Self-Reproduction and Darwinian Evolution in Autocatalytic Chemical Reaction Systems
Abstract
1. Introduction: Autocatalysis and the Emergence of Life
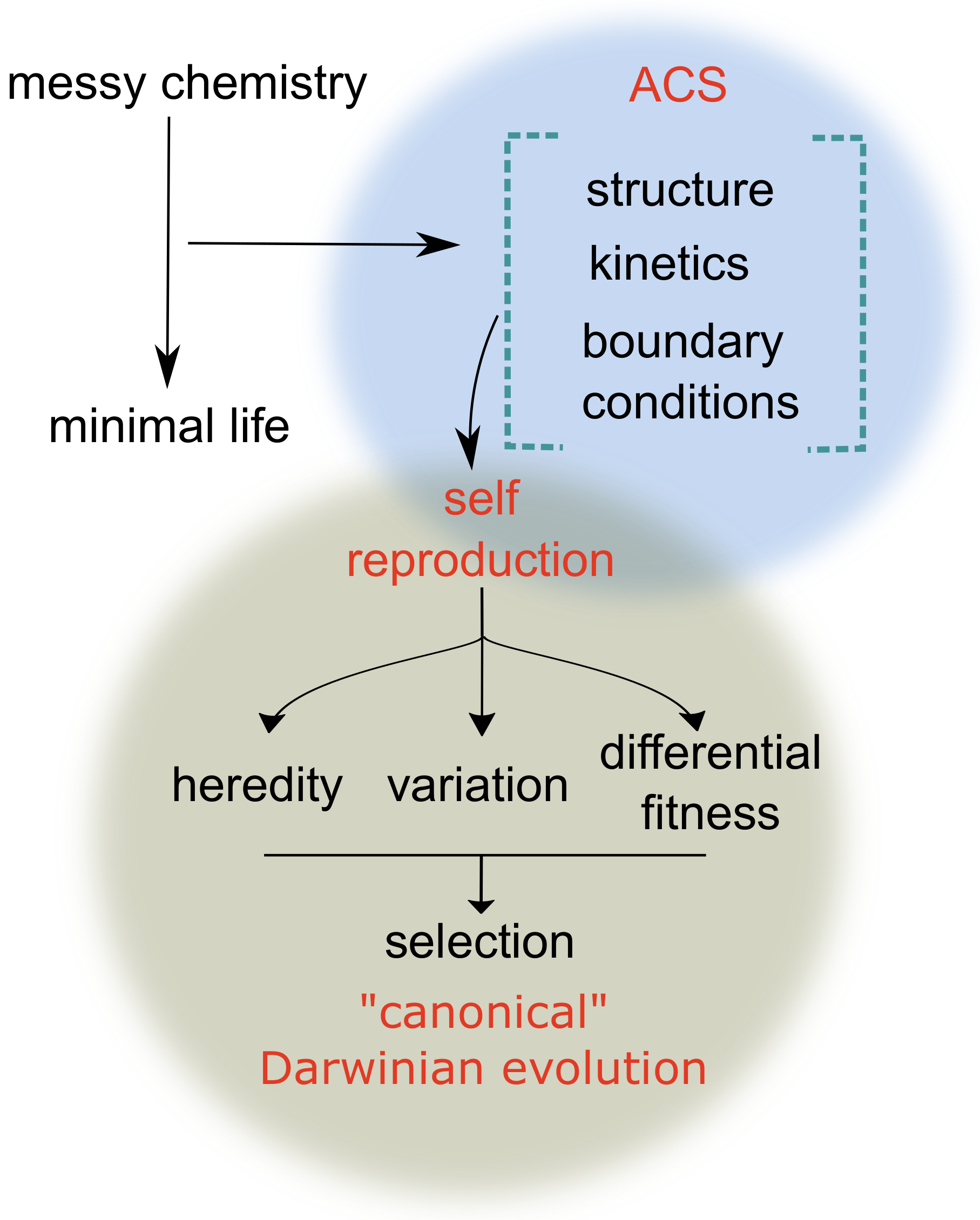
2. Theoretical Perspectives on the Emergence and Evolvability of Autocatalytic Systems
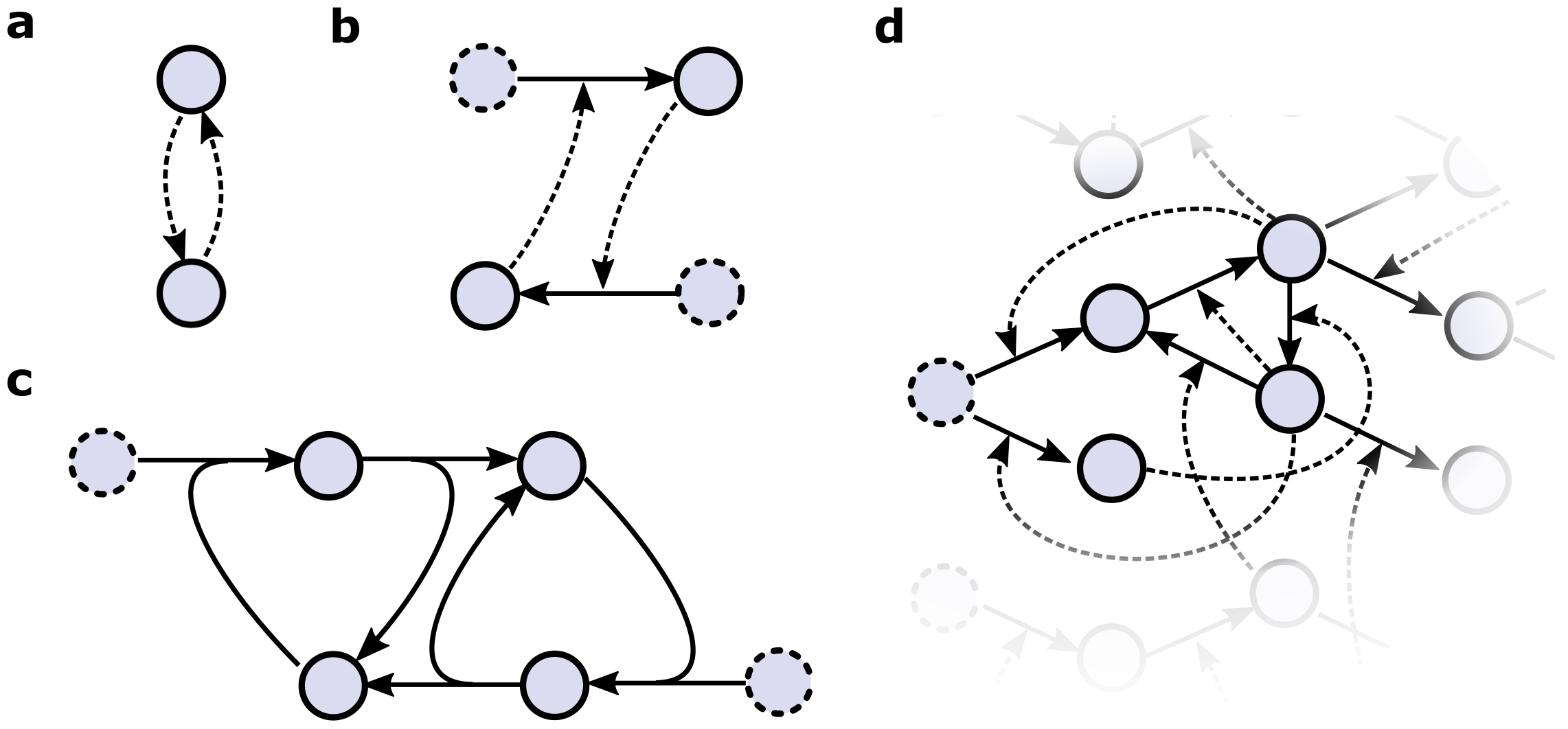
2.1. Defining Autocatalytic Chemical Reaction Networks
2.2. Self-Reproduction of Autocatalytic Systems
2.3. Spontaneous Emergence of Autocatalytic Systems
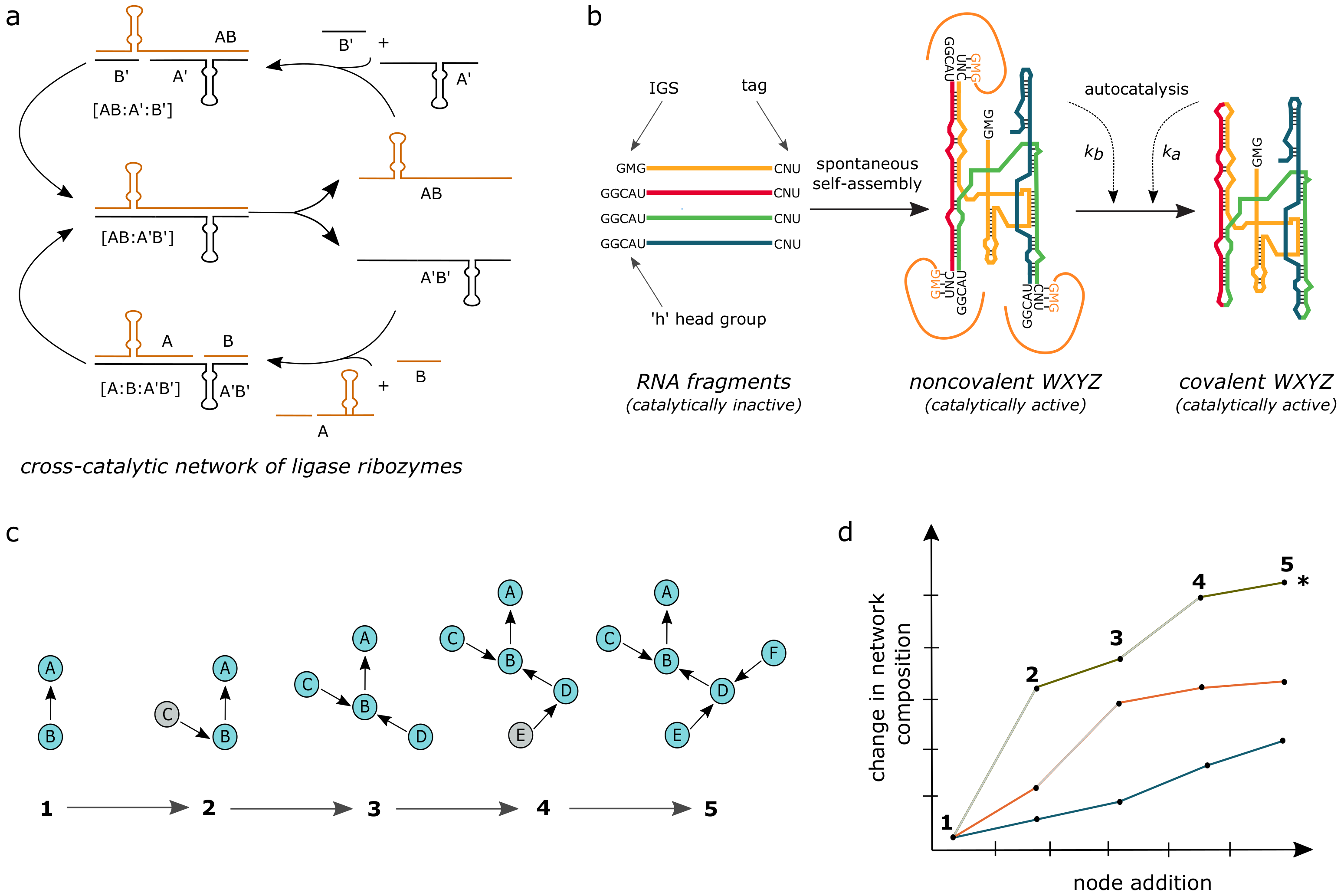
2.4. Darwinian Evolution of Autocatalytic Systems
3. Experimental Autocatalytic Systems
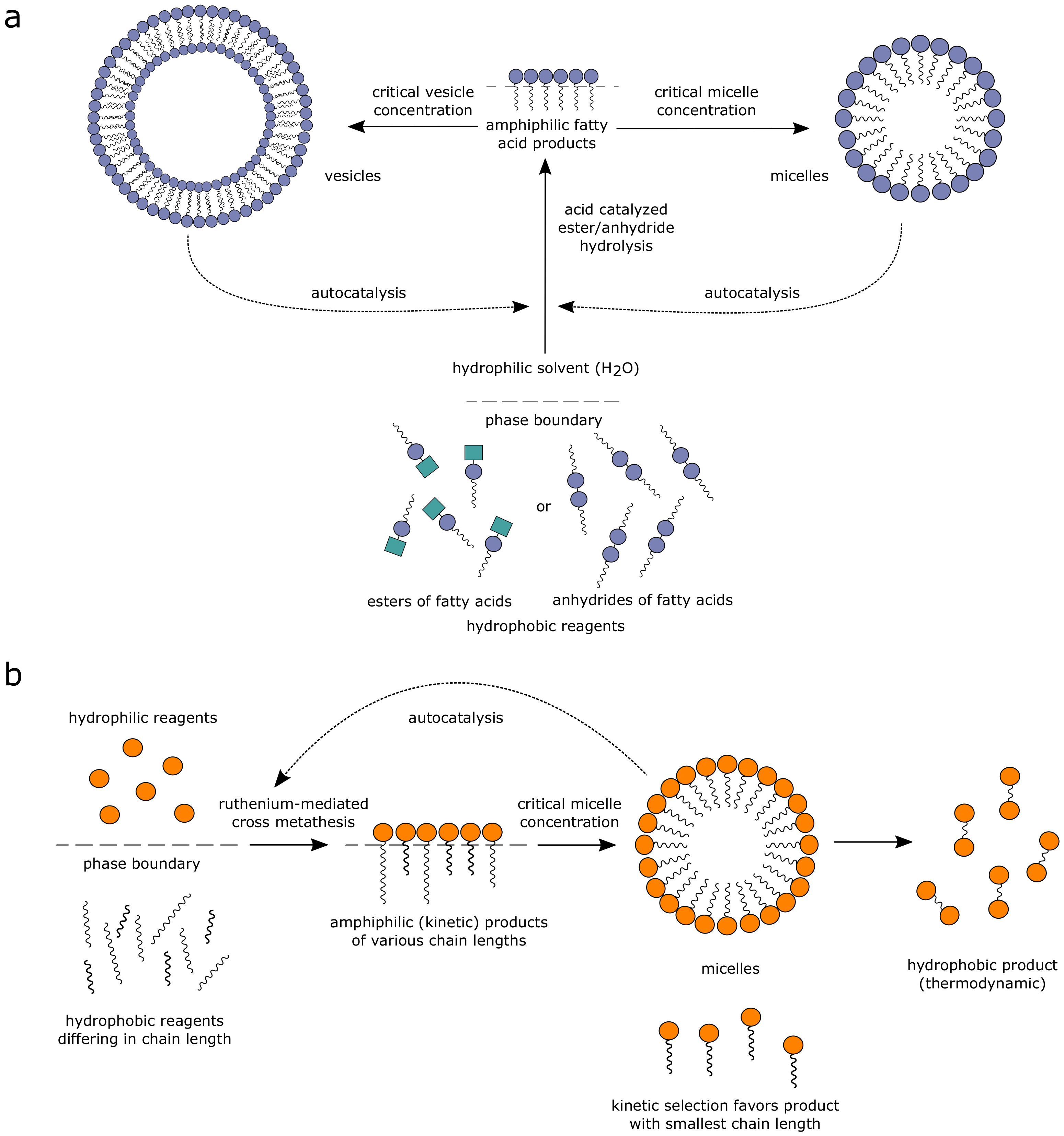
3.1. Lipid-Based Autocatalytic Systems
3.2. DNA-Based Chemistries
3.3. Inorganic Chemistries
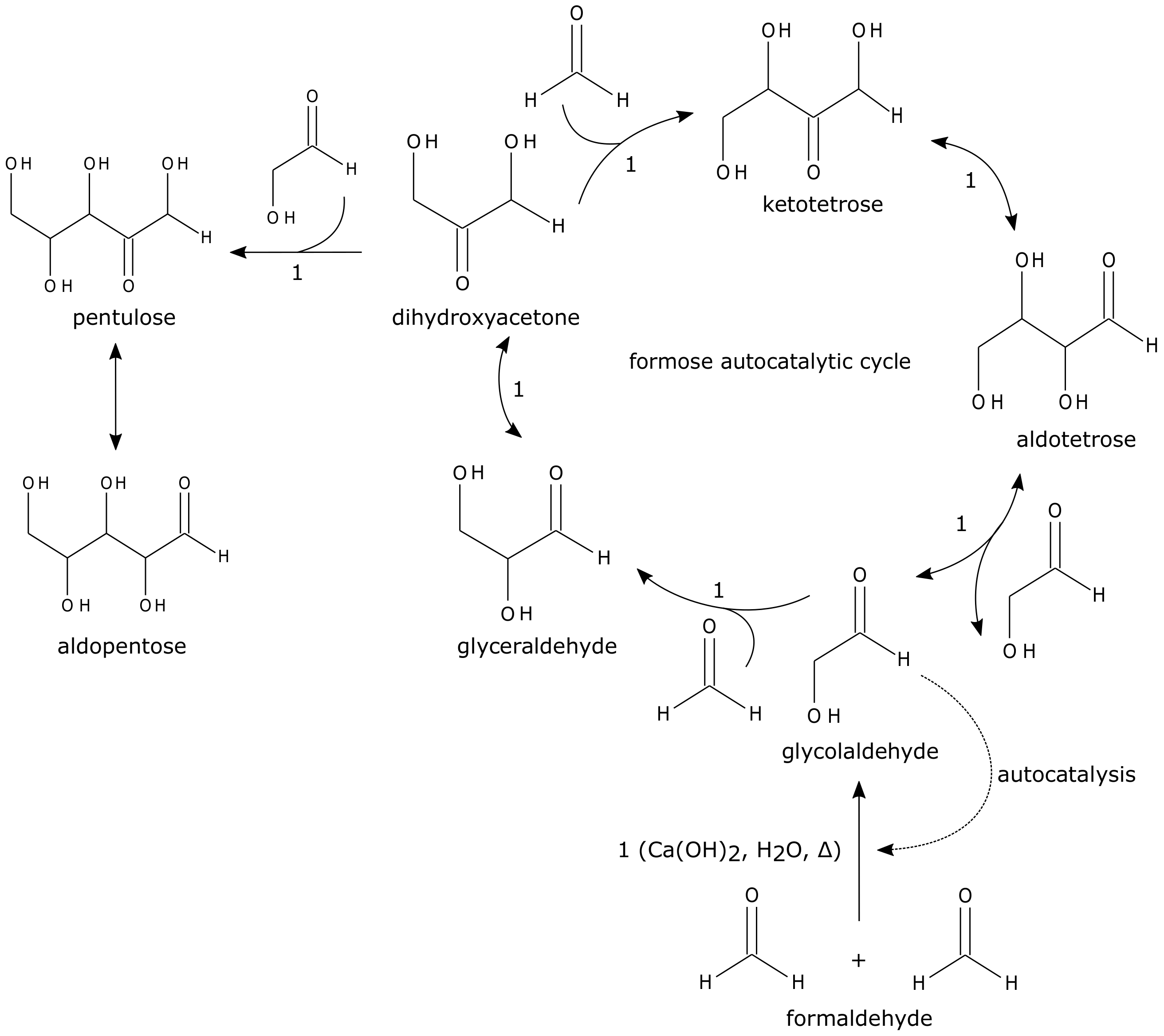
3.4. Sugars and Small Organic Molecule-Based Chemistries
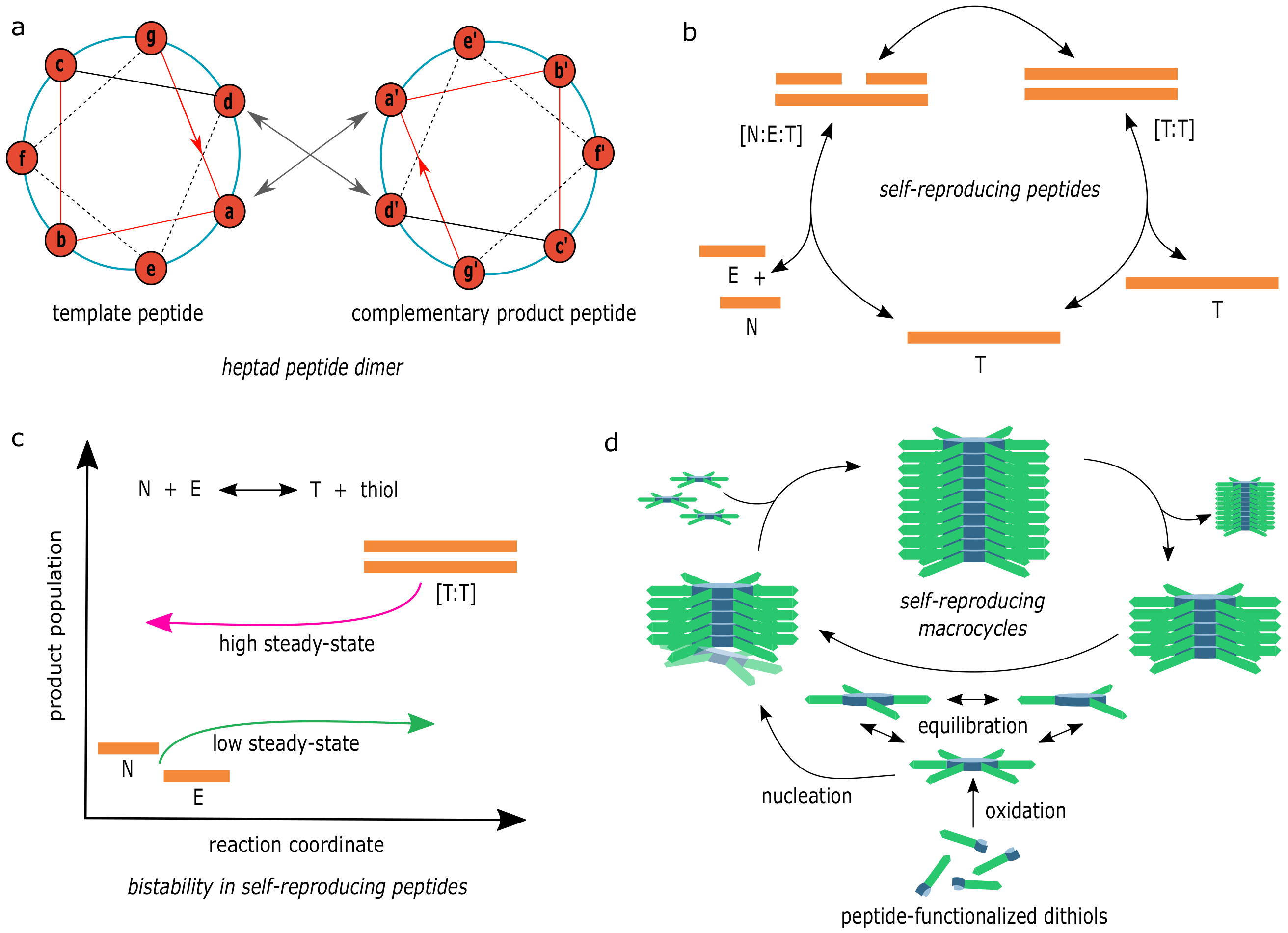
3.5. Peptide Chemistries
3.6. Macrocycle-Based Systems
3.7. RNA Chemistries
3.8. Summary of Experimental Systems
4. Future Perspectives
5. Glossary of Terms
- (Canonical) Darwinian evolution: The dynamics of a heritable variation being selected from among a population of reproducing units according to differential fitness. We define evolution more specifically in Section 2.4 in dynamical systems terminology, and we refer to it as Darwinian evolution in a “canonical” sense.
- Self-reproduction/replication: The self generation of a new copy of a unit/individual. In the present context, self-reproduction of a chemical system is realized by autocatalytic reactions. Although reproduction and replication are regarded as conceptually similar in general, in the present review, we only use “self-replication” for a generation of new copy via template-directed replication.
- Template-directed replication: A chemical reaction process where a sequence of a polymer is copied monomer-by-monomer, directed by another polymer as a template. The replication of DNA plays a major role in the present life to propagate information across generations.
- Metabolism-first scenarios: An origins-of-life hypothesis that surmises that metabolism proceeded template-directed replication, e.g., Oparin’s coacervate hypothesis (its counterpart are replication-first scenarios).
- (Chemical) composition: The abundance (or the number) of each species in a chemical system.
- Compositional heredity: The propagation of information via chemical composition, without template-directed replication (alternatively named as “composome”).
- Chemical reaction network: A network composed of chemical species (nodes) and reactions among them (edges); refer to the main text for a more precise meaning of “edges”.
- Dynamical system: A system describing the time dependence of variables (in the present context, chemical composition).
- Stochastic process: A process describing the time course of random variables (in the present context, chemical composition with random fluctuations). The dynamics of the chemical composition should be treated as a stochastic process when the system size (i.e., the number of molecules) is small.
- Catalyst: A molecule that accelerates the rate of a chemical reaction; the process is termed as catalysis. In particular, a catalytic protein is called an enzyme and a catalytic RNA is called a ribozyme.
- Autocatalytic reaction: A chemical reaction in which the catalytic molecules catalyze their own production; the process is called autocatalysis.
- Background reaction: In general, a chemical reaction that is not associated with autocatalytic reactions (or autocatalytic sets). Note that background reactions are context-dependent.
- Autocatalytic sets/cycle (ACS): A set (or cycle) of reactions such that the catalyst of the reactions is synthesized from the food molecules by a series of reactions in the set. There are several formal definitions for the ACS (e.g., RAF (Reflexively Autocatalytic and Food-generated) set; for details, refer to the main text Section 2.1).
- Food molecules (set): A molecule or a set of molecules that is/are externally supplied as substrates to an autocatalytic system.
- Network percolation: A transition from a network with disconnected clusters of nodes into a network with almost all nodes connected to each other due to the increase in the number of edges in a random network (graph).
- Continuous Stirred-Tank Reactor (CSTR): A boundary condition of a reaction system where food molecules are continuously supplied while all molecules continuously diffuse out (typically known as a chemostat in the biological context).
- Bi/multi-stability: A property of a dynamical system with multiple steady states. A large enough perturbation can cause transitions among such states.
- Dissipative structure: An emerging stable structure in a system with open boundary conditions, for which the characteristics have been studied in non-equilibrium physics since the pioneering works by Ilya Prigogine (also known as dynamic kinetic stability in systems chemistry).
- Quasi-species model: A model, introduced by Eigen, describing the population dynamics of replicative template (nucleotide) polymers.
- Hypercycle: A cooperative structure between template polymers proposed by Eigen, where the replication of template polymer is catalyzed by the another polymer.
- (Spontaneous) emergence of ACS: A transition process where an ACS first emerges from the food molecules (or “messy chemistry”), mediated by background reactions and stochastic fluctuations.
- Units of selection: Units that can be subjects of Darwinian evolution; according to the formulation by Lewontin, the necessary conditions for the population of such units are (i) variation, (ii) heredity, and (iii) differential fitness.
- (Autocatalytic) core network: A subset of the autocatalytic network, which is autocatalytic in itself, and independent from the rest of the network. Core networks could be units of selection and are hence significant for the evolvability of ACS.
- Amphiphile: A chemical compound with both hydrophilic (affinity for water) and hydrophobic (affinity for lipids or fats) properties.
- Micelle/Vesicle: Micelles are spontaneous spherical supramolecular arrangements of amphiphiles in aqueous solutions, formed due to amphiphilic interactions. Vesicles are similar supramolecular structures comprising of fluid enclosed by a bilayer of amphipathic molecules.
- Autopoiesis: An autocatalytic reaction that takes place within a closed boundary and contributes to the sustenance of the boundary. An illustrative example of an autopoietic system is the self-reproduction of lipid micelles/vesicles (see Section 3.1).
- Stopped-flow: A flow mixing setup used to investigate the kinetics of very fast reactions.
- Molybdenum clusters: Neutral or charged chemical compounds comprising of many metal (molybdenum) moieties, usually with significant metal–metal interactions.
- Dynamic Combinatorial Library (DCL): A library (or set) of simple chemical species (building blocks), from which a combinatorial variety of product molecules can be generated via reactions between the building blocks.
- Macrocycle: Chemical compounds with closed ring-like structures of 12 or more members.
- Oligomers/oligonucleotides: Short strands of polymers (for example, a 10-mer is an oligomer that is ten monomers long). In particular, oligomers of DNA or RNA molecules are called oligonucleotides.
- Coiled-coil: Protein structural elements composed of alpha helices () wrapped around together to form spiral structures.
- Ligation: Reactions involving covalent bond formation between two chemical species.
- Cross-catalysis: The process (chemical reaction) in which a molecule (catalyst) catalyzes the formation of a different molecule.
- Recombination: A reaction where polymers (DNA, RNA, or peptides) are cut and then re-joined/spliced together to generate new combinations.
- Polymerization: Reactions in which small molecules (monomers) are covalently linked to produce longer molecules (polymers).
- Seeding: A protocol of adding a molecule to a chemical reaction system in a concentration-dependent manner.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benner, S.A. Defining life. Astrobiology 2010, 10, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Segré, D.; Ben-Eli, D.; Lancet, D. Compositional genomes: Prebiotic information transfer in mutually catalytic noncovalent assemblies. Proc. Natl. Acad. Sci. USA 2000, 97, 4112–4117. [Google Scholar] [CrossRef]
- Adamski, P.; Eleveld, M.; Sood, A.; Kun, Á.; Szilágyi, A.; Czárán, T.; Szathmáry, E.; Otto, S. From self-replication to replicator systems en route to de novo life. Nat. Rev. Chem. 2020, 4, 386–403. [Google Scholar] [CrossRef]
- Yčas, M. A note on the origin of life. Proc. Natl. Acad. Sci. USA 1955, 41, 714. [Google Scholar] [CrossRef] [PubMed]
- Calvin, M. Chemical evolution and the origin of life. Am. Sci. 1956, 44, 248–263. [Google Scholar]
- Dyson, F. Origins of Life, 2nd ed.; Cambridge University Press: Cambridge, UK, 1999. [Google Scholar]
- Kauffman, S.A. Autocatalytic sets of proteins. J. Theor. Biol. 1986, 119, 1–24. [Google Scholar] [CrossRef]
- Jain, S.; Krishna, S. Autocatalytic sets and the growth of complexity in an evolutionary model. Phys. Rev. Lett. 1998, 81, 5684. [Google Scholar] [CrossRef]
- Ichihashi, N.; Usui, K.; Kazuta, Y.; Sunami, T.; Matsuura, T.; Yomo, T. Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment. Nat. Commun. 2013, 4, 1–7. [Google Scholar] [CrossRef]
- Gánti, T. The Principles of Life; Oxford University Press: Oxford, UK, 2003. [Google Scholar]
- Eigen, M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971, 58, 465–523. [Google Scholar] [CrossRef] [PubMed]
- Hordijk, W.; Steel, M.; Kauffman, S. The structure of autocatalytic sets: Evolvability, enablement, and emergence. Acta Biotheor. 2012, 60, 379–392. [Google Scholar] [CrossRef]
- Nghe, P.; Hordijk, W.; Kauffman, S.A.; Walker, S.I.; Schmidt, F.J.; Kemble, H.; Yeates, J.A.; Lehman, N. Prebiotic network evolution: Six key parameters. Mol. Biosyst. 2015, 11, 3206–3217. [Google Scholar] [CrossRef]
- Vasas, V.; Fernando, C.; Santos, M.; Kauffman, S.; Szathmáry, E. Evolution before genes. Biol. Direct 2012, 7, 1–14. [Google Scholar] [CrossRef]
- Cairns-Smith, A.G. Genetic Takeover and the Mineral Origins of Life; Cambridge University Press: Cambridge, UK, 1982. [Google Scholar]
- Dyson, F.J. A model for the origin of life. J. Mol. Evol. 1982, 18, 344–350. [Google Scholar] [CrossRef]
- Nowak, M.A.; Ohtsuki, H. Prevolutionary dynamics and the origin of evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 14924–14927. [Google Scholar] [CrossRef] [PubMed]
- Lewontin, R.C. The units of selection. Annu. Rev. Ecol. Syst. 1970, 1, 1–18. [Google Scholar] [CrossRef]
- Godfrey-Smith, P. Conditions for evolution by natural selection. J. Philos. 2007, 104, 489–516. [Google Scholar] [CrossRef]
- Farmer, J.D.; Kauffman, S.A.; Packard, N.H. Autocatalytic replication of polymers. Phys. D Nonlinear Phenom. 1986, 22, 50–67. [Google Scholar] [CrossRef]
- Jain, S.; Krishna, S. A model for the emergence of cooperation, interdependence, and structure in evolving networks. Proc. Natl. Acad. Sci. USA 2001, 98, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M.; Schuster, P. A principle of natural self-organization. Naturwissenschaften 1977, 64, 541–565. [Google Scholar] [CrossRef] [PubMed]
- Szathmáry, E. On the propagation of a conceptual error concerning hypercycles and cooperation. J. Syst. Chem. 2013, 4, 1–4. [Google Scholar] [CrossRef]
- Hordijk, W.; Steel, M. Detecting autocatalytic, self-sustaining sets in chemical reaction systems. J. Theor. Biol. 2004, 227, 451–461. [Google Scholar] [CrossRef] [PubMed]
- King, G.A.M. Autocatalysis. Chem. Soc. Rev. 1978, 7, 297–316. [Google Scholar] [CrossRef]
- Virgo, N.; Ikegami, T. Autocatalysis before enzymes: The emergence of prebiotic chain reactions. Adv. Artif. Life ECAL 2013, 12, 240–247. [Google Scholar]
- Barenholz, U.; Davidi, D.; Reznik, E.; Bar-On, Y.; Antonovsky, N.; Noor, E.; Milo, R. Design principles of autocatalytic cycles constrain enzyme kinetics and force low substrate saturation at flux branch points. eLife 2017, 6, e20667. [Google Scholar] [CrossRef]
- Blokhuis, A.; Lacoste, D.; Nghe, P. Universal motifs and the diversity of autocatalytic systems. Proc. Natl. Acad. Sci. USA 2020, 117, 25230–25236. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Plum, A.M.; Gagrani, P.; Baum, D.A. An ecological framework for the analysis of prebiotic chemical reaction networks. J. Theor. Biol. 2020, 507, 110451. [Google Scholar] [CrossRef]
- Xavier, J.C.; Hordijk, W.; Kauffman, S.; Steel, M.; Martin, W.F. Autocatalytic chemical networks at the origin of metabolism. Proc. R. Soc. B 2020, 287, 20192377. [Google Scholar] [CrossRef]
- Wołos, A.; Roszak, R.; Żądło-Dobrowolska, A.; Beker, W.; Mikulak-Klucznik, B.; Spólnik, G.; Dygas, M.; Szymkuć, S.; Grzybowski, B.A. Synthetic connectivity, emergence, and self-regeneration in the network of prebiotic chemistry. Science 2020, 369, eaaw1955. [Google Scholar] [CrossRef]
- Bornholdt, S.; Schuster, H.G. Handbook of Graphs and Networks: From Genome to the Internet; Willey-VCH (2003 Weinheim): Weinheim, Germany, 2001. [Google Scholar]
- Furusawa, C.; Kaneko, K. Zipf’s law in gene expression. Phys. Rev. Lett. 2003, 90, 088102. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S. Formation and destruction of autocatalytic sets in an evolving network model. arXiv 2004, arXiv:0403050. [Google Scholar]
- Awazu, A.; Kaneko, K. Self-organized criticality of a catalytic reaction network under flow. Phys. Rev. E 2009, 80, 010902. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; England, J.L. Design of conditions for self-replication. Phys. Rev. E 2019, 100, 022414. [Google Scholar] [CrossRef] [PubMed]
- Achlioptas, D.; D’Souza, R.M.; Spencer, J. Explosive percolation in random networks. Science 2009, 323, 1453–1455. [Google Scholar] [CrossRef]
- Erdos, P.; Rényi, A. On the evolution of random graphs. Publ. Math. Inst. Hung. Acad. Sci 1960, 5, 17–60. [Google Scholar]
- Schwartz, N.; Cohen, R.; Ben-Avraham, D.; Barabási, A.L.; Havlin, S. Percolation in directed scale-free networks. Phys. Rev. E 2002, 66, 015104. [Google Scholar] [CrossRef]
- Diestel, R. Graph Theory, 5th ed.; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Plasson, R.; Brandenburg, A.; Jullien, L.; Bersini, H. Autocatalyses. J. Phys. Chem. A 2011, 115, 8073–8085. [Google Scholar] [CrossRef] [PubMed]
- Schuster, P. What is special about autocatalysis? Monatshefte-Chem. Chem. Mon. 2019, 150, 763–775. [Google Scholar] [CrossRef]
- Draper, W.E.; Hayden, E.J.; Lehman, N. Mechanisms of covalent self-assembly of the Azoarcus ribozyme from four fragment oligonucleotides. Nucleic Acids Res. 2008, 36, 520–531. [Google Scholar] [CrossRef]
- Von Kiedrowski, G. Minimal replicator theory I: Parabolic versus exponential growth. In Bioorganic Chemistry Frontiers; Springer: Berlin/Heidelberg, Germany, 1993; pp. 113–146. [Google Scholar]
- Szathmáry, E.; Gladkih, I. Sub-exponential growth and coexistence of non-enzymatically replicating templates. J. Theor. Biol. 1989, 138, 55–58. [Google Scholar] [CrossRef]
- Toyabe, S.; Braun, D. Cooperative ligation breaks sequence symmetry and stabilizes early molecular replication. Phys. Rev. X 2019, 9, 011056. [Google Scholar] [CrossRef]
- Wagner, N.; Mukherjee, R.; Maity, I.; Kraun, S.; Ashkenasy, G. Programming Multistationarity in Chemical Replication Networks. ChemSystemsChem 2020, 2, e1900048. [Google Scholar] [CrossRef]
- Schrödinger, E. What is Life? Cambridge University Press: Cambridge, UK, 1944. [Google Scholar]
- Prigogine, I.; Nicolis, G. Self-organisation in nonequilibrium systems: Towards a dynamics of complexity. In Bifurcation Analysis; Springer: Dordrecht, The Netherlands, 1985; pp. 3–12. [Google Scholar]
- Schlögl, F. Chemical reaction models for non-equilibrium phase transitions. Z. Phys. 1972, 253, 147–161. [Google Scholar] [CrossRef]
- Wilhelm, T. The smallest chemical reaction system with bistability. BMC Syst. Biol. 2009, 3, 1–9. [Google Scholar] [CrossRef]
- Segré, D.; Lancet, D.; Kedem, O.; Pilpel, Y. Graded autocatalysis replication domain (GARD): Kinetic analysis of self-replication in mutually catalytic sets. Orig. Life Evol. Biosph. 1998, 28, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, N.; Manapat, M.L.; Chen, I.A.; Xulvi-Brunet, R.; Hayden, E.J.; Lehman, N. Spontaneous network formation among cooperative RNA replicators. Nature 2012, 491, 72–77. [Google Scholar] [CrossRef]
- Ghadiri, M. Self-Organized Autocatalytic Chemical Networks and Molecular Ecosystems: Do They Provide the Experimental Tools for Modeling the Transition from Inanimate to Animate Chemistry. In Recent Trends in Molecular Recognition; Springer: Berlin/Heidelberg, Germany, 1998; pp. 213–238. [Google Scholar]
- Giri, V.; Jain, S. The origin of large molecules in primordial autocatalytic reaction networks. PLoS ONE 2012, 7, e29546. [Google Scholar] [CrossRef]
- Oparin, A.I. The origin of life on the earth, 3rd ed.; Oliver & Boyd: Edinburgh, UK; London, UK, 1957. [Google Scholar]
- Bagley, R.J.; Farmer, J.D. Spontaneous Emergence of a Metabolism; Technical Report; Los Alamos National Lab.: Los Alamos, NM, USA, 1990. [Google Scholar]
- Bagley, R.J.; Farmer, J.D.; Fontana, W. Evolution of a metabolism. Artif. Life II 1992, 10, 141–158. [Google Scholar]
- Guseva, E.; Zuckermann, R.N.; Dill, K.A. Foldamer hypothesis for the growth and sequence differentiation of prebiotic polymers. Proc. Natl. Acad. Sci. USA 2017, 114, E7460–E7468. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, Y.J.; Kaneko, K. Optimal size for emergence of self-replicating polymer system. Phys. Rev. E 2016, 93, 032503. [Google Scholar] [CrossRef]
- Wu, M.; Higgs, P.G. Origin of self-replicating biopolymers: Autocatalytic feedback can jump-start the RNA world. J. Mol. Evol. 2009, 69, 541–554. [Google Scholar] [CrossRef]
- Tkachenko, A.V.; Maslov, S. Spontaneous emergence of autocatalytic information-coding polymers. J. Chem. Phys. 2015, 143, 045102. [Google Scholar] [CrossRef] [PubMed]
- Worst, E.G.; Zimmer, P.; Wollrab, E.; Kruse, K.; Ott, A. Unbounded growth patterns of reproducing, competing polymers—Similarities to biological evolution. New J. Phys. 2016, 18, 103003. [Google Scholar] [CrossRef]
- Kudella, P.W.; Tkachenko, A.V.; Salditt, A.; Maslov, S.; Braun, D. Structured sequences emerge from random pool when replicated by templated ligation. Proc. Natl. Acad. Sci. USA 2021, 118, e2018830118. [Google Scholar] [CrossRef]
- Vasas, V.; Fernando, C.; Szilágyi, A.; Zachár, I.; Santos, M.; Szathmáry, E. Primordial evolvability: Impasses and challenges. J. Theor. Biol. 2015, 381, 29–38. [Google Scholar] [CrossRef]
- Smith, J.M. Models of evolution. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1983, 219, 315–325. [Google Scholar]
- Vasas, V.; Szathmáry, E.; Santos, M. Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life. Proc. Natl. Acad. Sci. USA 2010, 107, 1470–1475. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Krishna, S. Crashes, recoveries, and “core shifts” in a model of evolving networks. Phys. Rev. E 2002, 65, 026103. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K. Recursiveness, switching, and fluctuations in a replicating catalytic network. Phys. Rev. E 2003, 68, 031909. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.W. Suggested model for prebiotic evolution: The use of chaos. Proc. Natl. Acad. Sci. USA 1983, 80, 3386–3390. [Google Scholar] [CrossRef]
- Stein, D.; Anderson, P. A model for the origin of biological catalysis. Proc. Natl. Acad. Sci. USA 1984, 81, 1751–1753. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, Y.J.; Kaneko, K. Kinetic selection of template polymer with complex sequences. Phys. Rev. Lett. 2018, 121, 118101. [Google Scholar] [CrossRef] [PubMed]
- Togashi, Y.; Kaneko, K. Transitions induced by the discreteness of molecules in a small autocatalytic system. Phys. Rev. Lett. 2001, 86, 2459. [Google Scholar] [CrossRef]
- Jafarpour, F.; Biancalani, T.; Goldenfeld, N. Noise-induced mechanism for biological homochirality of early life self-replicators. Phys. Rev. Lett. 2015, 115, 158101. [Google Scholar] [CrossRef]
- Kaneko, K. On recursive production and evolvabilty of cells: Catalytic reaction network approach. Adv. Chem. Phys. 2005, 130, 543. [Google Scholar]
- Van Kampen, N.G. Stochastic Processes in Physics and Chemistry, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Gardiner, C. Stochastic Methods, A Handbook for the Natural and Social Sciences, 4th ed.; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Bresch, C.; Niesert, U.; Harnasch, D. Hypercycles, parasites and packages. J. Theor. Biol. 1980, 85, 399–405. [Google Scholar] [CrossRef]
- Szathmáry, E.; Demeter, L. Group selection of early replicators and the origin of life. J. Theor. Biol. 1987, 128, 463–486. [Google Scholar] [CrossRef]
- Matsuura, T.; Yamaguchi, M.; Ko-Mitamura, E.P.; Shima, Y.; Urabe, I.; Yomo, T. Importance of compartment formation for a self-encoding system. Proc. Natl. Acad. Sci. USA 2002, 99, 7514–7517. [Google Scholar] [CrossRef]
- Matsumura, S.; Kun, Á.; Ryckelynck, M.; Coldren, F.; Szilágyi, A.; Jossinet, F.; Rick, C.; Nghe, P.; Szathmáry, E.; Griffiths, A.D. Transient compartmentalization of RNA replicators prevents extinction due to parasites. Science 2016, 354, 1293–1296. [Google Scholar] [CrossRef]
- Traulsen, A.; Nowak, M.A. Evolution of cooperation by multilevel selection. Proc. Natl. Acad. Sci. USA 2006, 103, 10952–10955. [Google Scholar] [CrossRef]
- Takeuchi, N.; Mitarai, N.; Kaneko, K. Scaling laws of multilevel selection: A striking difference between continuous-trait and binary-trait models. arXiv 2020, arXiv:2005.04421. [Google Scholar]
- Jain, S.; Krishna, S. Large extinctions in an evolutionary model: The role of innovation and keystone species. Proc. Natl. Acad. Sci. USA 2002, 99, 2055–2060. [Google Scholar] [CrossRef]
- Kaneko, K.; Yomo, T. On a kinetic origin of heredity: Minority control in a replicating system with mutually catalytic molecules. J. Theor. Biol. 2002, 214, 563–576. [Google Scholar] [CrossRef]
- Hordijk, W.; Naylor, J.; Krasnogor, N.; Fellermann, H. Population dynamics of autocatalytic sets in a compartmentalized spatial world. Life 2018, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Lancet, D.; Zidovetzki, R.; Markovitch, O. Systems protobiology: Origin of life in lipid catalytic networks. J. R. Soc. Interface 2018, 15, 20180159. [Google Scholar] [CrossRef] [PubMed]
- Derr, J.; Manapat, M.L.; Rajamani, S.; Leu, K.; Xulvi-Brunet, R.; Joseph, I.; Nowak, M.A.; Chen, I.A. Prebiotically plausible mechanisms increase compositional diversity of nucleic acid sequences. Nucleic Acids Res. 2012, 40, 4711–4722. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, A.; Kaneko, K. Negative scaling relationship between molecular diversity and resource abundances. Phys. Rev. E 2016, 93, 062419. [Google Scholar] [CrossRef] [PubMed]
- Mast, C.B.; Schink, S.; Gerland, U.; Braun, D. Escalation of polymerization in a thermal gradient. Proc. Natl. Acad. Sci. USA 2013, 110, 8030–8035. [Google Scholar] [CrossRef] [PubMed]
- Mizuuchi, R.; Blokhuis, A.; Vincent, L.; Nghe, P.; Lehman, N.; Baum, D. Mineral surfaces select for longer RNA molecules. Chem. Commun. 2019, 55, 2090–2093. [Google Scholar] [CrossRef]
- Kashiwagi, A.; Urabe, I.; Kaneko, K.; Yomo, T. Adaptive response of a gene network to environmental changes by fitness-induced attractor selection. PLoS ONE 2006, 1, e49. [Google Scholar] [CrossRef]
- Luisi, P.L. Enzymes hosted in reverse micelles in hydrocarbon solution. Angew. Chem. Int. Ed. 1985, 24, 439–450. [Google Scholar] [CrossRef]
- Luisi, P.L.; Varela, F.J. Self-replicating micelles—A chemical version of a minimal autopoietic system. Orig. Life Evol. Biosph. 1989, 19, 633–643. [Google Scholar] [CrossRef]
- Walde, P.; Wick, R.; Fresta, M.; Mangone, A.; Luisi, P.L. Autopoietic self-reproduction of fatty acid vesicles. J. Am. Chem. Soc. 1994, 116, 11649–11654. [Google Scholar] [CrossRef]
- Hordijk, W.; Steel, M. Autocatalytic sets and boundaries. J. Syst. Chem. 2015, 6, 1–5. [Google Scholar] [CrossRef]
- McMullin, B.; Varela, F. 30 years of computational autopoiesis: A review. Artif. Life 2004, 10, 277–295. [Google Scholar] [CrossRef]
- Morigaki, K.; Dallavalle, S.; Walde, P.; Colonna, S.; Luisi, P.L. Autopoietic self-reproduction of chiral fatty acid vesicles. J. Am. Chem. Soc. 1997, 119, 292–301. [Google Scholar] [CrossRef]
- Bissette, A.J.; Odell, B.; Fletcher, S.P. Physical autocatalysis driven by a bond-forming thiol–ene reaction. Nat. Commun. 2014, 5, 1–8. [Google Scholar] [CrossRef]
- Colomer, I.; Morrow, S.M.; Fletcher, S.P. A transient self-assembling self-replicator. Nat. Commun. 2018, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Morrow, S.M.; Colomer, I.; Fletcher, S.P. A chemically fuelled self-replicator. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Colomer, I.; Borissov, A.; Fletcher, S.P. Selection from a pool of self-assembling lipid replicators. Nat. Commun. 2020, 11, 1–9. [Google Scholar]
- Von Kiedrowski, G. A self-replicating hexadeoxynucleotide. Angew. Chem. Int. Ed. 1986, 25, 932–935. [Google Scholar] [CrossRef]
- Luther, A.; Brandsch, R.; Von Kiedrowski, G. Surface-promoted replication and exponential amplification of DNA analogues. Nature 1998, 396, 245–248. [Google Scholar] [CrossRef]
- Sievers, D.; Von Kiedrowski, G. Self-replication of complementary nucleotide-based oligomers. Nature 1994, 369, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Barge, L.M.; Cardoso, S.S.S.; Cartwright, J.H.E.; Cooper, G.J.T.; Cronin, L.; De Wit, A.; Doloboff, I.J.; Escribano, B.; Goldstein, R.E.; Haudin, F.; et al. From chemical gardens to chemobrionics. Chem. Rev. 2015, 115, 8652–8703. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, W. A review on inorganic nanostructure self-assembly. J. Nanosci. Nanotechnol. 2010, 10, 1563–1583. [Google Scholar] [CrossRef] [PubMed]
- Hazen, R.M.; Sverjensky, D.A. Mineral surfaces, geochemical complexities, and the origins of life. Cold Spring Harb. Perspect. Biol. 2010, 2, a002162. [Google Scholar] [CrossRef] [PubMed]
- Cairns-Smith, A.G. The origin of life and the nature of the primitive gene. J. Theor. Biol. 1966, 10, 53–88. [Google Scholar] [CrossRef]
- Wächtershäuser, G. Before enzymes and templates: Theory of surface metabolism. Microbiol. Rev. 1988, 52, 452. [Google Scholar] [CrossRef]
- Martin, W.; Russell, M.J. On the origin of biochemistry at an alkaline hydrothermal vent. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1887–1926. [Google Scholar] [CrossRef]
- Mulkidjanian, A.Y. On the origin of life in the zinc world: 1. Photosynthesizing, porous edifices built of hydrothermally precipitated zinc sulfide as cradles of life on Earth. Biol. Direct 2009, 4, 1–39. [Google Scholar] [CrossRef]
- Miras, H.N.; Mathis, C.; Xuan, W.; Long, D.L.; Pow, R.; Cronin, L. Spontaneous formation of autocatalytic sets with self-replicating inorganic metal oxide clusters. Proc. Natl. Acad. Sci. USA 2020, 117, 10699–10705. [Google Scholar] [CrossRef]
- Weber, A.L. The sugar model: Autocatalytic activity of the triose–ammonia reaction. Orig. Life Evol. Biosph. 2007, 37, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Breslow, R. On the mechanism of the formose reaction. Tetrahedron Lett. 1959, 1, 22–26. [Google Scholar] [CrossRef]
- Simonov, A.; Pestunova, O.; Matvienko, L.; Parmon, V.N. The nature of autocatalysis in the Butlerov reaction. Kinet. Catal. 2007, 48, 245–254. [Google Scholar] [CrossRef]
- Delidovich, I.V.; Simonov, A.N.; Taran, O.P.; Parmon, V.N. Catalytic formation of monosaccharides: From the formose reaction towards selective synthesis. ChemSusChem 2014, 7, 1833–1846. [Google Scholar] [CrossRef]
- Rebek, J. Molecular recognition and the development of self-replicating systems. Experientia 1991, 47, 1096–1104. [Google Scholar] [CrossRef]
- Wintner, E.; Rebek, J. Autocatalysis and the Generation of Self-Replicating Systems. Acta Chem. Scand. 1996, 50, 469–485. [Google Scholar] [CrossRef]
- Likholobov, V.; Weiss, A.; Sakharov, M. The use of temperature to simplify formose sugar composition. React. Kinet. Catal. Lett. 1978, 8, 155–166. [Google Scholar] [CrossRef]
- Ricardo, A.; Carrigan, M.; Olcott, A.; Benner, S. Borate minerals stabilize ribose. Science 2004, 303, 196. [Google Scholar] [CrossRef] [PubMed]
- Snytnikova, O.A.; Simonov, A.N.; Pestunova, O.P.; Parmon, V.N.; Tsentalovich, Y.P. Study of the photoinduced formose reaction by flash and stationary photolysis. Mendeleev Commun. 2006, 16, 9–10. [Google Scholar] [CrossRef]
- Meinert, C.; Myrgorodska, I.; De Marcellus, P.; Buhse, T.; Nahon, L.; Hoffmann, S.V.; d’Hendecourt, L.L.S.; Meierhenrich, U.J. Ribose and related sugars from ultraviolet irradiation of interstellar ice analogs. Science 2016, 352, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Michitaka, T.; Imai, T.; Hashidzume, A. Formose reaction controlled by a copolymer of n, n-dimethylacrylamide and 4-vinylphenylboronic acid. Polymers 2017, 9, 549. [Google Scholar] [CrossRef] [PubMed]
- Parry, D.A.; Squire, J.M. Fibrous Proteins: Structures and Mechanisms; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Hope, I.A.; Struhl, K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 1986, 46, 885–894. [Google Scholar] [CrossRef]
- Lee, D.H.; Granja, J.R.; Martinez, J.A.; Severin, K.; Ghadiri, M.R. A self-replicating peptide. Nature 1996, 382, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Severin, K.; Lee, D.H.; Kennan, A.J.; Ghadiri, M.R. A synthetic peptide ligase. Nature 1997, 389, 706–709. [Google Scholar] [CrossRef]
- Mukherjee, R.; Cohen-Luria, R.; Wagner, N.; Ashkenasy, G. A Bistable Switch in Dynamic Thiodepsipeptide Folding and Template-Directed Ligation. Angew. Chem. 2015, 127, 12629–12633. [Google Scholar] [CrossRef]
- Otto, S. Dynamic molecular networks: From synthetic receptors to self-replicators. Accounts Chem. Res. 2012, 45, 2200–2210. [Google Scholar] [CrossRef]
- Malakoutikhah, M.; Peyralans, J.J.P.; Colomb-Delsuc, M.; Fanlo-Virgós, H.; Stuart, M.C.; Otto, S. Uncovering the selection criteria for the emergence of multi-building-block replicators from dynamic combinatorial libraries. J. Am. Chem. Soc. 2013, 135, 18406–18417. [Google Scholar] [CrossRef]
- Colomb-Delsuc, M.; Mattia, E.; Sadownik, J.W.; Otto, S. Exponential self-replication enabled through a fibre elongation/breakage mechanism. Nat. Commun. 2015, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Malakoutikhah, M.; Leonetti, G.; Tezcan, M.; Colomb-Delsuc, M.; Nguyen, V.D.; van der Gucht, J.; Otto, S. Controlling the structure and length of self-synthesizing supramolecular polymers through nucleated growth and disassembly. Angew. Chem. Int. Ed. 2015, 127, 7963–7967. [Google Scholar] [CrossRef]
- Ashkenasy, G.; Jagasia, R.; Yadav, M.; Ghadiri, M.R. Design of a directed molecular network. Proc. Natl. Acad. Sci. USA 2004, 101, 10872–10877. [Google Scholar] [CrossRef]
- Kennan, A.J.; Haridas, V.; Severin, K.; Lee, D.H.; Ghadiri, M.R. A de novo designed peptide ligase: A mechanistic investigation. J. Am. Chem. Soc. 2001, 123, 1797–1803. [Google Scholar] [CrossRef]
- Brown, J.H.; Kim, K.H.; Jun, G.; Greenfield, N.J.; Dominguez, R.; Volkmann, N.; Hitchcock-DeGregori, S.E.; Cohen, C. Deciphering the design of the tropomyosin molecule. Proc. Natl. Acad. Sci. USA 2001, 98, 8496–8501. [Google Scholar] [CrossRef]
- Yao, S.; Ghosh, I.; Zutshi, R.; Chmielewski, J. Selective amplification by auto-and cross-catalysis in a replicating peptide system. Nature 1998, 396, 447–450. [Google Scholar] [CrossRef]
- Yao, S.; Ghosh, I.; Zutshi, R.; Chmielewski, J. A pH-modulated, self-replicating peptide. J. Am. Chem. Soc. 1997, 119, 10559–10560. [Google Scholar] [CrossRef]
- Yao, S.; Ghosh, I.; Zutshi, R.; Chmielewski, J. A self-replicating peptide under ionic control. Angew. Chem. Int. Ed. 1998, 37, 478–481. [Google Scholar] [CrossRef]
- Issac, R.; Chmielewski, J. Approaching exponential growth with a self-replicating peptide. J. Am. Chem. Soc. 2002, 124, 6808–6809. [Google Scholar] [CrossRef]
- Li, X.; Chmielewski, J. Peptide self-replication enhanced by a proline kink. J. Am. Chem. Soc. 2003, 125, 11820–11821. [Google Scholar] [CrossRef] [PubMed]
- Dadon, Z.; Samiappan, M.; Wagner, N.; Ashkenasy, G. Chemical and light triggering of peptide networks under partial thermodynamic control. Chem. Commun. 2012, 48, 1419–1421. [Google Scholar] [CrossRef]
- Dadon, Z.; Samiappan, M.; Shahar, A.; Zarivach, R.; Ashkenasy, G. A High-Resolution Structure that Provides Insight into Coiled-Coil Thiodepsipeptide Dynamic Chemistry. Angew. Chem. Int. Ed. 2013, 52, 9944–9947. [Google Scholar] [CrossRef]
- Dadon, Z.; Wagner, N.; Alasibi, S.; Samiappan, M.; Mukherjee, R.; Ashkenasy, G. Competition and cooperation in dynamic replication networks. Chemistry 2015, 21, 648–654. [Google Scholar] [CrossRef]
- Maity, I.; Wagner, N.; Mukherjee, R.; Dev, D.; Peacock-Lopez, E.; Cohen-Luria, R.; Ashkenasy, G. A chemically fueled non-enzymatic bistable network. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Severin, K.; Lee, D.H.; Martinez, J.A.; Vieth, M.; Ghadiri, M.R. Dynamic error correction in autocatalytic peptide networks. Angew. Chem. Int. Ed. 1998, 37, 126–128. [Google Scholar] [CrossRef]
- Buchanan, L.E.; Dunkelberger, E.B.; Tran, H.Q.; Cheng, P.N.; Chiu, C.C.; Cao, P.; Raleigh, D.P.; De Pablo, J.J.; Nowick, J.S.; Zanni, M.T. Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet. Proc. Natl. Acad. Sci. USA 2013, 110, 19285–19290. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nowak, P.; Otto, S. Dynamic combinatorial libraries: From exploring molecular recognition to systems chemistry. J. Am. Chem. Soc. 2013, 135, 9222–9239. [Google Scholar] [CrossRef] [PubMed]
- Santiago, G.M.; Liu, K.; Browne, W.R.; Otto, S. Emergence of light-driven protometabolism on recruitment of a photocatalytic cofactor by a self-replicator. Nat. Chem. 2020, 12, 603–607. [Google Scholar] [CrossRef]
- Wellings, D.A.; Atherton, E. [4] Standard Fmoc protocols. Methods Enzymol. 1997, 289, 44–67. [Google Scholar]
- Duim, H.; Otto, S. Towards open-ended evolution in self-replicating molecular systems. Beilstein J. Org. Chem. 2017, 13, 1189–1203. [Google Scholar] [CrossRef]
- Sadownik, J.W.; Mattia, E.; Nowak, P.; Otto, S. Diversification of self-replicating molecules. Nat. Chem. 2016, 8, 264–269. [Google Scholar] [CrossRef]
- Ekland, E.H.; Szostak, J.W.; Bartel, D.P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 1995, 269, 364–370. [Google Scholar] [CrossRef]
- Rogers, J.; Joyce, G.F. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA 2001, 7, 395–404. [Google Scholar] [CrossRef]
- Paul, N.; Joyce, G.F. A self-replicating ligase ribozyme. Proc. Natl. Acad. Sci. USA 2002, 99, 12733–12740. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, T.A.; Joyce, G.F. Self-sustained replication of an RNA enzyme. Science 2009, 323, 1229–1232. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004, 73, 791–836. [Google Scholar] [CrossRef] [PubMed]
- Reinhold-Hurek, B.; Shub, D.A. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature 1992, 357, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Hayden, E.J.; von Kiedrowski, G.; Lehman, N. Systems chemistry on ribozyme self-construction: Evidence for anabolic autocatalysis in a recombination network. Angew. Chem. Int. Ed. 2008, 120, 8552–8556. [Google Scholar] [CrossRef]
- Ameta, S.; Arsène, S.; Foulon, S.; Saudemont, B.; Clifton, B.E.; Griffiths, A.D.; Nghe, P. Darwinian properties and their trade-offs in autocatalytic RNA reaction networks. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yeates, J.A.; Hilbe, C.; Zwick, M.; Nowak, M.A.; Lehman, N. Dynamics of prebiotic RNA reproduction illuminated by chemical game theory. Proc. Natl. Acad. Sci. USA 2016, 113, 5030–5035. [Google Scholar] [CrossRef]
- Arsène, S.; Ameta, S.; Lehman, N.; Griffiths, A.D.; Nghe, P. Coupled catabolism and anabolism in autocatalytic RNA sets. Nucleic Acids Res. 2018, 46, 9660–9666. [Google Scholar] [CrossRef] [PubMed]
- Yeates, J.A.; Nghe, P.; Lehman, N. Topological and thermodynamic factors that influence the evolution of small networks of catalytic RNA species. RNA 2017, 23, 1088–1096. [Google Scholar] [CrossRef]
- Ameta, S.; Winz, M.L.; Previti, C.; Jäschke, A. Next-generation sequencing reveals how RNA catalysts evolve from random space. Nucleic Acids Res. 2014, 42, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Pressman, A.; Moretti, J.E.; Campbell, G.W.; Müller, U.F.; Chen, I.A. Analysis of in vitro evolution reveals the underlying distribution of catalytic activity among random sequences. Nucleic Acids Res. 2017, 45, 8167–8179. [Google Scholar] [CrossRef]
- Olea, C.; Gerald, F.J. Real-time detection of a self-replicating RNA Enzyme. Molecules 2016, 21, 1310. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Mizuuchi, R.; Lehman, N. Limited sequence diversity within a population supports prebiotic RNA reproduction. Life 2019, 9, 20. [Google Scholar] [CrossRef]
- Hordijk, W.; Steel, M.; Kauffman, S.A. Molecular diversity required for the formation of autocatalytic sets. Life 2019, 9, 23. [Google Scholar] [CrossRef]
- Johnston, W.K.; Unrau, P.J.; Lawrence, M.S.; Glasner, M.E.; Bartel, D.P. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 2001, 292, 1319–1325. [Google Scholar] [CrossRef]
- Wochner, A.; Attwater, J.; Coulson, A.; Holliger, P. Ribozyme-catalyzed transcription of an active ribozyme. Science 2011, 332, 209–212. [Google Scholar] [CrossRef]
- Horning, D.P.; Joyce, G.F. Amplification of RNA by an RNA polymerase ribozyme. Proc. Natl. Acad. Sci. USA 2016, 113, 9786–9791. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Szostak, J.W. RNA-catalysed synthesis of complementary-strand RNA. Nature 1989, 339, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Mutschler, H.; Wochner, A.; Holliger, P. Freeze–thaw cycles as drivers of complex ribozyme assembly. Nat. Chem. 2015, 7, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L. A production of amino acids under possible primitive earth conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef]
- Oró, J. Mechanism of synthesis of adenine from hydrogen cyanide under possible primitive Earth conditions. Nature 1961, 191, 1193–1194. [Google Scholar] [CrossRef] [PubMed]
- Powner, M.W.; Gerland, B.; Sutherland, J.D. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 2009, 459, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Schwendinger, M.G.; Rode, B.M. Possible role of copper and sodium chloride in prebiotic evolution of peptides. Anal. Sci. 1989, 5, 411–414. [Google Scholar] [CrossRef]
- Huber, C.; Wächtershäuser, G. Peptides by activation of amino acids with CO on (Ni, Fe) S surfaces: Implications for the origin of life. Science 1998, 281, 670–672. [Google Scholar] [CrossRef]
- Ferris, J.P.; Hill, A.R.; Liu, R.; Orgel, L.E. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 1996, 381, 59–61. [Google Scholar] [CrossRef]
- Orgel, L.E. The implausibility of metabolic cycles on the prebiotic Earth. PLoS Biol. 2008, 6, e18. [Google Scholar] [CrossRef]
- Hashizume, H. Role of clay minerals in chemical evolution and the origins of life. In Clay Minerals in Nature—Their Characterization, Modification and Application; BoD - Books on Demand: Norderstedt, Germany, 2012. [Google Scholar]
- Kitadai, N.; Maruyama, S. Origins of building blocks of life: A review. Geosci. Front. 2018, 9, 1117–1153. [Google Scholar] [CrossRef]
- Muchowska, K.B.; Varma, S.J.; Moran, J. Nonenzymatic metabolic reactions and life’s origins. Chem. Rev. 2020, 120, 7708–7744. [Google Scholar] [CrossRef] [PubMed]
- Danger, G.; d’Hendecourt, L.L.S.; Pascal, R. On the conditions for mimicking natural selection in chemical systems. Nat. Rev. Chem. 2020, 4, 102–109. [Google Scholar] [CrossRef]
- Adamala, K.; Szostak, J.W. Competition between model protocells driven by an encapsulated catalyst. Nat. Chem. 2013, 5, 495. [Google Scholar] [CrossRef] [PubMed]
- Adamala, K.; Szostak, J.W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science 2013, 342, 1098–1100. [Google Scholar] [CrossRef] [PubMed]
- Drobot, B.; Iglesias-Artola, J.M.; Le Vay, K.; Mayr, V.; Kar, M.; Kreysing, M.; Mutschler, H.; Tang, T.D. Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Poudyal, R.R.; Guth-Metzler, R.M.; Veenis, A.J.; Frankel, E.A.; Keating, C.D.; Bevilacqua, P.C. Template-directed RNA polymerization and enhanced ribozyme catalysis inside membraneless compartments formed by coacervates. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Zwicker, D.; Seyboldt, R.; Weber, C.A.; Hyman, A.A.; Jülicher, F. Growth and division of active droplets provides a model for protocells. Nat. Phys. 2017, 13, 408–413. [Google Scholar] [CrossRef]

| Chemical System | Reproducing Unit | Networks | Boundary Conditions | Variation | Heredity |
|---|---|---|---|---|---|
| Lipid-based [94,102] | Chemical composition 1 | N/A | CSTR | Food set-generated 2 | Protocol-mediated 3 |
| DNA-based [103,105] | Oligonucleotides | 1 network, 2 nodes [105] | Equilibrium | Reaction kinetics 4 | N/A |
| Inorganic-based [113] | Molybdenum clusters | N/A | Stopped-flow | N/A | N/A |
| Sugar-based [115,116] | Sugars (C2–C5) | N/A | CSTR [116] | Reaction kinetics 4 | N/A |
| Peptide-based [127,134,138] | Peptides | 1 network, >20 nodes [134] | Both 5 | Reaction kinetics 4 | Concentration-mediated 6 |
| Macrocycle-based [148] | Macrocycle assemblies | N/A | DCL | Reaction kinetics 4 | N/A |
| RNA-based [155,156,159] | Chemical composition 1 | >20,000 networks [160], >40 nodes [53] | Both 5 | Reaction kinetics 4 | Differential seeding 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ameta, S.; Matsubara, Y.J.; Chakraborty, N.; Krishna, S.; Thutupalli, S. Self-Reproduction and Darwinian Evolution in Autocatalytic Chemical Reaction Systems. Life 2021, 11, 308. https://doi.org/10.3390/life11040308
Ameta S, Matsubara YJ, Chakraborty N, Krishna S, Thutupalli S. Self-Reproduction and Darwinian Evolution in Autocatalytic Chemical Reaction Systems. Life. 2021; 11(4):308. https://doi.org/10.3390/life11040308
Chicago/Turabian StyleAmeta, Sandeep, Yoshiya J. Matsubara, Nayan Chakraborty, Sandeep Krishna, and Shashi Thutupalli. 2021. "Self-Reproduction and Darwinian Evolution in Autocatalytic Chemical Reaction Systems" Life 11, no. 4: 308. https://doi.org/10.3390/life11040308
APA StyleAmeta, S., Matsubara, Y. J., Chakraborty, N., Krishna, S., & Thutupalli, S. (2021). Self-Reproduction and Darwinian Evolution in Autocatalytic Chemical Reaction Systems. Life, 11(4), 308. https://doi.org/10.3390/life11040308






