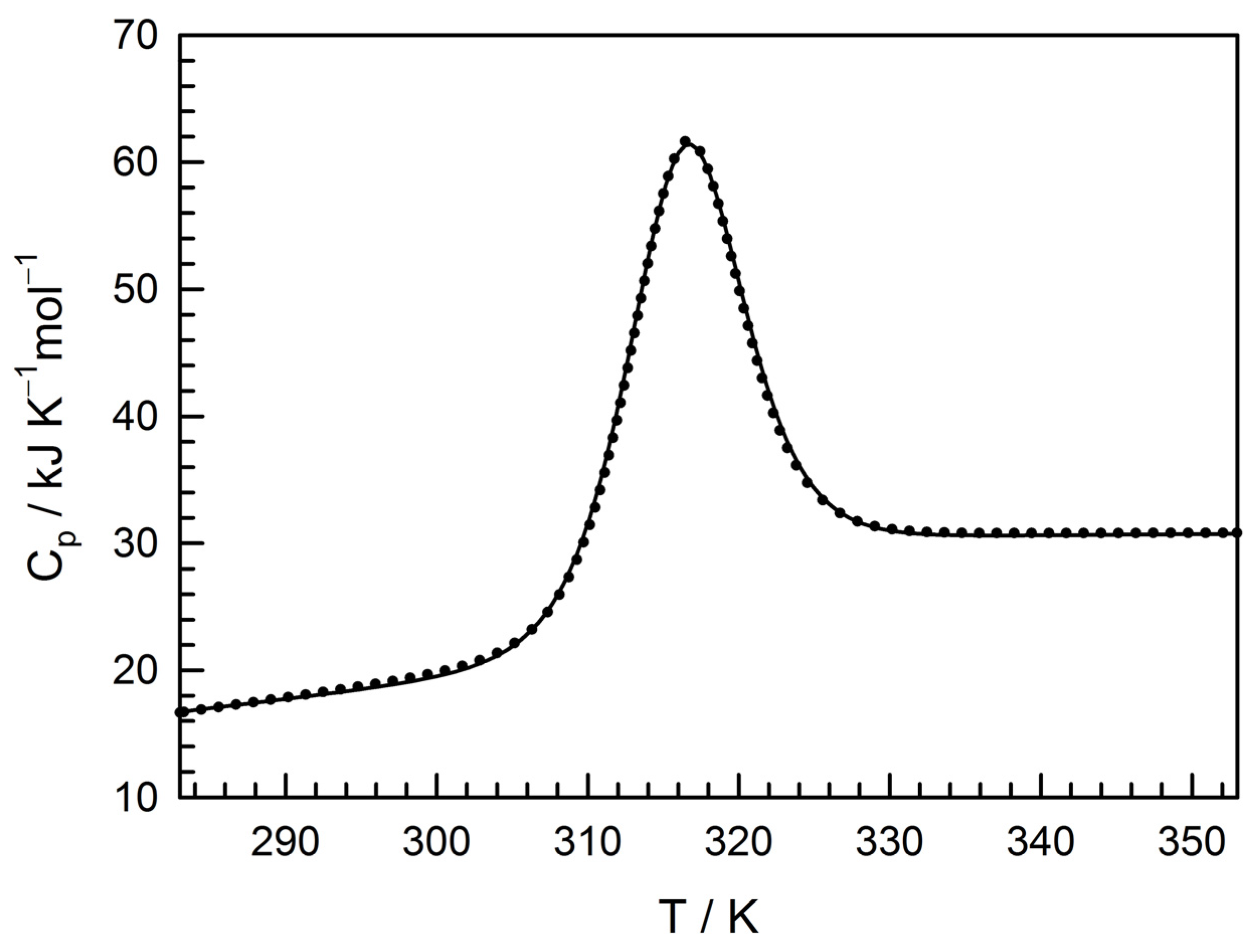
3.2. Physico-Chemical Characterization of the Recombinant Protein
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaper, J.B.; Morris, J.G.; Levine, M.M. Cholera. Clin. Microbiol. Rev. 1995, 8, 48–86. [Google Scholar] [CrossRef]
- Smirnova, N.I.; Goryaev, A.A.; Kutyrev, V.V. The evolution of the Vibrio cholerae genome during the modern period. Mol. Genet. Microbiol. Virol. 2010, 25, 148–157. [Google Scholar] [CrossRef]
- Hu, D.; Liu, B.; Feng, L.; Ding, P.; Guo, X.; Wang, M.; Cao, B.; Reeves, P.R.; Wang, L. Origins of the current seventh cholera pandemic. Proc. Natl. Acad. Sci. USA. 2016, 113, E7730–E7739. [Google Scholar] [CrossRef] [PubMed]
- Grim, C.J.; Hasan, N.A.; Taviani, E.; Haley, B.; Chun, J.; Brettin, T.S.; Bruce, D.C.; Detter, J.C.; Han, C.S.; Chertkov, O.; et al. Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J. Bacteriol. 2010, 192, 3524–3533. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, M.; Jennison, A.V.; Rathnayake, I.U.; Huygens, F. Evolution, distribution and genetics of atypical Vibrio cholerae—A review. Infect. Genet. Evol. 2021, 89, 104726. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kiaie, S.; Abtahi, H.; Mosayebi, G.; Alikhani, M.; Pakzad, I. Recombinant Toxin-Coregulated Pilus A (TcpA) as a candidate subunit cholera vaccine. Iran J. Microbiol. 2014, 6, 68–73. [Google Scholar]
- Molaee, N.; Mosayebi, G.; Amozande-Nobaveh, A.; Soleyman, M.R.; Abtahi, H. Evolution of the Immune response against recombinant proteins (TcpA, TcpB, and FlaA) as a candidate subunit cholera vaccine. J. Immunol. Res. 2017, 2017, 2412747. [Google Scholar] [CrossRef]
- Price, G.A.; Holmes, R.K. Immunizing adult female mice with a TcpA-A2-CTB chimera provides a high level of protection for their pups in the infant mouse model of cholera. PLoS Negl. Trop. Dis. 2014, 8, e3356. [Google Scholar] [CrossRef] [PubMed]
- Rollenhagen, J.E.; Kalsy, A.; Cerda, F.; John, M.; Harris, J.B.; LaRocque, R.C.; Qadri, F.; Calderwood, S.B.; Taylor, R.K.; Ryan, E.T. Transcutaneous Immunization with toxin-coregulated pilin a induces protective immunity against Vibrio cholerae O1 El tor challenge in mice. Infect. Immun. 2006, 74, 5834–5839. [Google Scholar] [CrossRef]
- Rhine, J.A.; Taylor, R.K. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol. Microbiol. 1994, 13, 1013–1020. [Google Scholar] [CrossRef]
- Herrington, D.A.; Hall, R.H.; Losonsky, G.; Mekalanos, J.J.; Taylor, R.K.; Levine, M.M. Toxin, toxin-coregulated pili, and the toxR regulon are essential for vibrio cholerae pathogenesis in humans. J. Exp. Med. 1988, 168, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Craig, L.; Pique, M.E.; Tainer, J.A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2004, 2, 363–378. [Google Scholar] [CrossRef]
- Krebs, S.J.; Taylor, R.K. Protection and attachment of vibrio cholerae mediated by the toxin-coregulated pilus in the infant mouse model. J. Bacteriol. 2011, 193, 5260–5270. [Google Scholar] [CrossRef] [PubMed]
- Hauke, C.A.; Taylor, R.K. Production of putative enhanced oral cholera vaccine strains that express toxin-coregulated pilus. PLoS ONE 2017, 12, e0175170. [Google Scholar] [CrossRef]
- Lim, M.S.; Ng, D.; Zong, Z.; Arvai, A.S.; Taylor, R.K.; Tainer, J.A.; Craig, L. Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation. Mol. Microbiol. 2010, 77, 755–770. [Google Scholar] [CrossRef]
- Klavdienko, E.V.; Tuchkov, I.V.; Polunina, T.A.; Guseva, N.P.; Smirnova, N.I. Construction of recombinant escherichia coli strain—Producer of basic subunit of Toxin-Coregulated Pilus of Adhesion (TCPA) of Vibrio cholerae biovar El Tor. Probl. Part. Danger. Infect. 2017, 4, 32–37. [Google Scholar] [CrossRef][Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-59259-890-8. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef]
- Micsonai, A.; Moussong, É.; Wien, F.; Boros, E.; Vadászi, H.; Murvai, N.; Lee, Y.-H.; Molnár, T.; Réfrégiers, M.; Goto, Y.; et al. BeStSel: Webserver for secondary structure and fold prediction for protein CD spectroscopy. Nucleic Acids Res. 2022, 50, W90–W98. [Google Scholar] [CrossRef]
- Böhm, G. CDNN: CD Spectra Deconvolution Software, Version 2.1; University of Halle-Wittenberg: Halle, Germany, 1997. [Google Scholar]
- Louis-Jeune, C.; Andrade-Navarro, M.A.; Perez-Iratxeta, C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins Struct. Funct. Bioinforma. 2012, 80, 374–381. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Sreerama, N.; Venyaminov, S.Y.U.; Woody, R.W. Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999, 8, 370–380. [Google Scholar] [CrossRef]
- Creighton, T.E. Electrophoretic analysis of the unfolding of proteins by urea. J. Mol. Biol. 1979, 129, 235–264. [Google Scholar] [CrossRef]
- Goldenberg, D.P. Transverse urea-gradient gel electrophoresis. Curr. Protoc. Protein Sci. 1996, 3, 7.4.1–7.4.13. [Google Scholar] [CrossRef]
- Senin, A.A.; Potekhin, S.A.; Tiktopulo, E.I.; Filomonov, V.V. Differential scanning microcalorimeter SCAL-1. J. Therm. Anal. Calorim. 2000, 62, 153–160. [Google Scholar] [CrossRef]
- Zahler, W.L.; Cleland, W.W. A specific and sensitive assay for disulfides. J. Biol. Chem. 1968, 243, 716–719. [Google Scholar] [CrossRef]
- Tanford, C. Protein Denaturation. Adv. Protein Chem. 1968, 23, 121–282. [Google Scholar]
- Pace, C.N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986, 131, 266–280. [Google Scholar]
- Myers, J.K.; Nick Pace, C.; Martin Scholtz, J. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995, 4, 2138–2148. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, V.V.; Martinez, J.C.; Mateo, P.L.; Prieto, J.; Serrano, L.; Bruix, M. Thermodynamic analysis of the chemotactic protein from Escherichia coli, CheY. Biochemistry 1993, 32, 12906–12921. [Google Scholar] [CrossRef]
- Wüthrich, K. NMR in Biological Research: Peptides and Proteins; North-Holland Pub. Co.: Amsterdam, The Netherlands, 1976. [Google Scholar]
- Padhiar, A.A.; Chanda, W.; Joseph, T.P.; Guo, X.; Liu, M.; Sha, L.; Batool, S.; Gao, Y.; Zhang, W.; Huang, M.; et al. Comparative study to develop a single method for retrieving wide class of recombinant proteins from classical inclusion bodies. Appl. Microbiol. Biotechnol. 2018, 102, 2363–2377. [Google Scholar] [CrossRef]
- Slouka, C.; Kopp, J.; Spadiut, O.; Herwig, C. Perspectives of inclusion bodies for bio-based products: Curse or blessing? Appl. Microbiol. Biotechnol. 2019, 103, 1143–1153. [Google Scholar] [CrossRef]
- Singhvi, P.; Saneja, A.; Srichandan, S.; Panda, A.K. Bacterial inclusion bodies: A treasure trove of bioactive proteins. Trends Biotechnol. 2020, 38, 474–486. [Google Scholar] [CrossRef]
- Eiberle, M.K.; Jungbauer, A. Technical refolding of proteins: Do we have freedom to operate? Biotechnol. J. 2010, 5, 547–559. [Google Scholar] [CrossRef]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact. 2015, 14, 41. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchenkov, V.; Dubovitskya, E.; Kotova, N.; Tuchkov, I.; Smirnova, N.; Marchenko, N.; Surin, A.; Filimonov, V.; Semisotnov, G. Recombinant Globular Domain of TcpA Pilin from Vibrio cholerae El Tor: Recovery from Inclusion Bodies and Structural Characterization. Life 2022, 12, 1802. https://doi.org/10.3390/life12111802
Marchenkov V, Dubovitskya E, Kotova N, Tuchkov I, Smirnova N, Marchenko N, Surin A, Filimonov V, Semisotnov G. Recombinant Globular Domain of TcpA Pilin from Vibrio cholerae El Tor: Recovery from Inclusion Bodies and Structural Characterization. Life. 2022; 12(11):1802. https://doi.org/10.3390/life12111802
Chicago/Turabian StyleMarchenkov, Victor, Elena Dubovitskya, Nina Kotova, Igor Tuchkov, Nina Smirnova, Natalia Marchenko, Alexey Surin, Vladimir Filimonov, and Gennady Semisotnov. 2022. "Recombinant Globular Domain of TcpA Pilin from Vibrio cholerae El Tor: Recovery from Inclusion Bodies and Structural Characterization" Life 12, no. 11: 1802. https://doi.org/10.3390/life12111802
APA StyleMarchenkov, V., Dubovitskya, E., Kotova, N., Tuchkov, I., Smirnova, N., Marchenko, N., Surin, A., Filimonov, V., & Semisotnov, G. (2022). Recombinant Globular Domain of TcpA Pilin from Vibrio cholerae El Tor: Recovery from Inclusion Bodies and Structural Characterization. Life, 12(11), 1802. https://doi.org/10.3390/life12111802








