Protein Secondary Structure Affects Glycan Clustering in Native Mass Spectrometry
Abstract
1. Introduction
2. Materials and Methods
2.1. Glycans
2.2. Proteins
2.3. Native Mass Spectrometry
2.4. Determination of Glycan Binding: Calculation of the Dissociation Constant (KD)
2.5. Titration Measurement of Glycan Binding on P Dimers
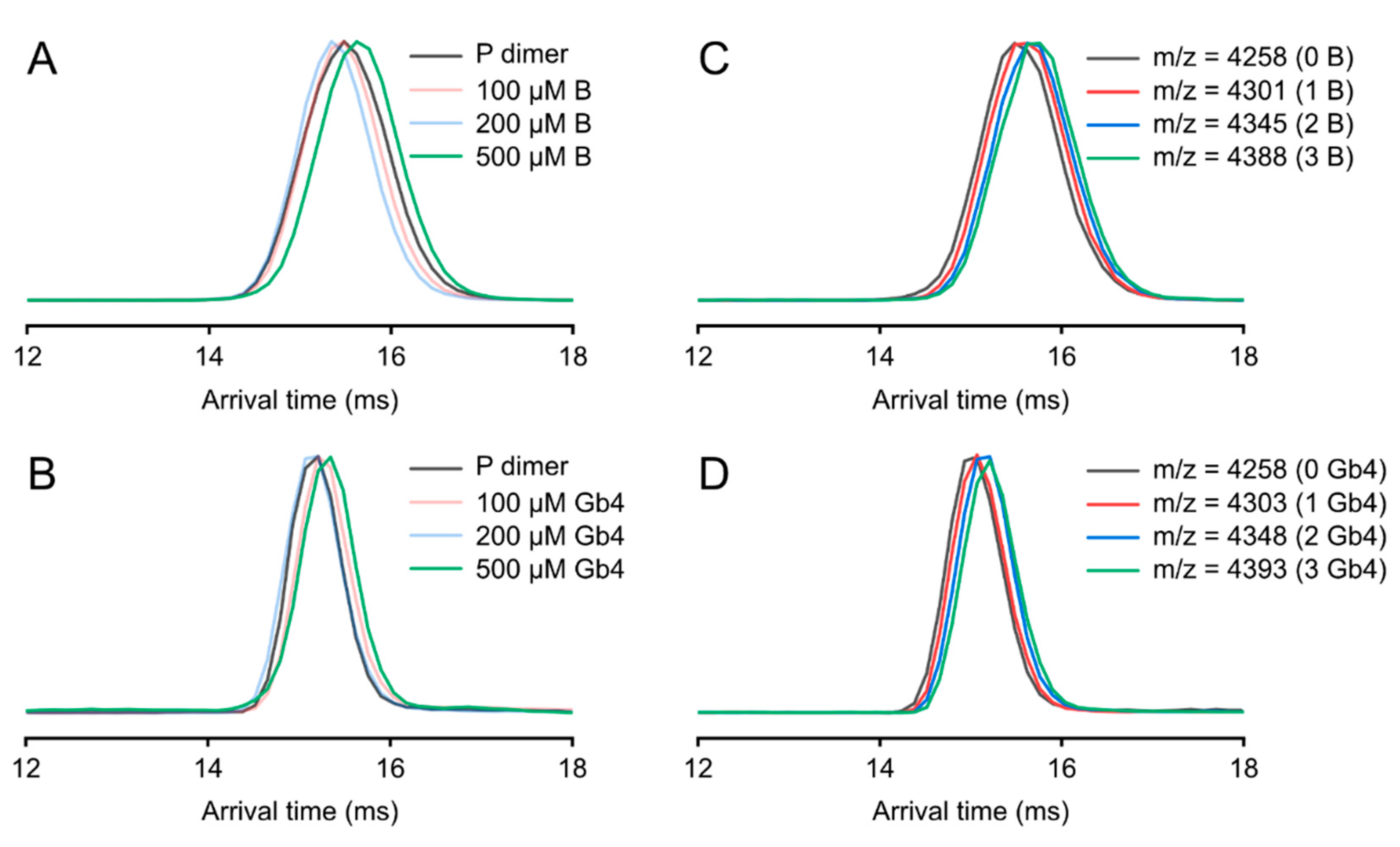
2.6. Ion Mobility Mass Spectrometry (IMMS)
3. Results
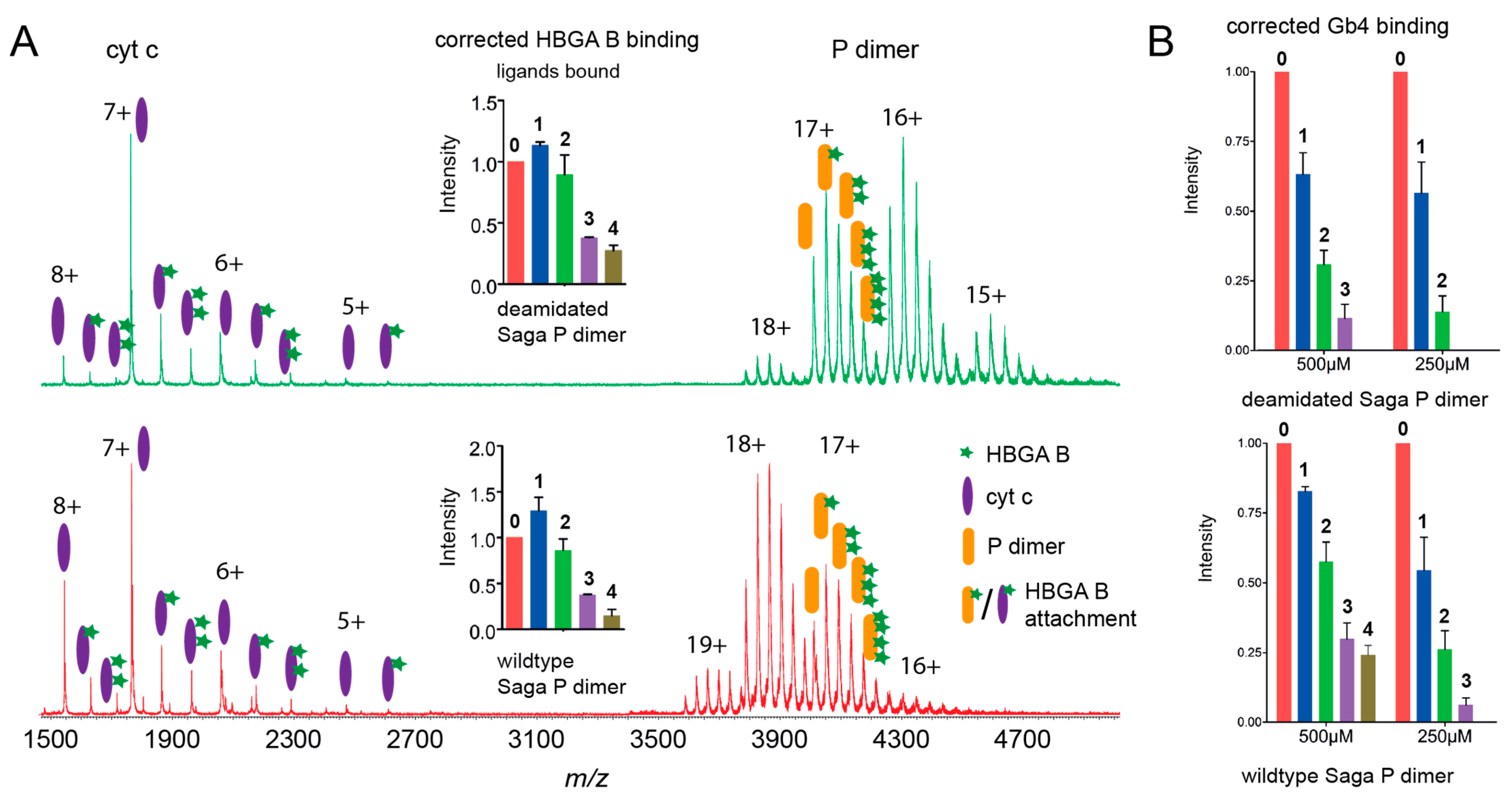
3.1. Glycan Clustering on Norovirus P Dimers
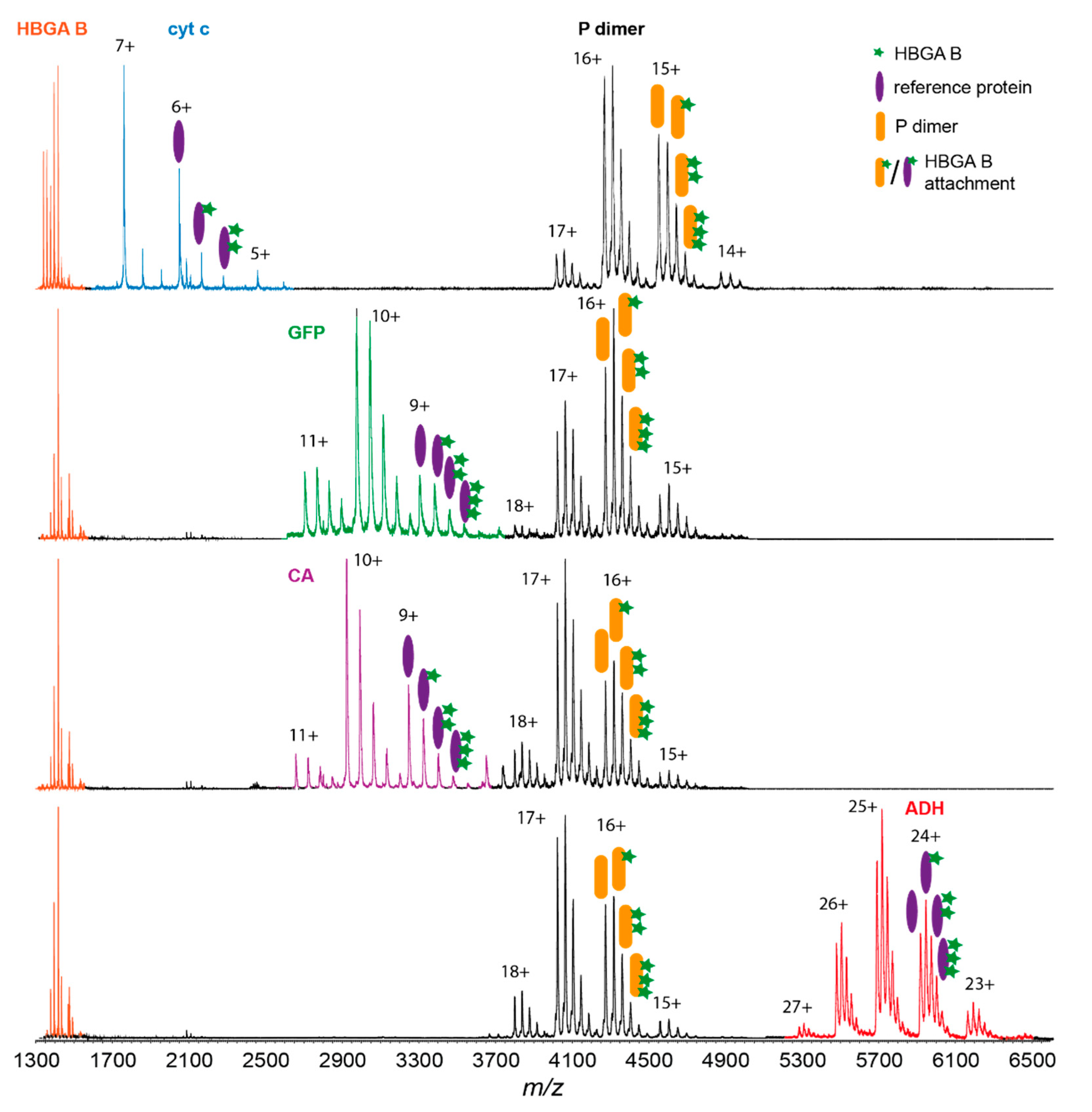
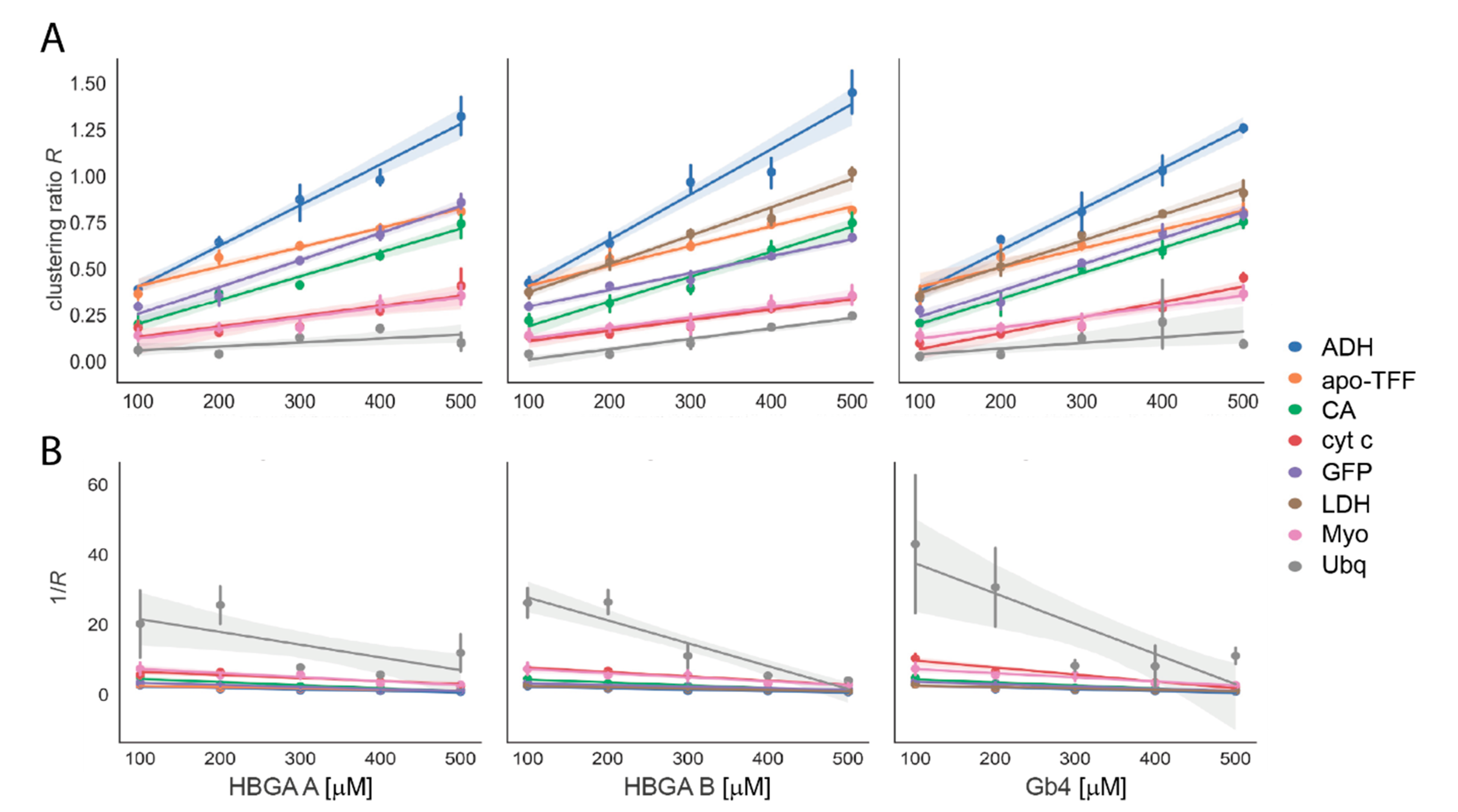
3.2. Glycan Clustering to Various Reference Proteins
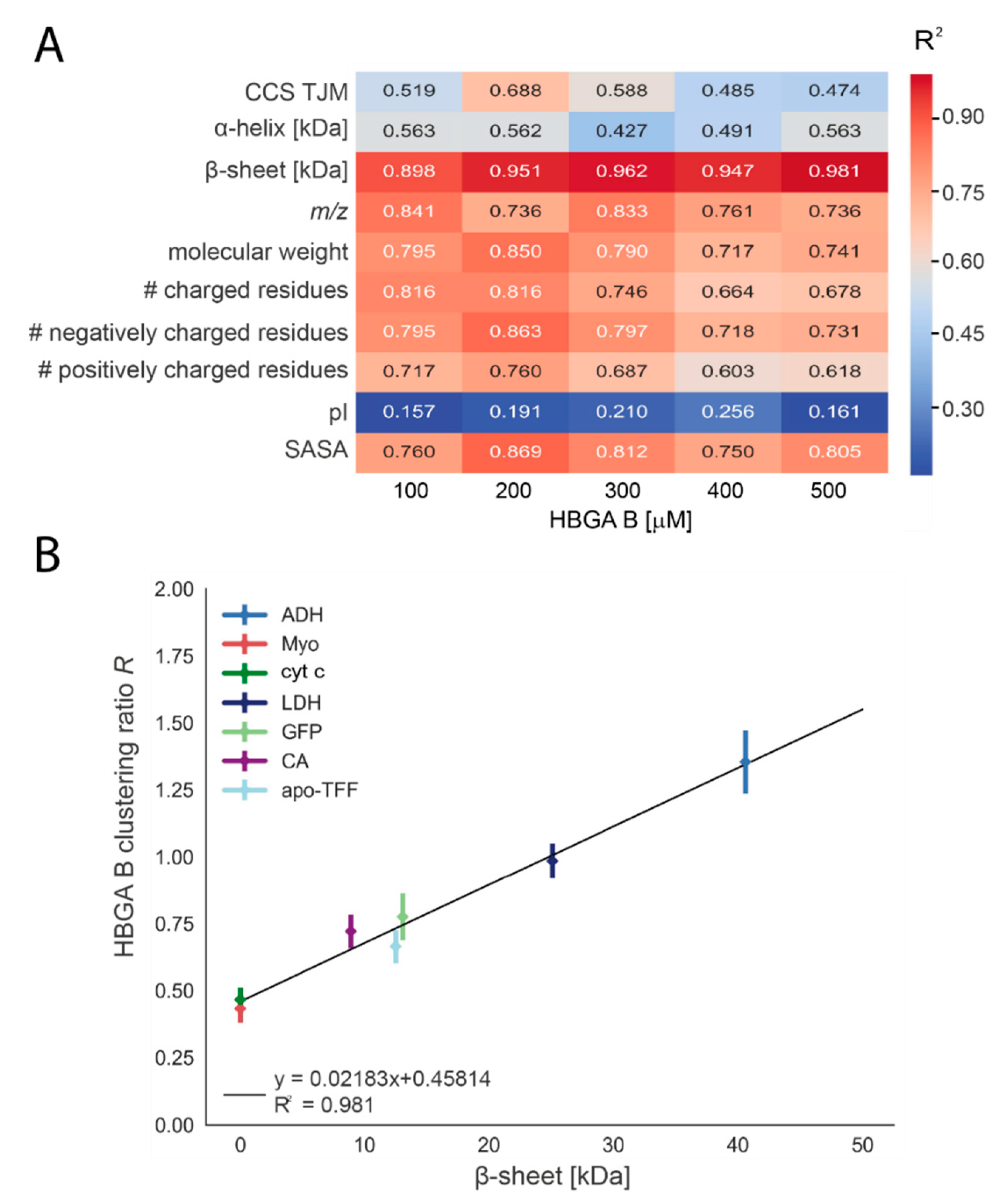
| Ref. Protein | Mass /kDa | PDB ID | Secondary Structure Composition | Glycan-Clustering Ratio R at 500 µM | KD for HBGA B /mM | |||
|---|---|---|---|---|---|---|---|---|
| % β-sheet | % α-helix | HBGA B | Gb4 | HBGA A | ||||
| Ubq | 8.9 | 1UBQ | 24.7 | 20.8 | 0.25 ± 0.00 | 0.09 ± 0.03 | 0.10 ± 0.05 | 0.20 ± 0.13 |
| Cyt c | 13.2 | 2N3B | 0 | 53.8 | 0.35 ± 0.03 | 0.45 ± 0.02 | 0.41 ± 0.09 | 0.16 ± 0.05 |
| Myo | 17.0 | 1AZI | 0 | 79.2 | 0.36 ± 0.05 | 0.37 ± 0.06 | 0.34 ± 0.03 | 0.25 ± 0.10 |
| GFP | 26.8 | 1BFP | 55.0 | 18.5 | 0.67 ± 0.02 | 0.80 ± 0.03 | 0.86 ± 0.04 | 0.90 ± 0.35 |
| CA | 29.1 | 1V9E | 35.8 | 15.8 | 0.75 ± 0.07 | 0.75 ± 0.03 | 0.74 ± 0.07 | 0.40 ± 0.10 |
| apo-TFF | 79.6 | 2HAU | 15.7 | 34.5 | 0.82 ± 0.03 | 0.80 ± 0.05 | 0.79 ± 0.03 | n.a. |
| LDH | 146.8 | 3H3F | 17.1 | 46.9 | 1.02 ± 0.04 | 0.91±0.06 | n.a. | n.a. |
| ADH | 147.0 | 4W6Z | 34.8 | 28.4 | 1.45 ± 0.11 | 1.26 ± 0.03 | 1.32 ± 0.10 | 24.00 ± 16.00 |
| SAGA P dimers | 68.1 | 4X7C | 32.5 | 6.6 | 1.55 ± 0.05 | 1.40 ± 0.21 | 1.44 ± 0.15 | n.a. |
3.3. Biophysical Properties Influence the Glycan Clustering
3.4. Ion Mobility on P Dimer Glycan Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cao, S.; Lou, Z.; Tan, M.; Chen, Y.; Liu, Y.; Zhang, Z.; Zhang, X.C.; Jiang, X.; Li, X.; Rao, Z. Structural Basis for the Recognition of Blood Group Trisaccharides by Norovirus. J. Virol. 2007, 81, 5949–5957. [Google Scholar] [CrossRef]
- Huang, P.; Farkas, T.; Marionneau, S.; Zhong, W.; Ruvoën-Clouet, N.; Morrow, A.L.; Altaye, M.; Pickering, L.K.; Newburg, D.S.; LePendu, J. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: Identification of 4 distinct strain-specific patterns. J. Infect. Dis. 2003, 188, 19–31. [Google Scholar] [CrossRef]
- Fiege, B.; Rademacher, C.; Cartmell, J.; Kitov, P.I.; Parra, F.; Peters, T. Molecular details of the recognition of blood group antigens by a human norovirus as determined by STD NMR spectroscopy. Angew. Chem. Int. Ed. 2012, 124, 952–956. [Google Scholar] [CrossRef]
- Mallagaray, A.; Creutznacher, R.; Dülfer, J.; Mayer, P.H.; Grimm, L.L.; Orduña, J.M.; Trabjerg, E.; Stehle, T.; Rand, K.D.; Blaum, B.S. A post-translational modification of human Norovirus capsid protein attenuates glycan binding. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Sun, J.; Kitova, E.N.; Wang, W.; Klassen, J.S. Method for distinguishing specific from nonspecific protein− ligand complexes in nanoelectrospray ionization mass spectrometry. Anal. Chem. 2006, 78, 3010–3018. [Google Scholar] [CrossRef]
- Pacholarz, K.J.; Garlish, R.A.; Taylor, R.J.; Barran, P.E. Mass spectrometry based tools to investigate protein-ligand interactions for drug discovery. Chem. Soc. Rev. 2012, 41, 4335–4355. [Google Scholar] [CrossRef]
- Jecklin, M.C.; Touboul, D.; Bovet, C.; Wortmann, A.; Zenobi, R. Which electrospray-based ionization method best reflects protein-ligand interactions found in solution? A comparison of ESI, nanoESI, and ESSI for the determination of dissociation constants with mass spectrometry. J. Am. Soc. Mass Spectrom. 2008, 19, 332–343. [Google Scholar] [CrossRef]
- Gulbakan, B.; Barylyuk, K.; Schneider, P.; Pillong, M.; Schneider, G.; Zenobi, R. Native Electrospray Ionization Mass Spectrometry Reveals Multiple Facets of Aptamer-Ligand Interactions: From Mechanism to Binding Constants. J. Am. Chem Soc. 2018, 140, 7486–7497. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Kitov, P.I.; Kitova, E.N.; Tan, M.; Wang, L.; Xia, M.; Jiang, X.; Klassen, J.S. Affinities of recombinant norovirus P dimers for human blood group antigens. Glycobiology 2013, 23, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Tan, M.; Xia, M.; Kitova, E.N.; Jiang, X.; Klassen, J.S. Gangliosides are ligands for human noroviruses. J. Am. Chem Soc. 2014, 136, 12631–12637. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zheng, R.; Richards, M.R.; Tan, M.; Kitova, E.N.; Jiang, X.; Klassen, J.S. Quantifying the binding stoichiometry and affinity of histo-blood group antigen oligosaccharides for human noroviruses. Glycobiology 2018, 28, 488–498. [Google Scholar] [CrossRef]
- Mallagaray, A.; Lockhauserbäumer, J.; Hansman, G.; Uetrecht, C.; Peters, T. Attachment of norovirus to histo blood group antigens: A cooperative multistep process. Angew. Chem. Int. Ed. 2015, 54, 12014–12019. [Google Scholar] [CrossRef] [PubMed]
- Bücher, K.S.; Yan, H.; Creutznacher, R.; Ruoff, K.; Mallagaray, A.; Grafmuller, A.; Dirks, J.S.; Kilic, T.; Weickert, S.; Rubailo, A.; et al. Fucose-Functionalized Precision Glycomacromolecules Targeting Human Norovirus Capsid Protein. Biomacromolecules 2018, 19, 3714–3724. [Google Scholar] [CrossRef]
- Wegener, H.; Mallagaray, A.; Schone, T.; Peters, T.; Lockhauserbaumer, J.; Yan, H.; Uetrecht, C.; Hansman, G.S.; Taube, S. Human norovirus GII.4(MI001) P dimer binds fucosylated and sialylated carbohydrates. Glycobiology 2017, 27, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Kitova, E.N.; Tan, M.; Jiang, X.; Pluvinage, B.; Boraston, A.B.; Klassen, J.S. Affinities of human histo-blood group antigens for norovirus capsid protein complexes. Glycobiology 2015, 25, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Jecklin, M.C.; Schauer, S.; Dumelin, C.E.; Zenobi, R. Label-free determination of protein-ligand binding constants using mass spectrometry and validation using surface plasmon resonance and isothermal titration calorimetry. J. Mol. Recognit. 2010, 22, 319–329. [Google Scholar] [CrossRef]
- Deng, L.; Kitova, E.N.; Klassen, J.S. Dissociation Kinetics of the Streptavidin–Biotin Interaction Measured Using Direct Electrospray Ionization Mass Spectrometry Analysis. J. Am. Soc. Mass Spectrom. 2013, 24, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Creutznacher, R.; Maass, T.; Ogrissek, P.; Wallmann, G.; Feldmann, C.; Peters, H.; Lingemann, M.; Taube, S.; Peters, T.; Mallagaray, A. NMR Experiments Shed New Light on Glycan Recognition by Human and Murine Norovirus Capsid Proteins. Viruses 2021, 13, 416. [Google Scholar] [CrossRef]
- Kesgin-Schaefer, S.; Heidemann, J.; Puchert, A.; Koelbel, K.; Yorke, B.A.; Huse, N.; Pearson, A.R.; Uetrecht, C.; Tidow, H. Crystal structure of a domain-swapped photoactivatable sfGFP variant provides evidence for GFP folding pathway. FEBS J. 2019, 286, 2329–2340. [Google Scholar] [CrossRef]
- Van den Heuvel, R.H.; Heck, A.J. Native protein mass spectrometry: From intact oligomers to functional machineries. Curr. Opinion Chem. Biol. 2004, 8, 519–526. [Google Scholar] [CrossRef]
- Creutznacher, R.; Schulze, E.; Wallmann, G.; Peters, T.; Stein, M.; Mallagaray, A. Chemical-Shift Perturbations Reflect Bile Acid Binding to Norovirus Coat Protein: Recognition Comes in Different Flavors. Chembiochem 2020, 21, 1007–1021. [Google Scholar] [CrossRef]
- Creutznacher, R.; Maaß, T.; Dülfer, J.; Feldmann, C.; Hartmann, V.; Knickmann, J.; Westermann, L.T.; Smith, T.J.; Uetrecht, C.; Mallagaray, A.; et al. Murine norovirus capsid plasticity–Glycochenodeoxycholic acid stabilizes P-domain dimers and triggers escape from antibody recognition. bioRxiv 2021. [Google Scholar] [CrossRef]
- Garcia-Alai, M.M.; Heidemann, J.; Skruzny, M.; Gieras, A.; Mertens, H.D.T.; Svergun, D.I.; Kaksonen, M.; Uetrecht, C.; Meijers, R. Epsin and Sla2 form assemblies through phospholipid interfaces. Nat. Commun. 2018, 9, 328. [Google Scholar] [CrossRef] [PubMed]
- Aliyari, E.; Konermann, L. Formation of Gaseous Proteins via the Ion Evaporation Model (IEM) in Electrospray Mass Spectrometry. Anal. Chem. 2020, 92, 10807–10814. [Google Scholar] [CrossRef]
- Seo, J.; Hoffmann, W.; Warnke, S.; Bowers, M.T.; Pagel, K.; von Helden, G. Retention of Native Protein Structures in the Absence of Solvent: A Coupled Ion Mobility and Spectroscopic Study. Angew. Chem. Int. Ed. Engl. 2016, 55, 14173–14176. [Google Scholar] [CrossRef] [PubMed]
- Testa, L.; Brocca, S.; Santambrogio, C.; D’Urzo, A.; Habchi, J.; Longhi, S.; Uversky, V.N.; Grandori, R. Extracting structural information from charge-state distributions of intrinsically disordered proteins by non-denaturing electrospray-ionization mass spectrometry. Intrinsically Disord. Proteins 2013, 1, e25068. [Google Scholar] [CrossRef] [PubMed]
- Dülfer, J.; Yan, H.; Brodmerkel, M.N.; Creutznacher, R.; Mallagaray, A.; Peters, T.; Caleman, C.; Marklund, E.G.; Uetrecht, C. Glycan-Induced Protein Dynamics in Human Norovirus P Dimers Depend on Virus Strain and Deamidation Status. Molecules 2021, 26, 2125. [Google Scholar] [CrossRef] [PubMed]
- Baez Bolivar, E.G.; Bui, D.T.; Kitova, E.N.; Han, L.; Zheng, R.B.; Luber, E.J.; Sayed, S.Y.; Mahal, L.K.; Klassen, J.S. Submicron Emitters Enable Reliable Quantification of Weak Protein-Glycan Interactions by ESI-MS. Anal. Chem. 2021, 93, 4231–4239. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Lockhauserbäumer, J.; Szekeres, G.P.; Mallagaray, A.; Creutznacher, R.; Taube, S.; Peters, T.; Pagel, K.; Uetrecht, C. Protein Secondary Structure Affects Glycan Clustering in Native Mass Spectrometry. Life 2021, 11, 554. https://doi.org/10.3390/life11060554
Yan H, Lockhauserbäumer J, Szekeres GP, Mallagaray A, Creutznacher R, Taube S, Peters T, Pagel K, Uetrecht C. Protein Secondary Structure Affects Glycan Clustering in Native Mass Spectrometry. Life. 2021; 11(6):554. https://doi.org/10.3390/life11060554
Chicago/Turabian StyleYan, Hao, Julia Lockhauserbäumer, Gergo Peter Szekeres, Alvaro Mallagaray, Robert Creutznacher, Stefan Taube, Thomas Peters, Kevin Pagel, and Charlotte Uetrecht. 2021. "Protein Secondary Structure Affects Glycan Clustering in Native Mass Spectrometry" Life 11, no. 6: 554. https://doi.org/10.3390/life11060554
APA StyleYan, H., Lockhauserbäumer, J., Szekeres, G. P., Mallagaray, A., Creutznacher, R., Taube, S., Peters, T., Pagel, K., & Uetrecht, C. (2021). Protein Secondary Structure Affects Glycan Clustering in Native Mass Spectrometry. Life, 11(6), 554. https://doi.org/10.3390/life11060554






