Consequences and Resolution of Transcription–Replication Conflicts
Abstract
1. Introduction
2. TRCs as a Potent Endogenous Source of DNA Damage and Genomic Instability
2.1. Artificial Reporter and Plasmid Constructs as Model Loci for TRC Induction
2.1.1. Prokaryotes
2.1.2. Budding Yeast
2.1.3. Episomal System in Mammalian Cells
2.2. TRC-Induced Genomic Instability at Endogenous Chromosomal Loci
2.2.1. Highly Expressed Genes Challenge Replisome Progression
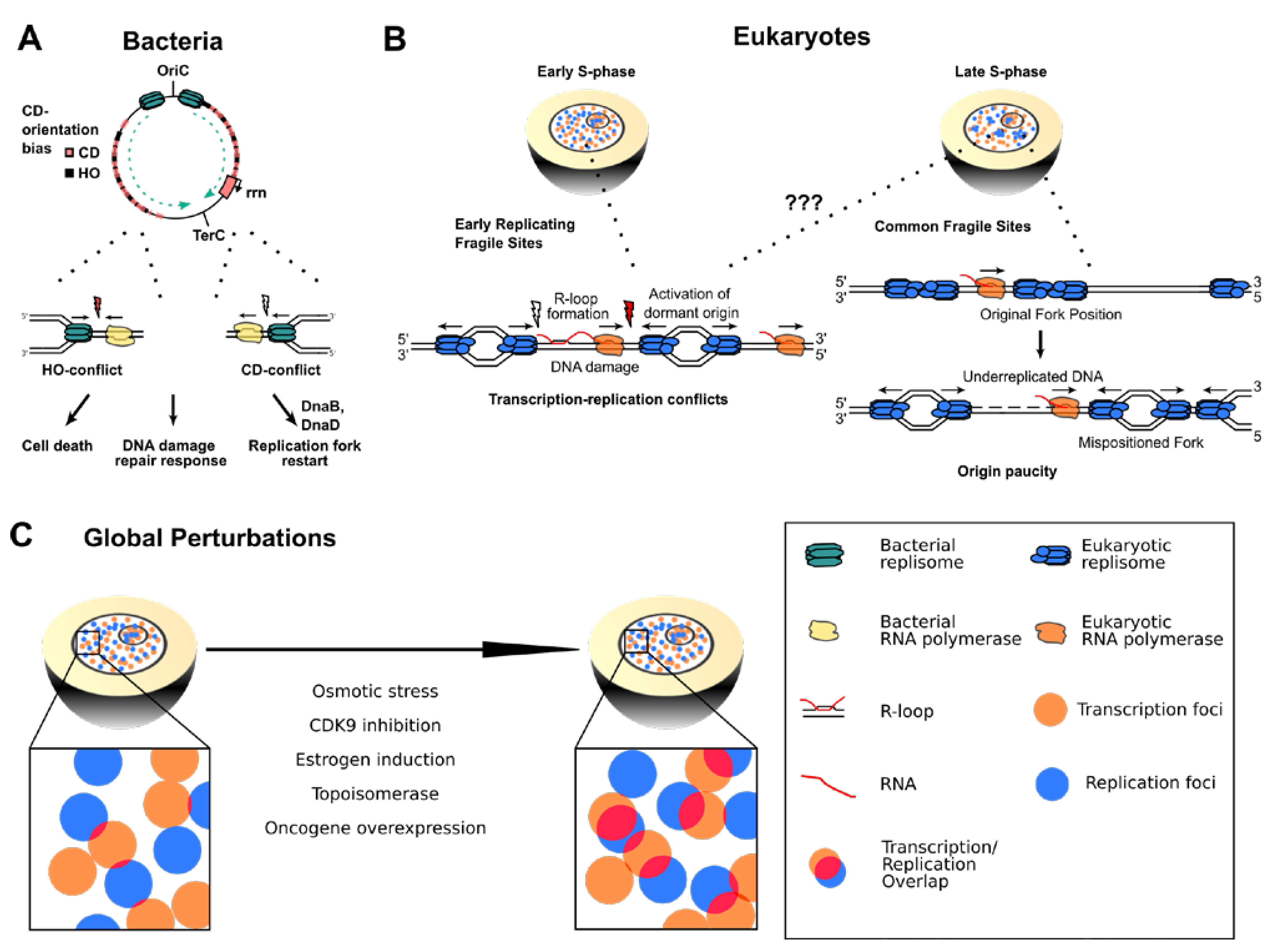
2.2.2. Common Fragile Sites and Early Replicating Fragile Sites
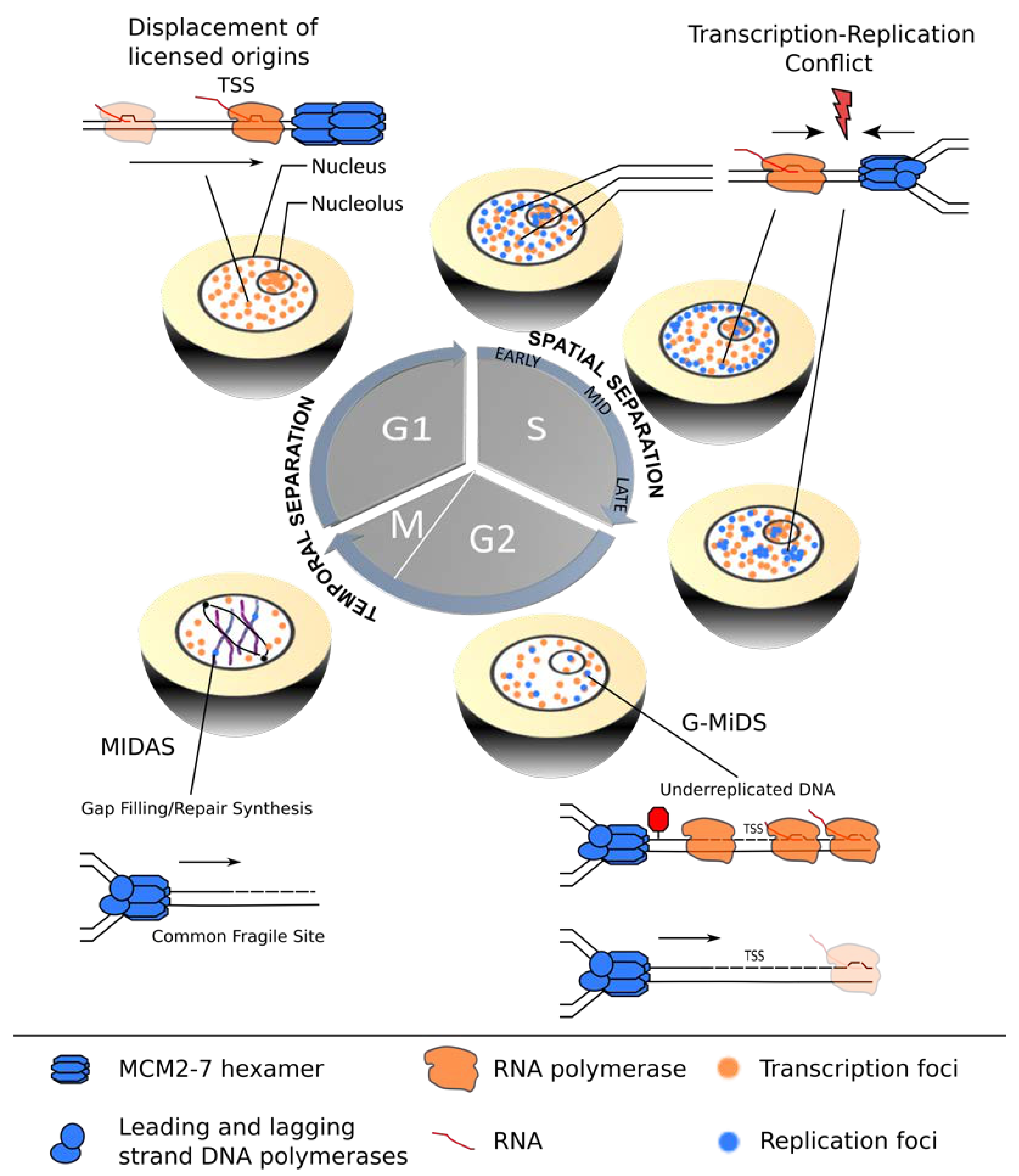
2.2.3. Global Perturbation of Transcription and Replication Programs in Eukaryotic Genomes
3. Crosstalk between the Chromatin Environment and TRCs
3.1. Histone PTMs as Regulators of Replication Fork Speed
3.2. Chromatin as an Insulator against Transcription-Induced DNA Damage and Replication Stress
3.3. Chromatin Compaction and Torsional Stress
4. Mechanisms to Resolve a TRC
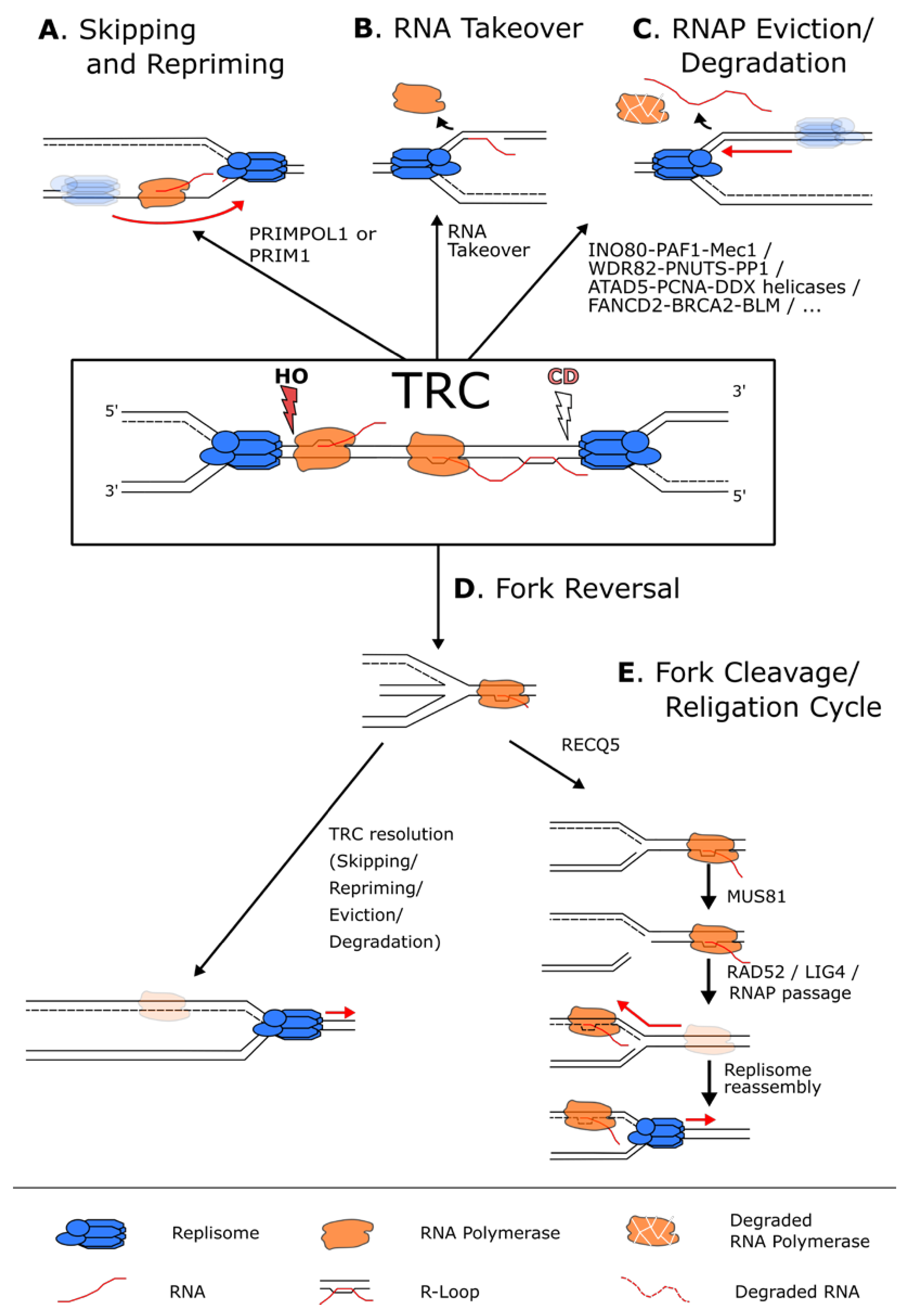
4.1. RNAP Skipping and Repriming
4.2. TRC-Induced Removal of RNA Polymerases
4.3. Processing R-Loops at Conflict Sites
4.4. Other Modes of RNAP Skipping at TRC Sites
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013, 9, 1003569. [Google Scholar] [CrossRef] [PubMed]
- Kurat, C.F.; Lambert, J.P.; van Dyk, D.; Tsui, K.; van Bakel, H.; Kaluarachchi, S.; Friesen, H.; Kainth, P.; Nislow, C.; Figeys, D.; et al. Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein. Genes Dev. 2011, 25, 2489–2501. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marzluff, W.F.; Wagner, E.J.; Duronio, R.J. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 2008, 9, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.K.; Boyd, J.R.; Grandy, R.A.; Medina, R.; Lian, J.B.; Stein, G.S.; Stein, J.L. Expression of Ribosomal RNA and Protein Genes in Human Embryonic Stem Cells Is Associated With the Activating H3K4me3 Histone Mark. J. Cell. Physiol. 2016, 231, 2007–2013. [Google Scholar] [CrossRef]
- Helmrich, A.; Ballarino, M.; Tora, L. Collisions between Replication and Transcription Complexes Cause Common Fragile Site Instability at the Longest Human Genes. Mol. Cell 2011, 44, 966–977. [Google Scholar] [CrossRef]
- Wei, X.; Samarabandu, J.; Devdhar, R.S.; Siegel, A.J.; Acharya, R.; Berezney, R. Segregation of transcription and replication sites into higher order domains. Science 1998, 281, 1502–1505. [Google Scholar] [CrossRef]
- Wansink, D.G.; Manders, E.E.; van der Kraan, I.; Aten, J.A.; van Driel, R.; de Jong, L. RNA polymerase II transcription is concentrated outside replication domains throughout S-phase. J. Cell Sci. 1994, 107, 1449–1456. [Google Scholar] [CrossRef]
- Hassan, A.B.; Errington, R.J.; White, N.S.; Jackson, D.A.; Cook, P.R. Replication and transcription sites are colocalized in human cells. J. Cell Sci. 1994, 107, 425–434. [Google Scholar] [CrossRef]
- Meryet-Figuiere, M.; Alaei-Mahabadi, B.; Ali, M.M.; Mitra, S.; Subhash, S.; Pandey, G.K.; Larsson, E.; Kanduri, C. Temporal separation of replication and transcription during S-phase progression. Cell Cycle 2014, 13, 3241–3248. [Google Scholar] [CrossRef]
- Petryk, N.; Kahli, M.; D’Aubenton-Carafa, Y.; Jaszczyszyn, Y.; Shen, Y.; Silvain, M.; Thermes, C.; Chen, C.L.; Hyrien, O. Replication landscape of the human genome. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Chen, Y.H.; Keegan, S.; Kahli, M.; Tonzi, P.; Fenyö, D.; Huang, T.T.; Smith, D.J. Transcription shapes DNA replication initiation and termination in human cells. Nat. Struct. Mol. Biol. 2019, 26, 67–77. [Google Scholar] [CrossRef]
- Prado, F.; Aguilera, A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005, 24, 1267–1276. [Google Scholar] [CrossRef]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 2017, 170, 774–786.e19. [Google Scholar] [CrossRef]
- Lang, K.S.; Hall, A.N.; Merrikh, C.N.; Ragheb, M.; Tabakh, H.; Pollock, A.J.; Woodward, J.J.; Dreifus, J.E.; Merrikh, H. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell 2017, 170, 787–799.e18. [Google Scholar] [CrossRef]
- Lang, K.S.; Merrikh, H. Topological stress is responsible for the detrimental outcomes of head-on replication-transcription conflicts. Cell Rep. 2021, 34. [Google Scholar] [CrossRef]
- Wang, J.; Rojas, P.; Mao, J.; Mustè Sadurnì, M.; Garnier, O.; Xiao, S.; Higgs, M.R.; Garcia, P.; Saponaro, M. Persistence of RNA transcription during DNA replication delays duplication of transcription start sites until G2/M. Cell Rep. 2021, 34. [Google Scholar] [CrossRef]
- Brewer, B.J.; Fangman, W.L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 1988, 55, 637–643. [Google Scholar] [CrossRef]
- Gros, J.; Kumar, C.; Lynch, G.; Yadav, T.; Whitehouse, I.; Remus, D. Post-licensing Specification of Eukaryotic Replication Origins by Facilitated Mcm2-7 Sliding along DNA. Mol. Cell 2015, 60, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.K.; MacAlpine, H.K.; Prinz, J.A.; Li, Y.; Belsky, J.A.; MacAlpine, D.M. Dynamic loading and redistribution of the Mcm2-7 helicase complex through the cell cycle. EMBO J. 2015, 34, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Ramanathan, S.; Okuno, Y.; Kumagai, C.; Shaikh, S.S.; Gilbert, D.M. The Chinese Hamster Dihydrofolate Reductase Replication Origin Decision Point Follows Activation of Transcription and Suppresses Initiation of Replication within Transcription Units. Mol. Cell. Biol. 2006, 26, 1051–1062. [Google Scholar] [CrossRef]
- Hyrien, O. Peaks cloaked in the mist: The landscape of mammalian replication origins. J. Cell Biol. 2015, 208, 147–160. [Google Scholar] [CrossRef]
- Czajkowsky, D.M.; Liu, J.; Hamlin, J.L.; Shao, Z. DNA Combing Reveals Intrinsic Temporal Disorder in the Replication of Yeast Chromosome VI. J. Mol. Biol. 2008, 375, 12–19. [Google Scholar] [CrossRef]
- Ekundayo, B.; Bleichert, F. Origins of DNA replication. PLoS Genet. 2019, 15, e1008320. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef]
- Brüning, J.-G.G.; Marians, K.J. Replisome bypass of transcription complexes and R-loops. Nucleic Acids Res. 2020, 48, 10353–10367. [Google Scholar] [CrossRef]
- Schauer, G.D.; Spenkelink, L.M.; Lewis, J.S.; Yurieva, O.; Mueller, S.H.; van Oijen, A.M.; O’Donnell, M.E. Replisome bypass of a protein-based R-loop block by Pif1. Proc. Natl. Acad. Sci. USA 2020, 117, 30354–30361. [Google Scholar] [CrossRef]
- Sankar, T.S.; Wastuwidyaningtyas, B.D.; Dong, Y.; Lewis, S.A.; Wang, J.D. The nature of mutations induced by replication-transcription collisions. Nature 2016, 535, 178–181. [Google Scholar] [CrossRef]
- García-Rubio, M.; Aguilera, P.; Lafuente-Barquero, J.; Ruiz, J.F.; Simon, M.N.; Geli, V.; Rondón, A.G.; Aguilera, A. Yra1-bound RNA–DNA hybrids cause orientation-independent transcription– replication collisions and telomere instability. Genes Dev. 2018, 32, 965–977. [Google Scholar] [CrossRef]
- Kim, N.; Abdulovic, A.L.; Gealy, R.; Lippert, M.J.; Jinks-Robertson, S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair 2007, 6, 1285–1296. [Google Scholar] [CrossRef]
- Fragkos, M.; Ganier, O.; Coulombe, P.; Méchali, M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015, 16, 360–374. [Google Scholar] [CrossRef]
- Brison, O.; El-Hilali, S.; Azar, D.; Koundrioukoff, S.; Schmidt, M.; Nähse, V.; Jaszczyszyn, Y.; Lachages, A.M.; Dutrillaux, B.; Thermes, C.; et al. Transcription-mediated organization of the replication initiation program across large genes sets common fragile sites genome-wide. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Paul, S.; Million-Weaver, S.; Chattopadhyay, S.; Sokurenko, E.; Merrikh, H. Accelerated gene evolution through replication-transcription conflicts. Nature 2013, 495, 512–515. [Google Scholar] [CrossRef]
- Srivatsan, A.; Tehranchi, A.; MacAlpine, D.M.; Wang, J.D. Co-Orientation of Replication and Transcription Preserves Genome Integrity. PLoS Genet. 2010, 6, e1000810. [Google Scholar] [CrossRef]
- Merrikh, H.; Zhang, Y.; Grossman, A.D.; Wang, J.D. Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol. 2012, 10, 449–458. [Google Scholar] [CrossRef]
- Mangiameli, S.M.; Merrikh, C.N.; Wiggins, P.A.; Merrikh, H. Transcription leads to pervasive replisome instability in bacteria. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Kogoma, T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 1997, 61, 212–238. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.L.T.; Pohl, T.J.; Chen, C.F.; Chan, A.; Pott, S.; Zakian, V.A. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Azvolinsky, A.; Giresi, P.G.; Lieb, J.D.; Zakian, V.A. Highly Transcribed RNA Polymerase II Genes Are Impediments to Replication Fork Progression in Saccharomyces cerevisiae. Mol. Cell 2009, 34, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Osmundson, J.S.; Kumar, J.; Yeung, R.; Smith, D.J. Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat. Struct. Mol. Biol. 2017, 24, 162–170. [Google Scholar] [CrossRef]
- Yeung, R.; Smith, D.J. Determinants of Replication-Fork Pausing at tRNA Genes in Saccharomyces cerevisiae. Genetics 2020, 214, 825–838. [Google Scholar] [CrossRef]
- Hecht, F.; Glover, T.W. Cancer chromosome breakpoints and common fragile sites induced by aphidicolin. Cancer Genet. Cytogenet. 1984, 13, 185–188. [Google Scholar] [CrossRef]
- Glover, T.W.; Berger, C.; Coyle, J.; Echo, B. DNA polymerase α inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 1984, 67, 136–142. [Google Scholar] [CrossRef]
- Oestergaard, V.H.; Lisby, M. Transcription-replication conflicts at chromosomal fragile sites—consequences in M phase and beyond. Chromosoma 2017, 126, 213–222. [Google Scholar] [CrossRef]
- Macheret, M.; Bhowmick, R.; Sobkowiak, K.; Padayachy, L.; Mailler, J.; Hickson, I.D.; Halazonetis, T.D. High-resolution mapping of mitotic DNA synthesis regions and common fragile sites in the human genome through direct sequencing. Cell Res. 2020, 30, 997–1008. [Google Scholar] [CrossRef]
- Helmrich, A.; Stout-Weider, K.; Hermann, K.; Schrock, E.; Heiden, T. Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome Res. 2006, 16, 1222–1230. [Google Scholar] [CrossRef]
- Pentzold, C.; Shah, S.A.; Hansen, N.R.; Le Tallec, B.; Seguin-Orlando, A.; Debatisse, M.; Lisby, M.; Oestergaard, V.H. FANCD2 binding identifies conserved fragile sites at large transcribed genes in avian cells. Nucleic Acids Res. 2018, 46, 1280–1294. [Google Scholar] [CrossRef]
- Wilson, T.E.; Arlt, M.F.; Park, S.H.; Rajendran, S.; Paulsen, M.; Ljungman, M.; Glover, T.W. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 2015, 25, 189–200. [Google Scholar] [CrossRef]
- Blin, M.; Le Tallec, B.; Nähse, V.; Schmidt, M.; Brossas, C.; Millot, G.A.; Prioleau, M.-N.; Debatisse, M. Transcription-dependent regulation of replication dynamics modulates genome stability. Nat. Struct. Mol. Biol. 2019, 26, 58–66. [Google Scholar] [CrossRef]
- Barlow, J.H.; Faryabi, R.B.; Callén, E.; Wong, N.; Malhowski, A.; Chen, H.T.; Gutierrez-Cruz, G.; Sun, H.W.; McKinnon, P.; Wright, G.; et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 2013, 152, 620–632. [Google Scholar] [CrossRef]
- Mortusewicz, O.; Herr, P.; Helleday, T. Early replication fragile sites: Where replication–transcription collisions cause genetic instability. EMBO J. 2013, 32, 493–495. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 2018, 555, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Duch, A.; Canal, B.; Barroso, S.I.; García-Rubio, M.; Seisenbacher, G.; Aguilera, A.; de Nadal, E.; Posas, F. Multiple signaling kinases target Mrc1 to prevent genomic instability triggered by transcription-replication conflicts. Nat. Commun. 2018, 9, 379. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.A.; McCulley, A.; Haarer, B.; Arnak, R.; Feng, W. Break-seq reveals hydroxyurea-induced chromosome fragility as a result of unscheduled conflict between DNA replication and transcription. Genome Res. 2015, 25, 402–412. [Google Scholar] [CrossRef]
- Shao, X.; Joergensen, A.M.; Howlett, N.G.; Lisby, M.; Oestergaard, V.H. A distinct role for recombination repair factors in an early cellular response to transcription-replication conflicts. Nucleic Acids Res. 2020, 48, 5467–5484. [Google Scholar] [CrossRef]
- Stork, C.T.; Bocek, M.; Crossley, M.P.; Sollier, J.; Sanz, L.A.; Chédin, F.; Swigut, T.; Cimprich, K.A. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 2016, 5. [Google Scholar] [CrossRef]
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 2015, 15, 276–280. [Google Scholar] [CrossRef]
- Donati, B.; Lorenzini, E.; Ciarrocchi, A. BRD4 and Cancer: Going beyond transcriptional regulation. Mol. Cancer 2018, 17, 1–13. [Google Scholar] [CrossRef]
- Neelsen, K.J.; Zanini, I.M.Y.; Herrador, R.; Lopes, M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell Biol. 2013, 200, 699–708. [Google Scholar] [CrossRef]
- Miron, K.; Golan-Lev, T.; Dvir, R.; Ben-David, E.; Kerem, B. Oncogenes create a unique landscape of fragile sites. Nat. Commun. 2015, 6, 1–7. [Google Scholar] [CrossRef]
- Kotsantis, P.; Silva, L.M.; Irmscher, S.; Jones, R.M.; Folkes, L.; Gromak, N.; Petermann, E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Georgakilas, A.G.; Tsantoulis, P.; Kotsinas, A.; Michalopoulos, I.; Townsend, P.; Gorgoulis, V.G. Are common fragile sites merely structural domains or highly organized “functional” units susceptible to oncogenic stress? Cell. Mol. Life Sci. 2014, 71, 4519–4544. [Google Scholar] [CrossRef]
- Tsantoulis, P.K.; Kotsinas, A.; Sfikakis, P.P.; Evangelou, K.; Sideridou, M.; Levy, B.; Mo, L.; Kittas, C.; Wu, X.R.; Papavassiliou, A.G.; et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene 2008, 27, 3256–3264. [Google Scholar] [CrossRef]
- Stewart-Morgan, K.R.; Petryk, N.; Groth, A. Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 2020, 22, 361–371. [Google Scholar] [CrossRef]
- Schlissel, G.; Rine, J. The nucleosome core particle remembers its position through DNA replication and RNA transcription. Proc. Natl. Acad. Sci. USA 2019, 116, 20605–20611. [Google Scholar] [CrossRef]
- Francis, N.J.; Sihou, D. Inheritance of Histone (H3/H4): A Binary Choice? Trends Biochem. Sci. 2021, 46, 5–14. [Google Scholar] [CrossRef]
- Stewart-Morgan, K.R.; Reverón-Gómez, N.; Groth, A. Erratum: Transcription Restart Establishes Chromatin Accessibility after DNA Replication (Molecular Cell (2019) 75(2) (284–297.e6), (S1097276519303521), (10.1016/j.molcel.2019.04.033)). Mol. Cell 2019, 75, 408–414. [Google Scholar] [CrossRef]
- Marchal, C.; Sima, J.; Gilbert, D.M. Control of DNA replication timing in the 3D genome. Nat. Rev. Mol. Cell Biol. 2019, 20, 721–737. [Google Scholar] [CrossRef]
- Ehara, H.; Kujirai, T.; Fujino, Y.; Shirouzu, M.; Kurumizaka, H.; Sekine, S.I. Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science 2019, 363, 744–747. [Google Scholar] [CrossRef]
- Crickard, J.B.; Lee, J.; Lee, T.H.; Reese, J.C. The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome. Nucleic Acids Res. 2017, 45, 6362–6374. [Google Scholar] [CrossRef]
- Ferrari, P.; Strubin, M. Uncoupling histone turnover from transcription-associated histone H3 modifications. Nucleic Acids Res. 2015, 43, 3972–3985. [Google Scholar] [CrossRef][Green Version]
- Jeronimo, C.; Poitras, C.; Robert, F. Histone Recycling by FACT and Spt6 during Transcription Prevents the Scrambling of Histone Modifications. Cell Rep. 2019, 28, 1206–1218.e8. [Google Scholar] [CrossRef]
- Gates, L.A.; Foulds, C.E.; O’Malley, B.W. Histone Marks in the ‘Driver’s Seat’: Functional Roles in Steering the Transcription Cycle. Trends Biochem. Sci. 2017, 42, 977–989. [Google Scholar] [CrossRef]
- Chong, S.Y.; Cutler, S.; Lin, J.J.; Tsai, C.H.; Tsai, H.K.; Biggins, S.; Tsukiyama, T.; Lo, Y.C.; Kao, C.F. H3K4 methylation at active genes mitigates transcription-replication conflicts during replication stress. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Frenkel, N.; Jonas, F.; Carmi, M.; Yaakov, G.; Barkai, N. Rtt109 slows replication speed by histone N-terminal acetylation. Genome Res. 2021, 31, 426–435. [Google Scholar] [CrossRef]
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.; Chédin, F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell 2016, 63, 167–178. [Google Scholar] [CrossRef]
- Almeida, R.; Fernández-Justel, J.M.; Santa-María, C.; Cadoret, J.C.; Cano-Aroca, L.; Lombraña, R.; Herranz, G.; Agresti, A.; Gómez, M. Chromatin conformation regulates the coordination between DNA replication and transcription. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Salas-Armenteros, I.; Pérez-Calero, C.; Bayona-Feliu, A.; Tumini, E.; Luna, R.; Aguilera, A. Human THO–Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J. 2017, 36, 3532–3547. [Google Scholar] [CrossRef]
- Herrera-Moyano, E.; Mergui, X.; García-Rubio, M.L.; Barroso, S.; Aguilera, A. The yeast and human FACT chromatinreorganizing complexes solve R-loopmediated transcription-replication conflicts. Genes Dev. 2014, 28, 735–748. [Google Scholar] [CrossRef]
- García-Pichardo, D.; Cañas, J.C.; García-Rubio, M.L.; Gómez-González, B.; Rondón, A.G.; Aguilera, A. Histone Mutants Separate R Loop Formation from Genome Instability Induction. Mol. Cell 2017, 66, 597–609.e5. [Google Scholar] [CrossRef]
- Achar, Y.J.; Adhil, M.; Choudhary, R.; Gilbert, N.; Foiani, M. Negative supercoil at gene boundaries modulates gene topology. Nature 2020, 577, 701–705. [Google Scholar] [CrossRef] [PubMed]
- Boque-Sastre, R.; Soler, M.; Oliveira-Mateos, C.; Portela, A.; Moutinho, C.; Sayols, S.; Villanueva, A.; Esteller, M.; Guil, S. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. USA 2015, 112, 5785–5790. [Google Scholar] [CrossRef] [PubMed]
- Castellano-Pozo, M.; Santos-Pereira, J.; Rondón, A.G.; Barroso, S.; Andújar, E.; Pérez-Alegre, M.; García-Muse, T.; Aguilera, A. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell 2013, 52, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Colak, D.; Zaninovic, N.; Cohen, M.S.; Rosenwaks, Z.; Yang, W.Y.; Gerhardt, J.; Disney, M.D.; Jaffrey, S.R. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 2014, 343, 1002–1005. [Google Scholar] [CrossRef] [PubMed]
- Groh, M.; Lufino, M.M.P.; Wade-Martins, R.; Gromak, N. R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Loomis, E.W.; Sanz, L.A.; Chédin, F.; Hagerman, P.J. Transcription-Associated R-Loop Formation across the Human FMR1 CGG-Repeat Region. PLoS Genet. 2014, 10, e1004294. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Kamieniarz-Gdula, K.; Proudfoot, N.J. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 2014, 516, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Thys, R.G.; Lehman, E.C.; Pierce, C.T.L.; Wang, Y.L. DNA Secondary Structure at Chromosomal Fragile Sites in Human Disease. Curr. Genom. 2015, 16, 60–70. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Voon, H.P.J.; Xella, B.; Scott, C.; Clynes, D.; Babbs, C.; Ayyub, H.; Kerry, J.; Sharpe, J.A.; Sloane-Stanley, J.A.; et al. The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 2017, 18, 914–928. [Google Scholar] [CrossRef]
- Chen, P.B.; Chen, H.V.; Acharya, D.; Rando, O.J.; Fazzio, T.G. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 2015, 22, 999–1007. [Google Scholar] [CrossRef]
- Promonet, A.; Padioleau, I.; Liu, Y.; Sanz, L.; Biernacka, A.; Schmitz, A.-L.; Skrzypczak, M.; Sarrazin, A.; Mettling, C.; Rowicka, M.; et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 2020, 11, 3940. [Google Scholar] [CrossRef]
- Clark, D.J.; Felsenfeld, G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell 1992, 71, 11–22. [Google Scholar] [CrossRef]
- Kaczmarczyk, A.; Meng, H.; Ordu, O.; van Noort, J.; Dekker, N.H. Chromatin fibers stabilize nucleosomes under torsional stress. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Tsai, S.; Fournier, L.-A.; Chang, E.Y.; Wells, J.P.; Minaker, S.W.; Zhu, Y.D.; Wang, A.Y.-H.; Wang, Y.; Huntsman, D.G.; Stirling, P.C. ARID1A regulates R-loop associated DNA replication stress. PLOS Genet. 2021, 17, e1009238. [Google Scholar] [CrossRef]
- Bayona-Feliu, A.; Barroso, S.; Muñoz, S.; Aguilera, A. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription–replication conflicts. Nat. Genet. 2021, 2021, 1–14. [Google Scholar] [CrossRef]
- Gómez-González, B.; Aguilera, A. Transcription-mediated replication hindrance: A major driver of genome instability. Genes Dev. 2019, 33, 1008–1026. [Google Scholar] [CrossRef]
- Pomerantz, R.T.; O’Donnell, M.; O’Donnell, M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 2008, 456, 762–766. [Google Scholar] [CrossRef]
- Conti, B.A.; Smogorzewska, A. Mechanisms of direct replication restart at stressed replisomes. DNA Repair 2020, 95, 102947. [Google Scholar] [CrossRef]
- García-Gómez, S.; Reyes, A.; Martínez-Jiménez, M.I.; Chocrón, E.S.; Mourón, S.; Terrados, G.; Powell, C.; Salido, E.; Méndez, J.; Holt, I.J.; et al. PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 2013, 52, 541–553. [Google Scholar] [CrossRef]
- Mourón, S.; Rodriguez-Acebes, S.; Martínez-Jiménez, M.I.; García-Gómez, S.; Chocrón, S.; Blanco, L.; Méndez, J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 2013, 20, 1383–1389. [Google Scholar] [CrossRef]
- Bianchi, J.; Rudd, S.G.; Jozwiakowski, S.K.; Bailey, L.J.; Soura, V.; Taylor, E.; Stevanovic, I.; Green, A.J.; Stracker, T.H.; Lindsay, H.D.; et al. PrimPol Bypasses UV Photoproducts during Eukaryotic Chromosomal DNA Replication. Mol. Cell 2013, 52, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Guilliam, T.A.; Tsuda, M.; Yamamoto, J.; Bailey, L.J.; Iwai, S.; Takeda, S.; Doherty, A.J.; Hirota, K. Repriming by PrimPol is critical for DNA replication restart downstream of lesions and chain-terminating nucleosides. Cell Cycle 2016, 15, 1997–2008. [Google Scholar] [CrossRef] [PubMed]
- Wessel, S.R.; Mohni, K.N.; Luzwick, J.W.; Dungrawala, H.; Cortez, D. Functional Analysis of the Replication Fork Proteome Identifies BET Proteins as PCNA Regulators. Cell Rep. 2019, 28, 3497–3509.e4. [Google Scholar] [CrossRef] [PubMed]
- Šviković, S.; Crisp, A.; Tan-Wong, S.M.; Guilliam, T.A.; Doherty, A.J.; Proudfoot, N.J.; Guilbaud, G.; Sale, J.E. R-loop formation during S phase is restricted by PrimPol-mediated repriming. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, R.T.; O’Donnell, M. What happens when replication and transcription complexes collide? Cell Cycle 2010, 9, 2537–2543. [Google Scholar] [CrossRef]
- Huang, J.; Liu, S.; Bellani, M.A.; Thazhathveetil, A.K.; Ling, C.; de Winter, J.P.; Wang, Y.; Wang, W.; Seidman, M.M. The DNA Translocase FANCM/MHF Promotes Replication Traverse of DNA Interstrand Crosslinks. Mol. Cell 2013, 52, 434–446. [Google Scholar] [CrossRef]
- Trakselis, M.A.; Seidman, M.M.; Brosh, R.M. Mechanistic insights into how CMG helicase facilitates replication past DNA roadblocks. DNA Repair 2017, 55, 76–82. [Google Scholar] [CrossRef]
- Sparks, J.L.; Chistol, G.; Gao, A.O.; Räschle, M.; Larsen, N.B.; Mann, M.; Duxin, J.P.; Walter, J.C. The CMG Helicase Bypasses DNA-Protein Cross-Links to Facilitate Their Repair. Cell 2019, 176, 167–181.e21. [Google Scholar] [CrossRef]
- Hawkins, M.; Dimude, J.U.; Howard, J.A.L.; Smith, A.J.; Dillingham, M.S.; Savery, N.J.; Rudolph, C.J.; McGlynn, P. Direct removal of RNA polymerase barriers to replication by accessory replicative helicases. Nucleic Acids Res. 2019, 47, 5100–5113. [Google Scholar] [CrossRef]
- Ivessa, A.S.; Lenzmeier, B.A.; Bessler, J.B.; Goudsouzian, L.K.; Schnakenberg, S.L.; Zakian, V.A. The Saccharomyces cerevisiae Helicase Rrm3p Facilitates Replication Past Nonhistone Protein-DNA Complexes. Mol. Cell 2003, 12, 1525–1536. [Google Scholar] [CrossRef]
- Lang, K.S.; Merrikh, H. The Clash of Macromolecular Titans: Replication-Transcription Conflicts in Bacteria. Annu. Rev. Microbiol. 2018, 72, 71–88. [Google Scholar] [CrossRef]
- Felipe-Abrio, I.; Lafuente-Barquero, J.; García-Rubio, M.L.; Aguilera, A. RNA polymerase II contributes to preventing transcription-mediated replication fork stalls. EMBO J. 2015, 34, 236–250. [Google Scholar] [CrossRef]
- Lam, F.C.; Kong, Y.W.; Huang, Q.; Vu Han, T.L.; Maffa, A.D.; Kasper, E.M.; Yaffe, M.B. BRD4 prevents the accumulation of R-loops and protects against transcription–replication collision events and DNA damage. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Edwards, D.S.; Maganti, R.; Tanksley, J.P.; Luo, J.; Park, J.J.H.; Balkanska-Sinclair, E.; Ling, J.; Floyd, S.R. BRD4 Prevents R-Loop Formation and Transcription-Replication Conflicts by Ensuring Efficient Transcription Elongation. Cell Rep. 2020, 32, 108166. [Google Scholar] [CrossRef]
- Gaillard, H.; Aguilera, A. Transcription coupled repair at the interface between transcription elongation and mRNP biogenesis. Biochim. Biophys. Acta—Gene Regul. Mech. 2013, 1829, 141–150. [Google Scholar] [CrossRef]
- Hobson, D.J.; Wei, W.; Steinmetz, L.M.; Svejstrup, J.Q. RNA polymerase II collision interrupts convergent transcription. Mol. Cell 2012, 48, 365–374. [Google Scholar] [CrossRef]
- Wilson, M.D.; Harreman, M.; Taschner, M.; Reid, J.; Walker, J.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Proteasome-mediated processing of Def1, a critical step in the cellular response to transcription stress. Cell 2013, 154, 983–995. [Google Scholar] [CrossRef]
- McKay, B.C.; Becerril, C.; Spronck, J.C.; Ljungman, M. Ultraviolet light-induced apoptosis is associated with S-phase in primary human fibroblasts. DNA Repair 2002, 1, 811–820. [Google Scholar] [CrossRef]
- Zaratiegui, M.; Castel, S.E.; Irvine, D.V.; Kloc, A.; Ren, J.; Li, F.; de Castro, E.; Marín, L.; Chang, A.-Y.; Goto, D.; et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature 2011, 479, 135–138. [Google Scholar] [CrossRef]
- Castel, S.E.; Ren, J.; Bhattacharjee, S.; Chang, A.-Y.; Sánchez, M.; Valbuena, A.; Antequera, F.; Martienssen, R.A. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 2014, 159, 572–583. [Google Scholar] [CrossRef]
- Poli, J.; Gerhold, C.-B.; Tosi, A.; Hustedt, N.; Seeber, A.; Sack, R.; Herzog, F.; Pasero, P.; Shimada, K.; Hopfner, K.-P.; et al. Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 2016, 30, 337–354. [Google Scholar] [CrossRef]
- Landsverk, H.B.; Sandquist, L.E.; Bay, L.T.E.; Steurer, B.; Campsteijn, C.; Landsverk, O.J.B.; Marteijn, J.A.; Petermann, E.; Trinkle-Mulcahy, L.; Syljuåsen, R.G. WDR82/PNUTS-PP1 Prevents Transcription-Replication Conflicts by Promoting RNA Polymerase II Degradation on Chromatin. Cell Rep. 2020, 33, 108469. [Google Scholar] [CrossRef]
- Ciurciu, A.; Duncalf, L.; Jonchere, V.; Lansdale, N.; Vasieva, O.; Glenday, P.; Rudenko, A.; Vissi, E.; Cobbe, N.; Alphey, L.; et al. PNUTS/PP1 Regulates RNAPII-Mediated Gene Expression and Is Necessary for Developmental Growth. PLOS Genet. 2013, 9, e1003885. [Google Scholar] [CrossRef]
- Lee, J.-H.; You, J.; Dobrota, E.; Skalnik, D.G. Identification and Characterization of a Novel Human PP1 Phosphatase Complex. J. Biol. Chem. 2010, 285, 24466–24476. [Google Scholar] [CrossRef]
- Huertas, P.; Aguilera, A. Cotranscriptionally Formed DNA:RNA Hybrids Mediate Transcription Elongation Impairment and Transcription-Associated Recombination. Mol. Cell 2003, 12, 711–721. [Google Scholar] [CrossRef]
- Bermejo, R.; Capra, T.; Jossen, R.; Colosio, A.; Frattini, C.; Carotenuto, W.; Cocito, A.; Doksani, Y.; Klein, H.; Gómez-González, B.; et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 2011, 146, 233–246. [Google Scholar] [CrossRef]
- Gómez-González, B.; García-Rubio, M.; Bermejo, R.; Gaillard, H.; Shirahige, K.; Marín, A.; Foiani, M.; Aguilera, A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011, 30, 3106–3119. [Google Scholar] [CrossRef]
- Singh, S.; Ahmed, D.; Dolatshad, H.; Tatwavedi, D.; Schulze, U.; Sanchi, A.; Ryley, S.; Dhir, A.; Carpenter, L.; Watt, S.M.; et al. SF3B1 mutations induce R-loop accumulation and DNA damage in MDS and leukemia cells with therapeutic implications. Leukemia 2020, 34, 2525–2530. [Google Scholar] [CrossRef] [PubMed]
- Salas-Armenteros, I.; Barroso, S.I.; Rondón, A.G.; Pérez, M.; Andújar, E.; Luna, R.; Aguilera, A. Depletion of the MFAP1/SPP381 Splicing Factor Causes R-Loop-Independent Genome Instability. Cell Rep. 2019, 28, 1551–1563.e7. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, J.-Y.; Huang, Y.-J.; Gu, Y.; Qiu, J.; Qian, H.; Shao, C.; Zhang, X.; Hu, J.; Li, H.; et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol. Cell 2018, 69, 412–425.e6. [Google Scholar] [CrossRef] [PubMed]
- El Hage, A.; French, S.L.; Beyer, A.L.; Tollervey, D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010, 24, 1546–1558. [Google Scholar] [CrossRef]
- Tuduri, S.; Crabbé, L.; Conti, C.; Tourrière, H.; Holtgreve-Grez, H.; Jauch, A.; Pantesco, V.; De Vos, J.; Thomas, A.; Theillet, C.; et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009, 11, 1315–1324. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J.-Y.; Zhang, X.; Gu, Y.; Xiao, R.; Shao, C.; Tang, P.; Qian, H.; Luo, D.; Li, H.; et al. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol. Cell 2017, 68, 745–757.e5. [Google Scholar] [CrossRef]
- Sollier, J.; Stork, C.T.; García-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef]
- Sakasai, R.; Isono, M.; Wakasugi, M.; Hashimoto, M.; Sunatani, Y.; Matsui, T.; Shibata, A.; Matsunaga, T.; Iwabuchi, K. Aquarius is required for proper CtIP expression and homologous recombination repair. Sci. Rep. 2017, 7, 13808. [Google Scholar] [CrossRef]
- Groh, M.; Albulescu, L.O.; Cristini, A.; Gromak, N. Senataxin: Genome Guardian at the Interface of Transcription and Neurodegeneration. J. Mol. Biol. 2017, 429, 3181–3195. [Google Scholar] [CrossRef]
- Alzu, A.; Bermejo, R.; Begnis, M.; Lucca, C.; Piccini, D.; Carotenuto, W.; Saponaro, M.; Brambati, A.; Cocito, A.; Foiani, M.; et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 2012, 151, 835–846. [Google Scholar] [CrossRef]
- Silva, B.; Pentz, R.; Figueira, A.M.; Arora, R.; Lee, Y.W.; Hodson, C.; Wischnewski, H.; Deans, A.J.; Azzalin, C.M. FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat. Commun. 2019, 10, 2253. [Google Scholar] [CrossRef]
- Chang, E.Y.-C.; Novoa, C.A.; Aristizabal, M.J.; Coulombe, Y.; Segovia, R.; Chaturvedi, R.; Shen, Y.; Keong, C.; Tam, A.S.; Jones, S.J.M.; et al. RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability. J. Cell Biol. 2017, 216, 3991–4005. [Google Scholar] [CrossRef]
- Brambati, A.; Zardoni, L.; Achar, Y.J.; Piccini, D.; Galanti, L.; Colosio, A.; Foiani, M.; Liberi, G. Dormant origins and fork protection mechanisms rescue sister forks arrested by transcription. Nucleic Acids Res. 2018, 46, 1227–1239. [Google Scholar] [CrossRef]
- Appanah, R.; Lones, E.C.; Aiello, U.; Libri, D.; De, G.; Correspondence, P. Sen1 Is Recruited to Replication Forks via Ctf4 and Mrc1 and Promotes Genome Stability. Cell Rep. 2020. [Google Scholar] [CrossRef]
- Hodroj, D.; Recolin, B.; Serhal, K.; Martinez, S.; Tsanov, N.; Abou Merhi, R.; Maiorano, D. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017, 36, 1182–1198. [Google Scholar] [CrossRef]
- Song, C.; Hotz-Wagenblatt, A.; Voit, R.; Grummt, I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017, 31, 1370–1381. [Google Scholar] [CrossRef]
- Talwar, T.; Vidhyasagar, V.; Qing, J.; Guo, M.; Kariem, A.; Lu, Y.; Singh, R.S.; Lukong, K.E.; Wu, Y. The DEAD-box protein DDX43 (HAGE) is a dual RNA-DNA helicase and has a K-homology domain required for full nucleic acid unwinding activity. J. Biol. Chem. 2017, 292, 10429–10443. [Google Scholar] [CrossRef]
- Yu, Z.; Mersaoui, S.Y.; Guitton-Sert, L.; Coulombe, Y.; Song, J.; Masson, J.-Y.; Richard, S. DDX5 resolves R-loops at DNA double-strand breaks to promote DNA repair and avoid chromosomal deletions. NAR Cancer 2020, 2. [Google Scholar] [CrossRef]
- Villarreal, O.D.; Mersaoui, S.Y.; Yu, Z.; Masson, J.-Y.; Richard, S. Genome-wide R-loop analysis defines unique roles for DDX5, XRN2, and PRMT5 in DNA/RNA hybrid resolution. Life Sci. Alliance 2020, 3, e202000762. [Google Scholar] [CrossRef]
- Okamoto, Y.; Abe, M.; Itaya, A.; Tomida, J.; Ishiai, M.; Takaori-Kondo, A.; Taoka, M.; Isobe, T.; Takata, M. FANCD2 protects genome stability by recruiting RNA processing enzymes to resolve R-loops during mild replication stress. FEBS J. 2019, 286, 139–150. [Google Scholar] [CrossRef]
- Sridhara, S.C.; Carvalho, S.; Grosso, A.R.; Gallego-Paez, L.M.; Carmo-Fonseca, M.; de Almeida, S.F. Transcription Dynamics Prevent RNA-Mediated Genomic Instability through SRPK2-Dependent DDX23 Phosphorylation. Cell Rep. 2017, 18, 334–343. [Google Scholar] [CrossRef]
- Chakraborty, P.; Grosse, F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair 2011, 10, 654–665. [Google Scholar] [CrossRef]
- Patel, P.S.; Abraham, K.J.; Guturi, K.K.N.; Halaby, M.-J.; Khan, Z.; Palomero, L.; Ho, B.; Duan, S.; St-Germain, J.; Algouneh, A.; et al. RNF168 regulates R-loop resolution and genomic stability in BRCA1/2-deficient tumors. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Pérez-Calero, C.; Bayona-Feliu, A.; Xue, X.; Barroso, S.I.; Muñoz, S.; González-Basallote, V.M.; Sung, P.; Aguilera, A. UAP56/DDX39B is a major cotranscriptional RNA–DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 2020, 34, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Monckton, E.A.; Godbout, R. A role for DEAD box 1 at DNA double-strand breaks. Mol. Cell. Biol. 2008, 28, 6413–6425. [Google Scholar] [CrossRef] [PubMed]
- Abakir, A.; Giles, T.C.; Cristini, A.; Foster, J.M.; Dai, N.; Starczak, M.; Rubio-Roldan, A.; Li, M.; Eleftheriou, M.; Crutchley, J.; et al. N(6)-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat. Genet. 2020, 52, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Cheon, N.Y.; Park, H.; Jeong, G.W.; Ye, B.J.; Yoo, E.J.; Lee, J.H.; Hur, J.-H.; Lee, E.-A.; Kim, H.; et al. TonEBP recognizes R-loops and initiates m6A RNA methylation for R-loop resolution. Nucleic Acids Res. 2021, 49, 269–284. [Google Scholar] [CrossRef]
- Madireddy, A.; Kosiyatrakul, S.T.; Boisvert, R.A.; Herrera-Moyano, E.; García-Rubio, M.L.; Gerhardt, J.; Vuono, E.A.; Owen, N.; Yan, Z.; Olson, S.; et al. FANCD2 Facilitates Replication through Common Fragile Sites. Mol. Cell 2016, 64, 388–404. [Google Scholar] [CrossRef]
- Kim, S.; Kang, N.; Park, S.H.; Wells, J.; Hwang, T.; Ryu, E.; Kim, B.-G.; Hwang, S.; Kim, S.-J.; Kang, S.; et al. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res. 2020, 48, 7218–7238. [Google Scholar] [CrossRef]
- Lee, K.-Y.; Yang, K.; Cohn, M.A.; Sikdar, N.; D’Andrea, A.D.; Myung, K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J. Biol. Chem. 2010, 285, 10362–10369. [Google Scholar] [CrossRef]
- Lee, K.; Fu, H.; Aladjem, M.I.; Myung, K. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J. Cell Biol. 2013, 200, 31–44. [Google Scholar] [CrossRef]
- Park, S.H.; Kang, N.; Song, E.; Wie, M.; Lee, E.A.; Hwang, S.; Lee, D.; Ra, J.S.; Park, I.B.; Park, J.; et al. ATAD5 promotes replication restart by regulating RAD51 and PCNA in response to replication stress. Nat. Commun. 2019, 10, 5718. [Google Scholar] [CrossRef]
- Quinet, A.; Lemaçon, D.; Vindigni, A. Replication Fork Reversal: Players and Guardians. Mol. Cell 2017, 68, 830–833. [Google Scholar] [CrossRef]
- Chappidi, N.; Nascakova, Z.; Boleslavska, B.; Zellweger, R.; Isik, E.; Andrs, M.; Menon, S.; Dobrovolna, J.; Balbo Pogliano, C.; Matos, J.; et al. Fork Cleavage-Religation Cycle and Active Transcription Mediate Replication Restart after Fork Stalling at Co-transcriptional R-Loops. Mol. Cell 2020, 77, 528–541.e8. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lalonde, M.; Trauner, M.; Werner, M.; Hamperl, S. Consequences and Resolution of Transcription–Replication Conflicts. Life 2021, 11, 637. https://doi.org/10.3390/life11070637
Lalonde M, Trauner M, Werner M, Hamperl S. Consequences and Resolution of Transcription–Replication Conflicts. Life. 2021; 11(7):637. https://doi.org/10.3390/life11070637
Chicago/Turabian StyleLalonde, Maxime, Manuel Trauner, Marcel Werner, and Stephan Hamperl. 2021. "Consequences and Resolution of Transcription–Replication Conflicts" Life 11, no. 7: 637. https://doi.org/10.3390/life11070637
APA StyleLalonde, M., Trauner, M., Werner, M., & Hamperl, S. (2021). Consequences and Resolution of Transcription–Replication Conflicts. Life, 11(7), 637. https://doi.org/10.3390/life11070637





