An Electrochemical Approach to Follow and Evaluate the Kinetic Catalysis of Ricin on hsDNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Ricin Purification
2.2. Electrochemical Assays
2.3. Measurement of Released Adenine by HPLC
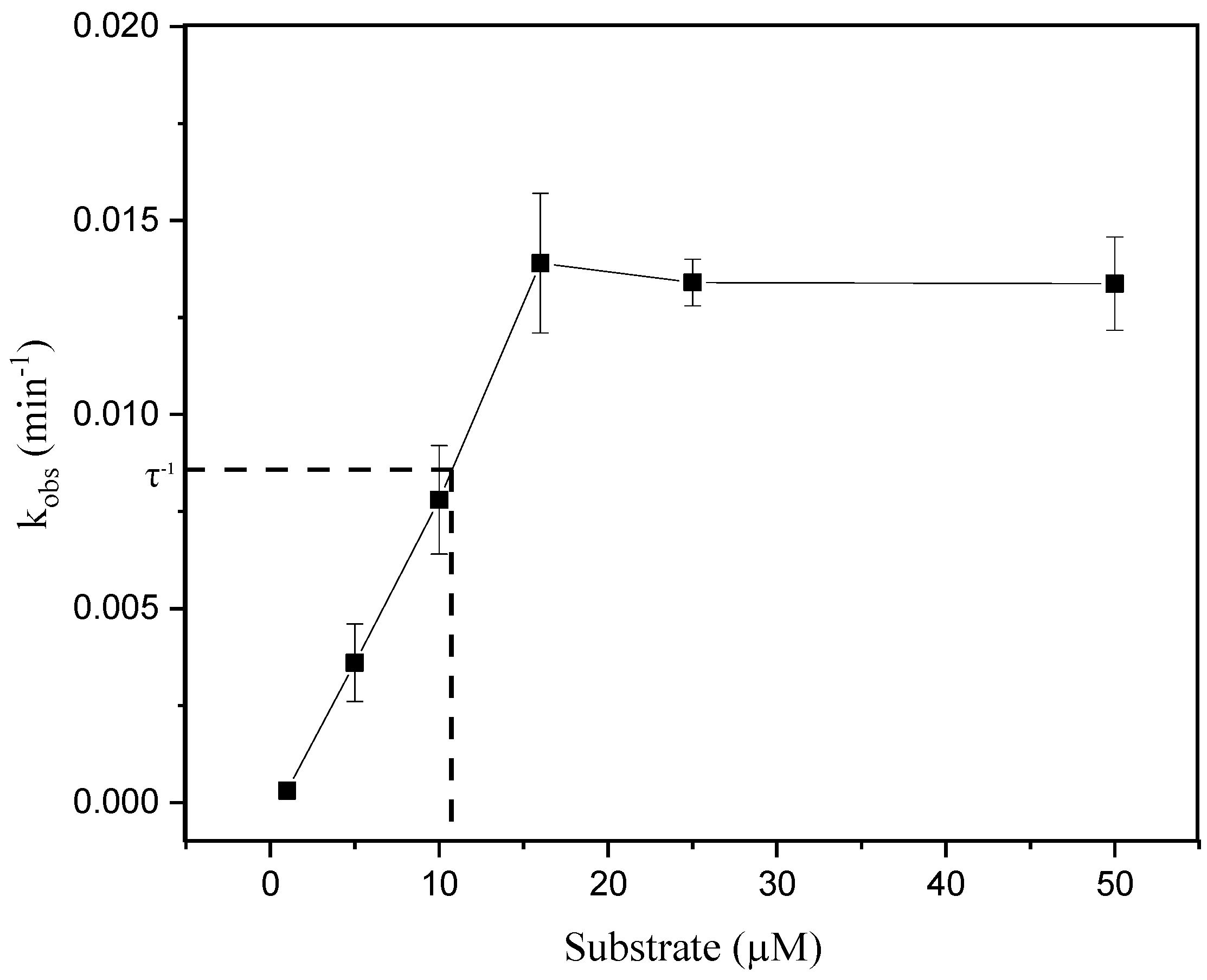
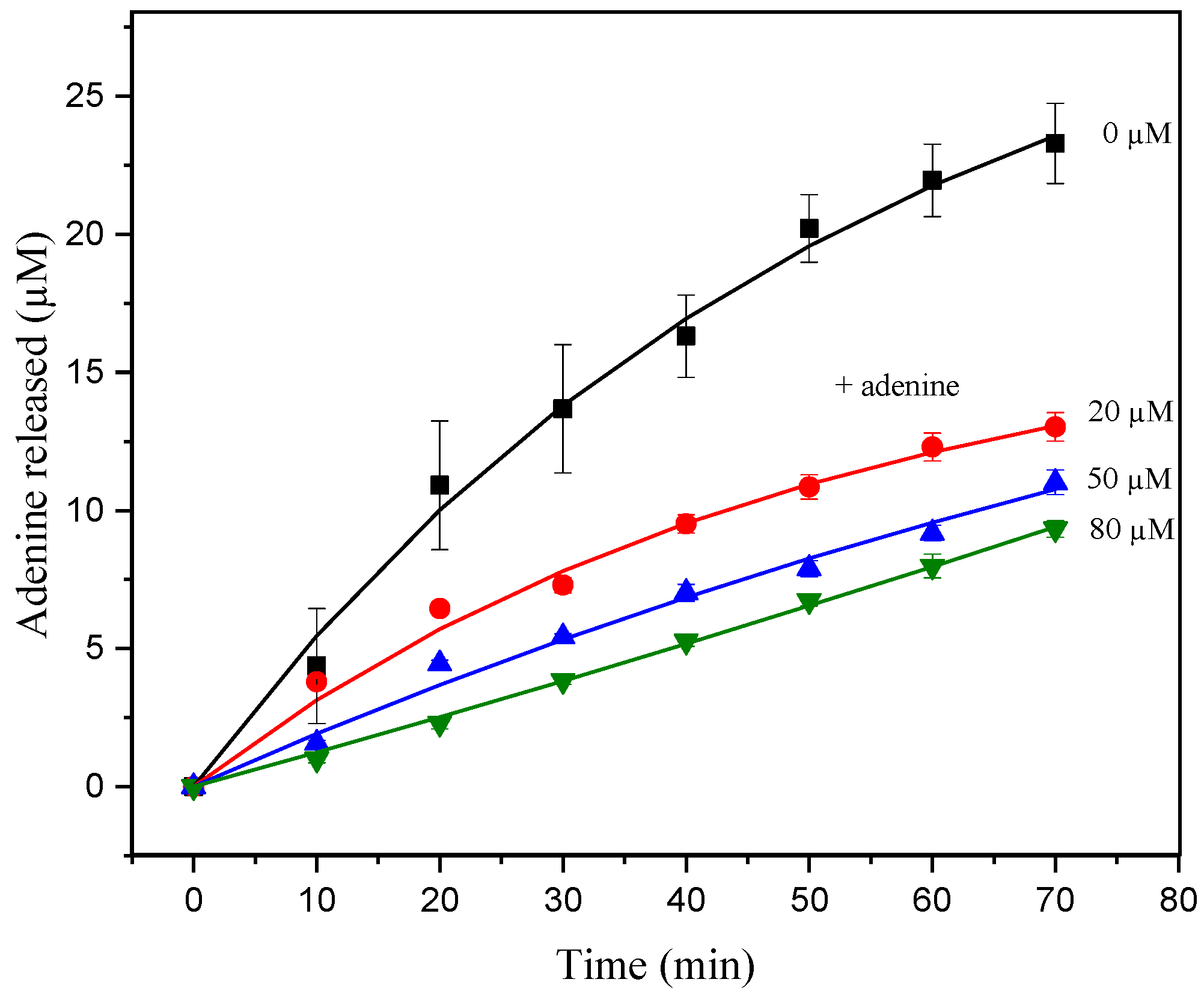
3. Results and Discussion
3.1. Ricin
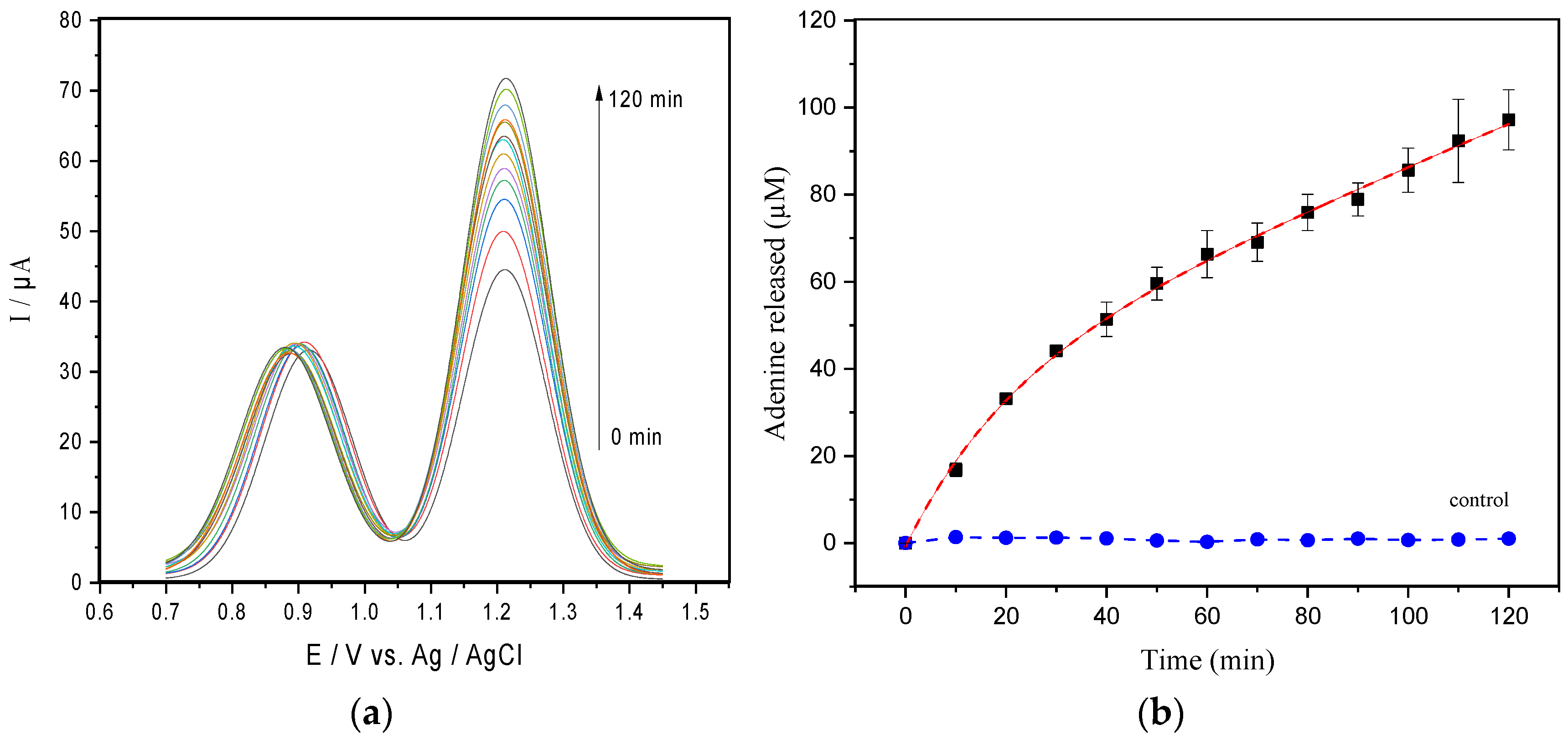
3.2. Enzymatic Catalysis of Ricin
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moshiri, M.; Hamid, F.; Etemad, L. Ricin Toxicity: Clinical and Molecular Aspects. Rep. Biochem. Mol. Biol. 2016, 4, 60–65. [Google Scholar] [PubMed]
- Vance, D.J.; Mantis, N.J. Progress and Challenges Associated with the Development of Ricin Toxin Subunit Vaccines. Expert Rev. Vaccines 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Lopez Nunez, O.F.; Pizon, A.F.; Tamama, K. Ricin Poisoning after Oral Ingestion of Castor Beans: A Case Report and Review of the Literature and Laboratory Testing. J. Emerg. Med. 2017, 53, e67–e71. [Google Scholar] [CrossRef]
- Polito, L.; Bortolotti, M.; Battelli, M.G.; Calafato, G.; Bolognesi, A. Ricin: An ancient story for a timeless plant toxin. Toxins 2019, 11, 324. [Google Scholar] [CrossRef]
- Bozza, W.P.; Tolleson, W.H.; Rivera Rosado, L.A.; Zhang, B. Ricin detection: Tracking active toxin. Biotechnol. Adv. 2015, 33, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Singh, J. Classification, Causes, Control Measures and Acts of Bioterrorism. Int. J. Appl. Biol. Pharm. Technol. 2016, 7, 342–355. [Google Scholar]
- Argent, R.H.; Roberts, L.M.; Wales, R.; Robertus, J.D.; Lord, J.M. Introduction of a disulfide bond into ricin A chain decreases the cytotoxicity of the ricin holotoxin. J. Biol. Chem. 1994, 269, 26705–26710. [Google Scholar] [CrossRef]
- Doan, L.G. Ricin: Mechanism of toxicity, clinical manifestations, and vaccine development. A review. J. Toxicol. Clin. Toxicol. 2004, 42, 201–208. [Google Scholar] [CrossRef]
- Rudolph, M.J.; Vance, D.J.; Cassidy, M.S.; Rong, Y.; Mantis, N.J. Structural analysis of single domain antibodies bound to a second neutralizing hot spot on ricin toxin’s enzymatic subunit. J. Biol. Chem. 2017, 292, 872–883. [Google Scholar] [CrossRef]
- Saito, R.; Pruet, J.M.; Manzano, L.A.; Jasheway, K.; Monzingo, P.A.W.; Kamat, I.; Anslyn, E.V.; Robertus, J.D. Peptide-conjugated pterins as inhibitors of Ricin Toxin A. J. Med. Chem. 2008, 23, 1–7. [Google Scholar] [CrossRef]
- Tan, Q.Q.; Dong, D.X.; Yin, X.W.; Sun, J.; Ren, H.J.; Li, R.X. Comparative analysis of depurination catalyzed by ricin A-chain on synthetic 32mer and 25mer oligoribonucleotides mimicking the sarcin/ricin domain of the rat 28S rRNA and E. coli 23S rRNA. J. Biotechnol. 2009, 139, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lei, S.; Tang, Y.; Zou, L.; Hu, H.; Wang, S. Drug effect of thulium(III)-Arsenazo III complex on herring sperm DNA. J. Chem. 2018, 2018, 1–9. [Google Scholar] [CrossRef]
- Wang, P.; Wu, H.; Dai, Z.; Zou, X. Simultaneous detection of guanine, adenine, thymine and cytosine at choline monolayer supported multiwalled carbon nanotubes film. Biosens. Bioelectron. 2011, 26, 3339–3345. [Google Scholar] [CrossRef]
- Manikandan, R.; Deepa, P.N.; Narayanan, S.S. Simultaneous electrochemical determination of adenine and guanine using poly 2-naphthol orange film–modified electrode. Ionics 2019, 26, 1475–1482. [Google Scholar] [CrossRef]
- Bevilacqua, V.L.H.; Nilles, J.M.; Rice, J.S.; Connell, T.R.; Schenning, A.M.; Reilly, L.M.; Durst, H.D. Ricin Activity Assay by Direct Analysis in Real Time Mass Spectrometry Detection of Adenine Release. Anal. Chem. 2010, 82, 798–800. [Google Scholar] [CrossRef]
- Heisler, I.; Keller, J.; Tauber, R.; Sutherland, M.; Fuchs, H. A Colorimetric Assay for the Quantitation of Free Adenine Applied to Determine the Enzymatic Activity of Ribosome-Inactivating Proteins. Anal. Biochem. 2002, 302, 114–122. [Google Scholar] [CrossRef]
- Sturm, M.B.; Schramm, V.L. Detecting Ricin: Sensitive Luminescent Assay for Ricin A-Chain Ribosome Depurination Kinetics. Anal. Chem. 2009, 81, 2847–2853. [Google Scholar] [CrossRef]
- Chen, X.Y.; Link, T.M.; Schramm, V.L. Ricin A-chain: Kinetics, mechanism, and RNA stem-loop inhibitors. Biochemistry 1998, 37, 11605–11613. [Google Scholar] [CrossRef] [PubMed]
- Jankowska-Śliwińska, J.; Dawgul, M.; Kruk, J.; Pijanowska, D.G. Comparison of electrochemical determination of purines and pyrimidines by means of carbon, graphite and gold paste electrodes. Int. J. Electrochem. Sci. 2017, 12, 2329–2343. [Google Scholar] [CrossRef]
- Ngo, N.T.; Nguyen, T.T.; Bui, T.T.D.; Hoang, T.M.N. Effects of ricin extracted from seeds of the castor bean (ricinus communis) on cytotoxicity and tumorigenesis of melanoma cells. Biomed. Res. Ther. 2016, 3, 633–644. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wei, H.; Sun, R.; Tian, Z.; Zheng, X. Rapid method for protein quantitation by Bradford assay after elimination of the interference of polysorbate 80. Anal. Biochem. 2016, 494, 37–39. [Google Scholar] [CrossRef]
- Leonardi, W.; Zilbermintz, L.; Cheng, L.W.; Zozaya, J.; Tran, S.H.; Elliott, J.H.; Polukhina, K.; Manasherob, R.; Li, A.; Chi, X.; et al. Bithionol blocks pathogenicity of bacterial toxins, ricin, and Zika virus. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Dyer, P.D.R.; Kotha, A.K.; Gollings, A.S.; Shorter, S.A.; Shepherd, T.R.; Pettit, M.W.; Alexander, B.D.; Getti, G.T.M.; El-Daher, S.; Baillie, L.; et al. An in vitro evaluation of epigallocatechin gallate (eGCG) as a biocompatible inhibitor of ricin toxin. Biochim. Biophys. Acta-Gen. Subj. 2016, 1860, 1541–1550. [Google Scholar] [CrossRef]
- Hines, H.B.; Brueggemann, E.E.; Hale, M.L. High-performance liquid chromatography-mass selective detection assay for adenine released from a synthetic RNA substrate by ricin A chain. Anal. Biochem. 2004, 330, 119–122. [Google Scholar] [CrossRef]
- Woodward, J. Enzyme Technology. Biochem. Educ. 1990, 18, 106. [Google Scholar] [CrossRef]
- Sehgal, P.; Khan, M.; Kumar, O.; Vijayaraghavan, R. Purification, characterization and toxicity profile of ricin isoforms from castor beans. Food Chem. Toxicol. 2010, 48, 3171–3176. [Google Scholar] [CrossRef]
- Copeland, R.A. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis; John Wiley & Sons, Inc.: New York, NY, USA, 2000; Volume 53, ISBN 0471359297. [Google Scholar]
- Approach, A.M. Enzyme kinetics: A modern approach. Choice Rev. Online 2003, 40, 40. [Google Scholar] [CrossRef]
- Selwyn, M.J. A simple test for inactivation of an enzyme during assay. Biochim. Biophys. Acta-Enzymol. Biol. Oxid. 1965, 105, 193–195. [Google Scholar] [CrossRef]
- Cao, W.; De La Cruz, E.M. Quantitative full time course analysis of nonlinear enzyme cycling kinetics. Sci. Rep. 2013, 3, 3–7. [Google Scholar] [CrossRef]
- Aitbakieva, V.R.; Ahmad, R.; Singh, S.; Domashevskiy, A.V. Inhibition of ricin A-chain (RTA) catalytic activity by a viral genome-linked protein (VPg). Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Punekar, N.S. ENZYMES: Catalysis, Kinetics and Mechanisms; Springer: Singapore, 2018; ISBN 978-981-13-0784-3. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, G.; Schneedorf, J.M. An Electrochemical Approach to Follow and Evaluate the Kinetic Catalysis of Ricin on hsDNA. Life 2021, 11, 405. https://doi.org/10.3390/life11050405
Oliveira G, Schneedorf JM. An Electrochemical Approach to Follow and Evaluate the Kinetic Catalysis of Ricin on hsDNA. Life. 2021; 11(5):405. https://doi.org/10.3390/life11050405
Chicago/Turabian StyleOliveira, George, and José Maurício Schneedorf. 2021. "An Electrochemical Approach to Follow and Evaluate the Kinetic Catalysis of Ricin on hsDNA" Life 11, no. 5: 405. https://doi.org/10.3390/life11050405
APA StyleOliveira, G., & Schneedorf, J. M. (2021). An Electrochemical Approach to Follow and Evaluate the Kinetic Catalysis of Ricin on hsDNA. Life, 11(5), 405. https://doi.org/10.3390/life11050405






