Effect of Five Polymorphisms on Percentage of Oleic Acid in Beef and Investigation of Linkage Disequilibrium to Confirm the Locations of Quantitative Trait Loci on BTA19 in Japanese Black Cattle
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Genotype and Allele Frequency
3.2. Association between C18:1 and Gene Markers
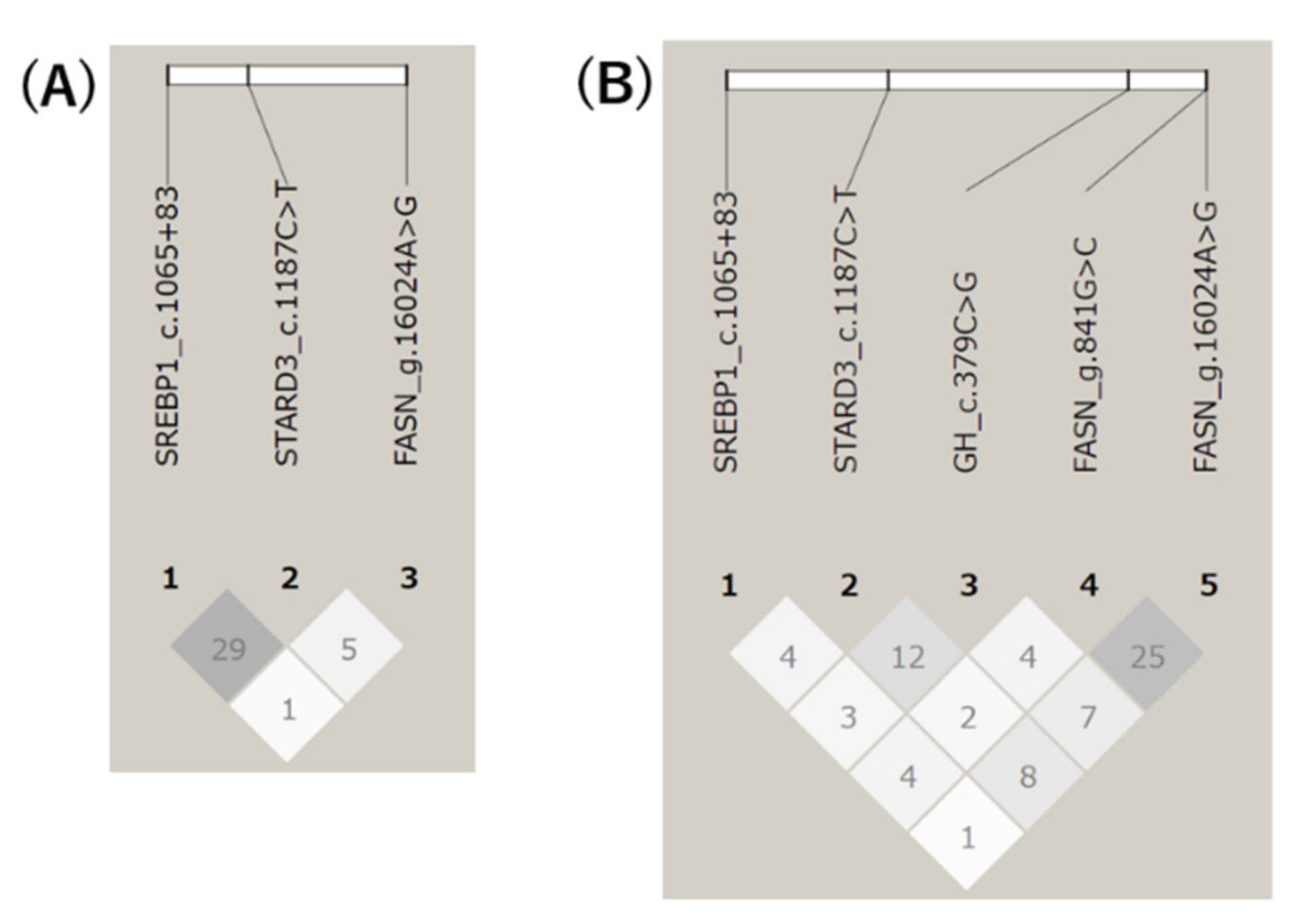
3.3. LD among Five Gene Polymorphisms
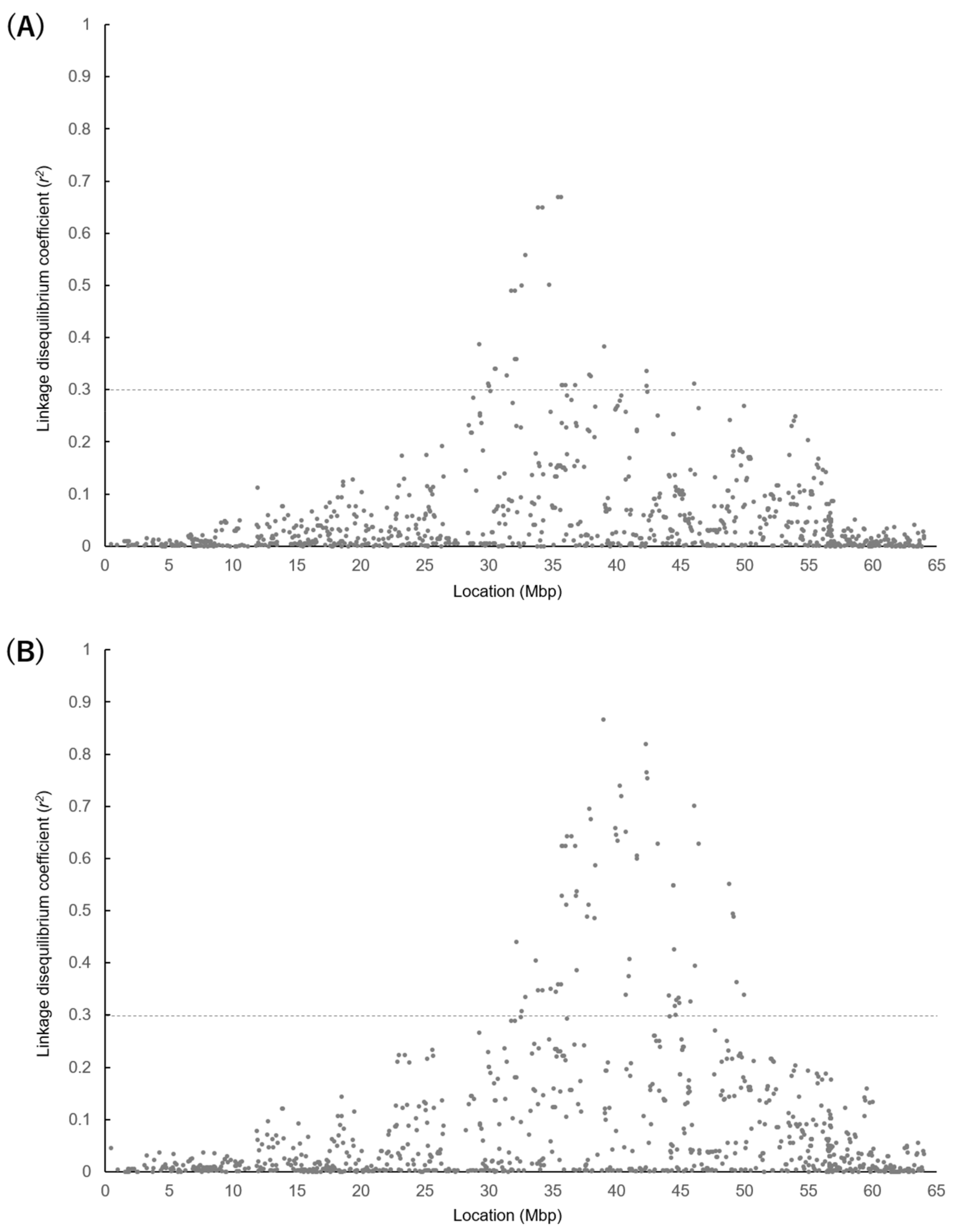
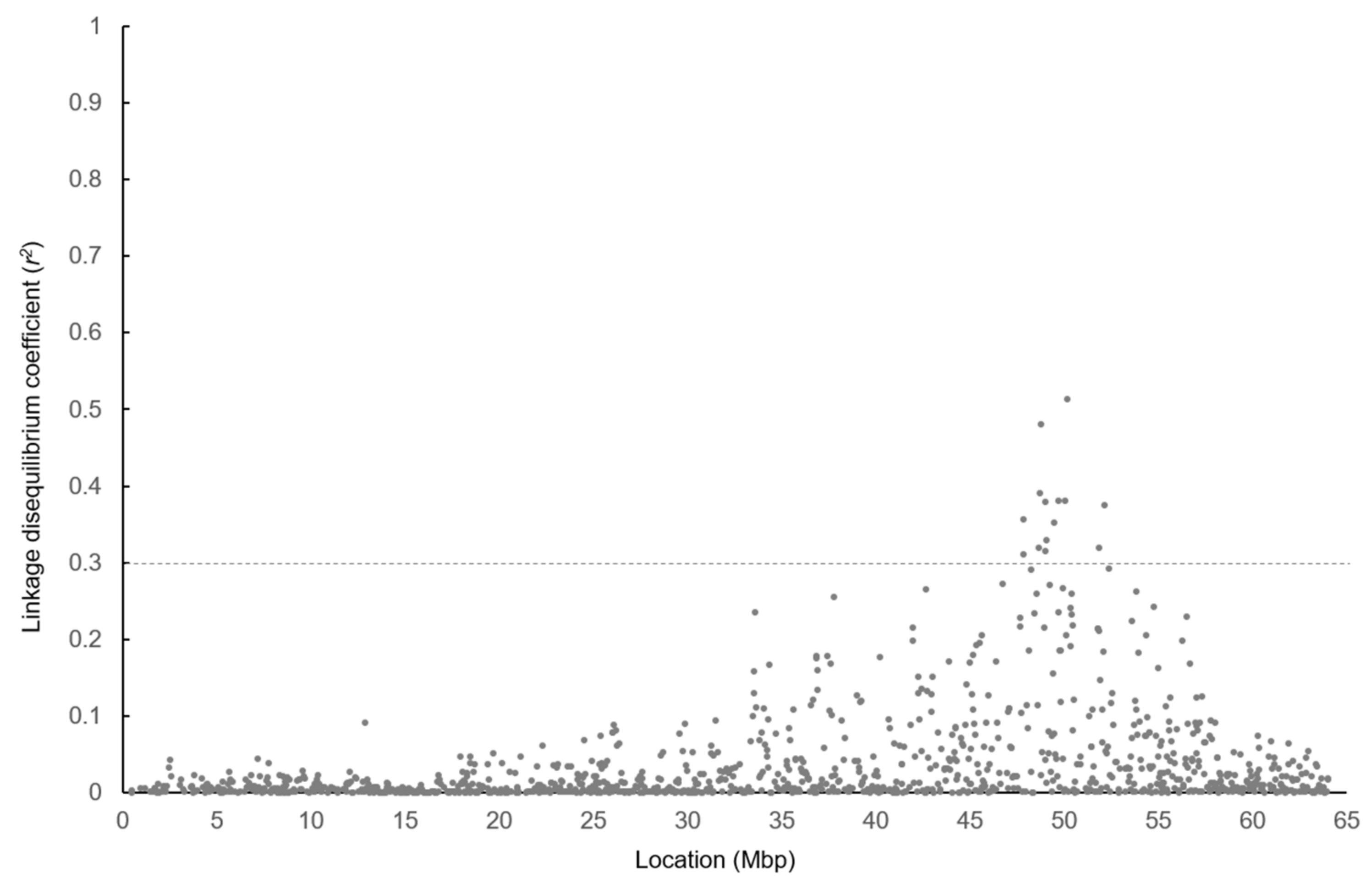
3.4. LD between 50K SNPs and Gene Polymorphisms
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, A.; Larsen, T.W.; Powell, V.H.; Tume, R.K. A comparison of fat composition of Japanese and long-term grain-fed Australian steers. Meat Sci. 1999, 51, 1–9. [Google Scholar] [CrossRef]
- Wood, J.D.; Richardson, R.I.; Nute, G.R.; Fisher, A.V.; Campo, M.M.; Kasapidou, E.; Sheard, P.R.; Enser, M. Effects of fatty acids on meat quality: A review. Meat Sci. 2003, 66, 21–32. [Google Scholar] [CrossRef]
- Abe, T.; Saburi, J.; Hasebe, H.; Nakagawa, T.; Misumi, S.; Nade, T.; Nakajima, H.; Shoji, N.; Kobayashi, M.; Kobayashi, E. Novel mutations of the FASN gene and their effect of fatty acid composition in Japanese Black beef. Biochem. Genet. 2009, 47, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Bartoň, L.; Bureš, D.; Kott, T.; Řehák, D. Associations of polymorphisms in bovine DGAT1, FABP4, FASN, and PPARGC1A genes with intramuscular fat content and the fatty acid composition of muscle and subcutaneous fat in Fleckvieh bulls. Meat Sci. 2016, 114, 18–23. [Google Scholar] [CrossRef]
- Hayakawa, K.; Sakamoto, T.; Ihii, A.; Yamaji, K.; Uemoto, Y.; Sasago, N.; Kobayashi, E.; Kobayashi, N.; Matsuhashi, T.; Maruyama, S.; et al. The g.841G>C SNP of FASN gene is associated with fatty acid composition in beef cattle. Anim. Sci. J. 2015, 86, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Knight, T.J.; Reecy, J.M.; Beitz, D.C. DNA polymorphisms in bovine fatty acid synthase are associated with beef fatty acid composition. Anim. Genet. 2008, 39, 62–70. [Google Scholar] [CrossRef]
- Hoashi, S.; Ashida, N.; Ohsaki, H.; Utsugi, T.; Sasazaki, S.; Taniguchi, M.; Oyama, K.; Mukai, F.; Mannen, H. Genotype of bovine sterol regulatory element binding protein-1 (SREBP-1) is associated with fatty acid composition in Japanese Black cattle. Mamm. Genome 2007, 18, 880–886. [Google Scholar] [CrossRef]
- Kigoshi, H.; Kawaguchi, F.; Oyama, K.; Mannen, H.; Sasazaki, S. Effect of STARD3 gene polymorphism on carcass traits and fatty acid composition in Japanese Black cattle. J. Anim. Genet. 2019, 47, 37–45. [Google Scholar] [CrossRef][Green Version]
- Matsuhashi, T.; Maruyama, S.; Uemoto, Y.; Kobayashi, N.; Mannen, H.; Abe, T.; Sakaguchi, S.; Kobayashi, E. Effects of bovine fatty acid synthase, stearoyl-coenzyme A desaturase, sterol regulatory element-binding protein 1, and growth hormone gene polymorphisms on fatty acid composition and carcass traits in Japanese Black cattle. J. Anim. Sci. 2011, 89, 12–22. [Google Scholar] [CrossRef]
- Chakravarty, B.; Gu, Z.; Chirala, S.S.; Wakil, S.J.; Quiocho, F.A. Human fatty acid synthase: Structure and substrate selectivity of the thioesterase domain. Proc. Natl. Acad. Sci. USA 2004, 101, 15567–15572. [Google Scholar] [CrossRef]
- Ordovás, L.; Roy, R.; Pampín, S.; Zaragoza, P.; Osta, R.; Rodríguez-Rey, J.C.; Rodellar, C. The g.763G>C SNP of the bovine FASN gene affects its promoter activity via Sp-mediated regulation: Implications for the bovine lactating mammary gland. Physiol. Genom. 2008, 34, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Shimano, H. Sterol regulatory element-binding proteins (SREBPs): Transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 2001, 40, 439–452. [Google Scholar] [CrossRef]
- Gamarra, D.; Aldai, N.; Arakawa, A.; de Pancorbo, M.M.; Taniguchi, M. Effect of a genetic polymorphism in SREBP1 on fatty acid composition and related gene expression in subcutaneous fat tissue of beef cattle breeds. Anim. Sci. J. 2021, 92, e13521. [Google Scholar] [CrossRef] [PubMed]
- Aply, F.; Stoeckel, M.E.; Dierich, A.; Escola, J.M.; Wendling, C.; Chenard, M.P.; Vanier, M.T.; Gruenberg, J.; Tomasetto, C.; Rio, M.C. The steroidogenic acute regulatory protein homolog MLN64, a late endosomal cholesterol-binging protein. J. Biol. Chem. 2001, 276, 4261–4269. [Google Scholar]
- Wilhelm, L.P.; Wendling, C.; Védie, B.; Kobayashi, T.; Chenard, M.P.; Tomasetto, C.; Drin, G.; Alpy, F. STARD3 mediates endoplasmic reticulum-to-endosome cholesterol trans port at membrane contact sites. EMBO J. 2017, 36, 1412–1433. [Google Scholar] [CrossRef] [PubMed]
- Hocquette, J.F. Endocrine and metabolic regulation of muscle growth and body composition in cattle. Animal 2010, 4, 1797–1809. [Google Scholar] [CrossRef][Green Version]
- Troike, K.M.; Henry, B.E.; Jensen, E.A.; Young, J.A.; List, E.O.; Kopchick, J.J.; Berryman, D.E. Impact of growth hormone on regulation of adipose tissue. Compr. Physiol. 2017, 7, 819–840. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Ookura, K.; Akiyama, T.; Yoshida, E.; Fukushima, M.; Iwamoto, E.; Oka, A.; Matsumoto, H.; Sasazaki, S.; Oyama, K.; Mannen, H. Effects of genes on economically important traits of Japanese Black cattle in Hyogo population. Nihon Chikusan Gakkaiho 2013, 84, 157–162. [Google Scholar] [CrossRef][Green Version]
- Chikuni, K.; Nagatsuma, T.; Tabata, T.; Monma, M.; Saito, M.; Ozawa, S.; Ozutsumi, K. Genetic variations of the growth hormone gene in Japanese cattle. Anim. Sci. Technol. 1994, 65, 340–346. [Google Scholar]
- Ishii, A.; Yamaji, K.; Uemoto, Y.; Sasago, N.; Kobayashi, E.; Kobayashi, N.; Matsuhashi, T.; Maruyama, S.; Matsumoto, H.; Sasazaki, S.; et al. Genome-wide association study for fatty acid composition in Japanese Black cattle. Anim. Sci. J. 2013, 84, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Nomura, T.; Fukushima, M.; Mukai, F. Genetic diversity of a closed population of Japanese Black cattle in Hyogo Prefecture. Anim. Sci. J. 2001, 72, 378–385. [Google Scholar] [CrossRef]
- Nomura, T.; Honda, T.; Mukai, F. Inbreeding and effective population size of Japanese Black cattle. J. Anim. Sci. 2001, 79, 366–370. [Google Scholar] [CrossRef]
- Jia, P.; Cai, C.; Qu, K.; Chen, N.; Jia, Y.; Hanif, Q.; Liu, J.; Zhang, J.; Che, H.; Huang, B.; et al. Four novel SNPs of MYO1A gene associated with heat-tolerance in Chinese cattle. Animals 2019, 9, 964. [Google Scholar] [CrossRef]
- Weikard, R.; Kühn, C.; Goldammer, T.; Freyer, G.; Schwerin, M. The bovine PPARGC1A gene: Molecular characterization and association of an SNP with variation of milk fat synthesis. Physiol. Genom. 2005, 21, 1–13. [Google Scholar] [CrossRef]
- Ohsaki, H.; Tanaka, A.; Hoashi, S.; Sasazaki, S.; Oyama, K.; Taniguchi, M.; Mukai, F.; Mannen, H. Effect of SCD and SREBP genotypes on fatty acid composition in adipose tissue of Japanese Black cattle herds. Anim. Sci. J. 2009, 80, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, L.P.; Yuan, Z.R.; Guo, L.P.; Zhu, M.; Gao, X.; Gao, H.J.; Li, J.Y.; Xu, S.Z. Polymorphism of SREBP1 is associated with beef fatty acid composition in Simmental bulls. Genet. Mol. Res. 2013, 12, 5802–5809. [Google Scholar] [CrossRef]

| Hyogo Population | Gifu Population | ||
|---|---|---|---|
| n | Total | 441 | 443 |
| steers/heifers | 352/89 | 443/0 | |
| Sires | 7 | 57 | |
| Age at slaughter (months) | Mean ± SD | 31.69 ± 1.24 | 28.32 ± 1.10 |
| Max | 35.87 | 34.62 | |
| Min | 28.44 | 25.74 | |
| C18:1 percentage (%) | Mean ± SD | 54.76 ± 3.01 | 51.92 ± 3.29 |
| Max | 64.01 | 60.54 | |
| Min | 45.45 | 43.67 |
| Location | ID | Gene | Position | Polymorphism | Amino Acid | Reference |
|---|---|---|---|---|---|---|
| 35,244,514 | rs133958066 | SREBP1 | Intron 5 | c.1065+83 (84bp indel) | - | [7] |
| 40,687,937 | rs134877666 | STARD3 | Exon 14 | c.1187 C > T | S396L | [8] |
| 48,768,916 | rs41923484 | GH | Exon 5 | c.379 C > G | L127V | [21] |
| 51,384,984 | rs41920005 | FASN | Exon 1 | g.841 G>C | - | [5] |
| 51,400,139 | rs480320793 | FASN | Exon 34 | g.16024 A>G | T1950A | [3] |
| Gene Marker | Method | Sequence (from 5′ to 3′) | AT | PL | RE | Reference |
|---|---|---|---|---|---|---|
| SREBP1 c.1065 + 83 | PCR | F: CCA CAA CGC CAT CGA GAA ACG CTA C | 60 | 348/432 | - | [7] |
| R: GGC CTT CCC TGA CCA CCC AAC TTA G | ||||||
| STARD3 c.1187 C > T | PCR-RFLP | F: AGG AGG ATT TGA GCA CCC CAT | 60 | 351 | MscI | [8] |
| R: CAA GGT CAC ACA GCA CAC TCC | ||||||
| GH c.379 C > G | PCR-RFLP | F: CTT AGC CAG GAG AAT GCA CG | 66 | 605 | AlwNI | - |
| R: ATG CCT GCT ATT GTC TTC CC | ||||||
| Taq-Man | F: CAA ATT TGT CAT AGG TCT GCT TG | 64 | 228 | - | - | |
| R: CCC TCT TTC TAG CAG TCC AG | ||||||
| P1: FAM- CAT CTT CCA GCT CCT GCC A -BHQ | ||||||
| P2: HEX- CAT CTT CCA CCT CCT GCC A -BHQ | ||||||
| FASN g.841 G > C | Taq-Man | F: ACA CTC CAT CCT CGC TC | 60 | 102 | - | [5] |
| R: TCC CGA CTC GCA ACT TC | ||||||
| P1: FAM- ACA GCC GCC CGC G -BHQ | ||||||
| P2: HEX- ACA GCC CCC CGC GC -BHQ |
| Polymorphism | A Allele | B Allele | Genotype Frequency | Allele Frequency | p-Value | Means ± SE of C18:1 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | A | B | AA | AB | BB | ||||
| SREBP1 c.1065 + 83 | L | S | 210 | 180 | 51 | 0.79 | 0.21 | 4.00 × 10−4 | 55.28 ± 0.20 | 54.56 ± 0.21 | 53.32 ± 0.47 |
| STARD3 c.1187 C > T | C | T | 141 | 206 | 94 | 0.55 | 0.45 | 1.68 × 10−5 | 55.88 ± 0.25 | 54.65 ± 0.19 | 53.29 ± 0.29 |
| GH c.379 C > G | G | C | 48 | 0 | 0 | 1.00 | 0.00 | - | - | - | - |
| FASN g.841 G > C | G | C | 44 | 0 | 0 | 1.00 | 0.00 | - | - | - | - |
| FASN g.16024 A > G | A | G | 360 | 73 | 8 | 0.90 | 0.10 | 0.614 | 54.63 ± 0.16 | 55.33 ± 0.34 | 55.09 ± 1.22 |
| Polymorphism | A Allele | B Allele | Genotype Frequency | Allele Frequency | p-Value | Means ± SE of C18:1 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | A | B | AA | AB | BB | ||||
| SREBP1 c.1065 + 83 | L | S | 159 | 218 | 66 | 0.61 | 0.39 | 0.060 | 52.06 ± 0.26 | 52.09 ± 0.22 | 51.02 ± 0.37 |
| STARD3 c.1187 C > T | C | T | 110 | 227 | 106 | 0.51 | 0.49 | 0.143 | 51.60 ± 0.33 | 51.83 ± 0.21 | 52.42 ± 0.31 |
| GH c.379 C > G | G | C | 189 | 208 | 46 | 0.66 | 0.34 | 4.00 × 10−3 | 52.58 ± 0.23 | 51.42 ± 0.24 | 51.46 ± 0.42 |
| FASN g.841 G > C | G | C | 362 | 78 | 3 | 0.91 | 0.09 | 8.24 × 10−5 | 52.23 ± 0.17 | 50.44 ± 0.37 | 52.79 ± 3.65 |
| FASN g.16024 A > G | A | G | 305 | 126 | 12 | 0.83 | 0.17 | 1.00 × 10−3 | 52.25 ± 0.19 | 51.25 ± 0.28 | 50.53 ± 1.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawaguchi, F.; Kakiuchi, F.; Oyama, K.; Mannen, H.; Sasazaki, S. Effect of Five Polymorphisms on Percentage of Oleic Acid in Beef and Investigation of Linkage Disequilibrium to Confirm the Locations of Quantitative Trait Loci on BTA19 in Japanese Black Cattle. Life 2021, 11, 597. https://doi.org/10.3390/life11070597
Kawaguchi F, Kakiuchi F, Oyama K, Mannen H, Sasazaki S. Effect of Five Polymorphisms on Percentage of Oleic Acid in Beef and Investigation of Linkage Disequilibrium to Confirm the Locations of Quantitative Trait Loci on BTA19 in Japanese Black Cattle. Life. 2021; 11(7):597. https://doi.org/10.3390/life11070597
Chicago/Turabian StyleKawaguchi, Fuki, Fuka Kakiuchi, Kenji Oyama, Hideyuki Mannen, and Shinji Sasazaki. 2021. "Effect of Five Polymorphisms on Percentage of Oleic Acid in Beef and Investigation of Linkage Disequilibrium to Confirm the Locations of Quantitative Trait Loci on BTA19 in Japanese Black Cattle" Life 11, no. 7: 597. https://doi.org/10.3390/life11070597
APA StyleKawaguchi, F., Kakiuchi, F., Oyama, K., Mannen, H., & Sasazaki, S. (2021). Effect of Five Polymorphisms on Percentage of Oleic Acid in Beef and Investigation of Linkage Disequilibrium to Confirm the Locations of Quantitative Trait Loci on BTA19 in Japanese Black Cattle. Life, 11(7), 597. https://doi.org/10.3390/life11070597





