Brown Fat Dnmt3b Deficiency Ameliorates Obesity in Female Mice
Abstract
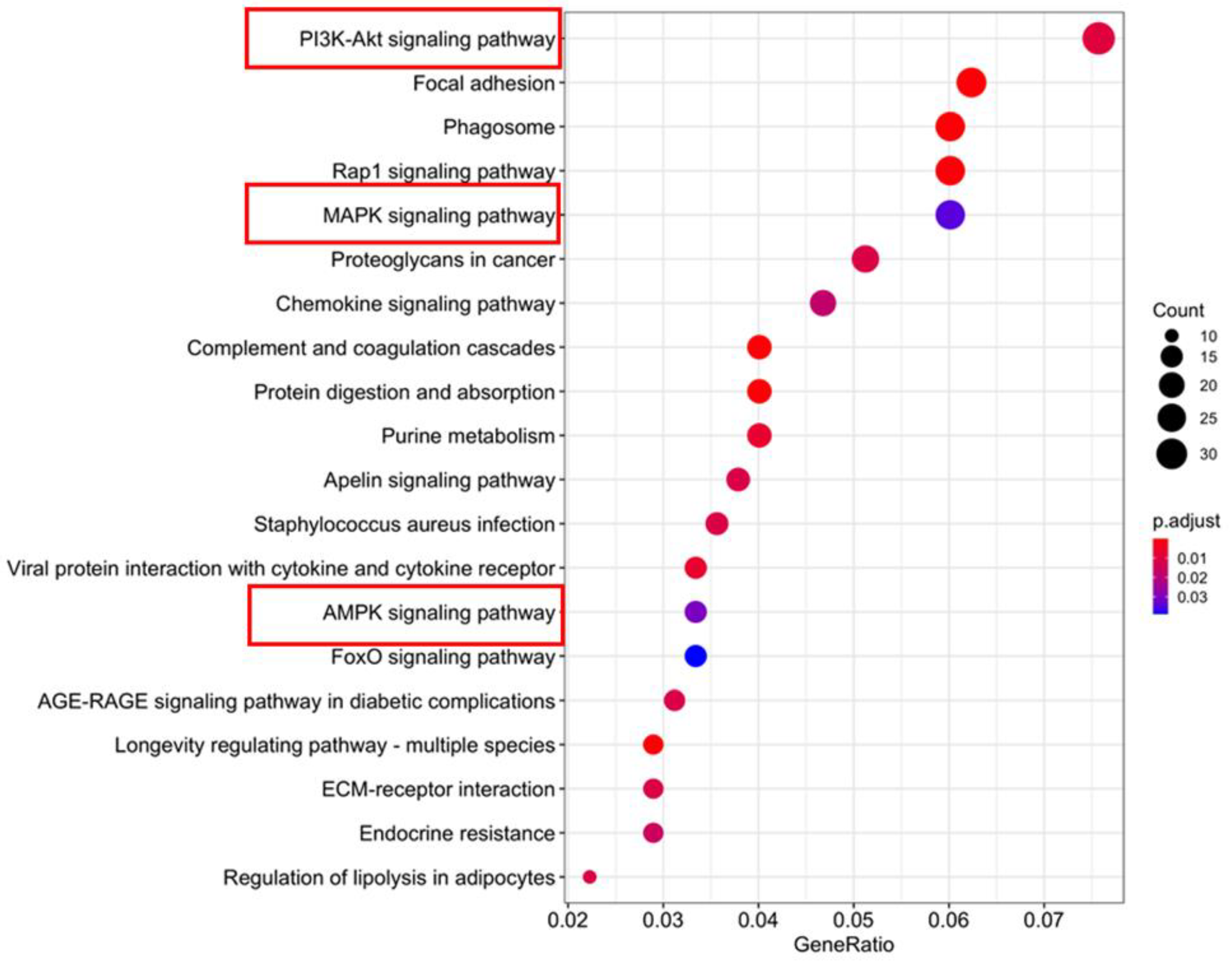
1. Introduction
2. Materials and Methods
3. Results
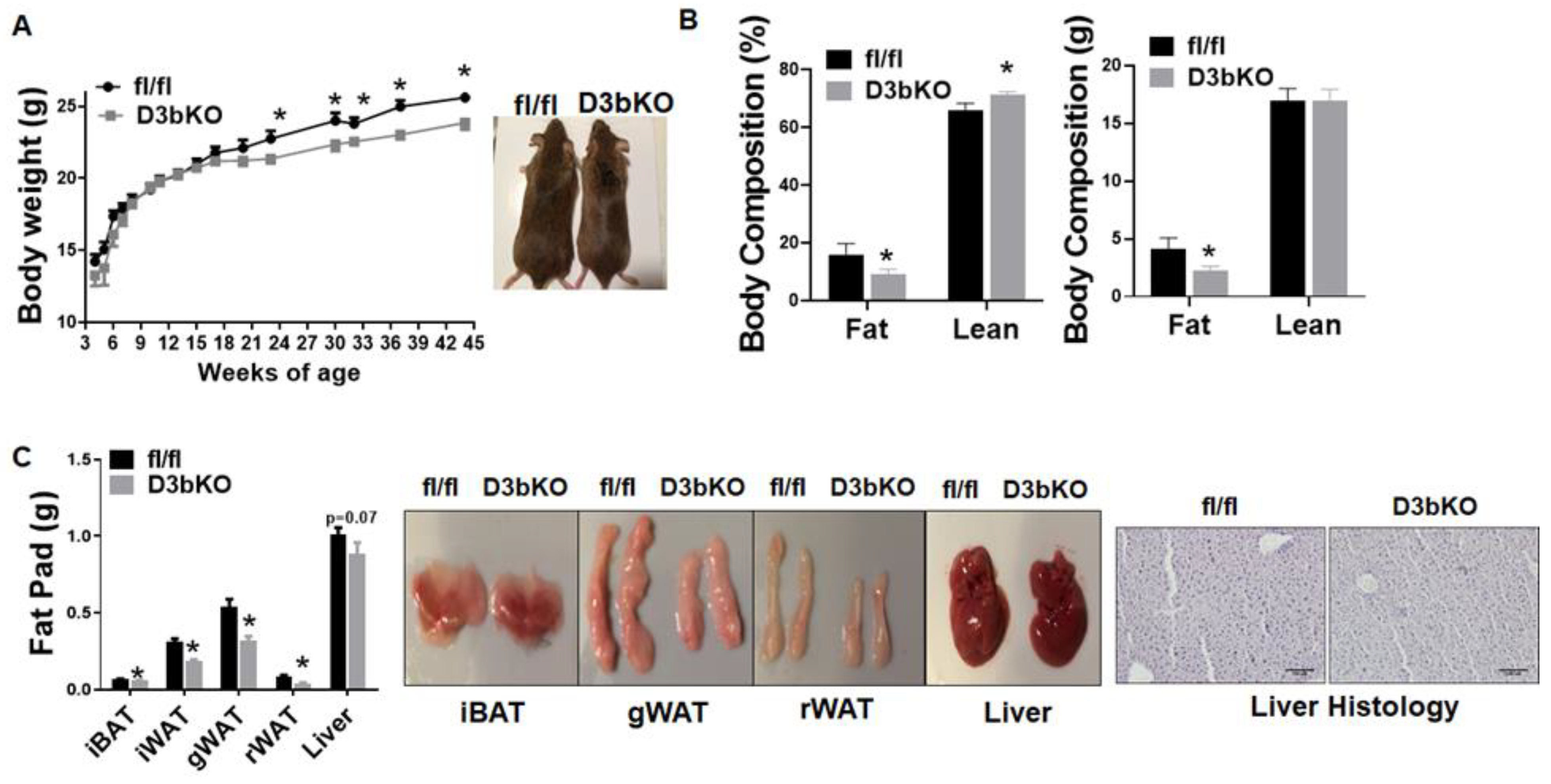
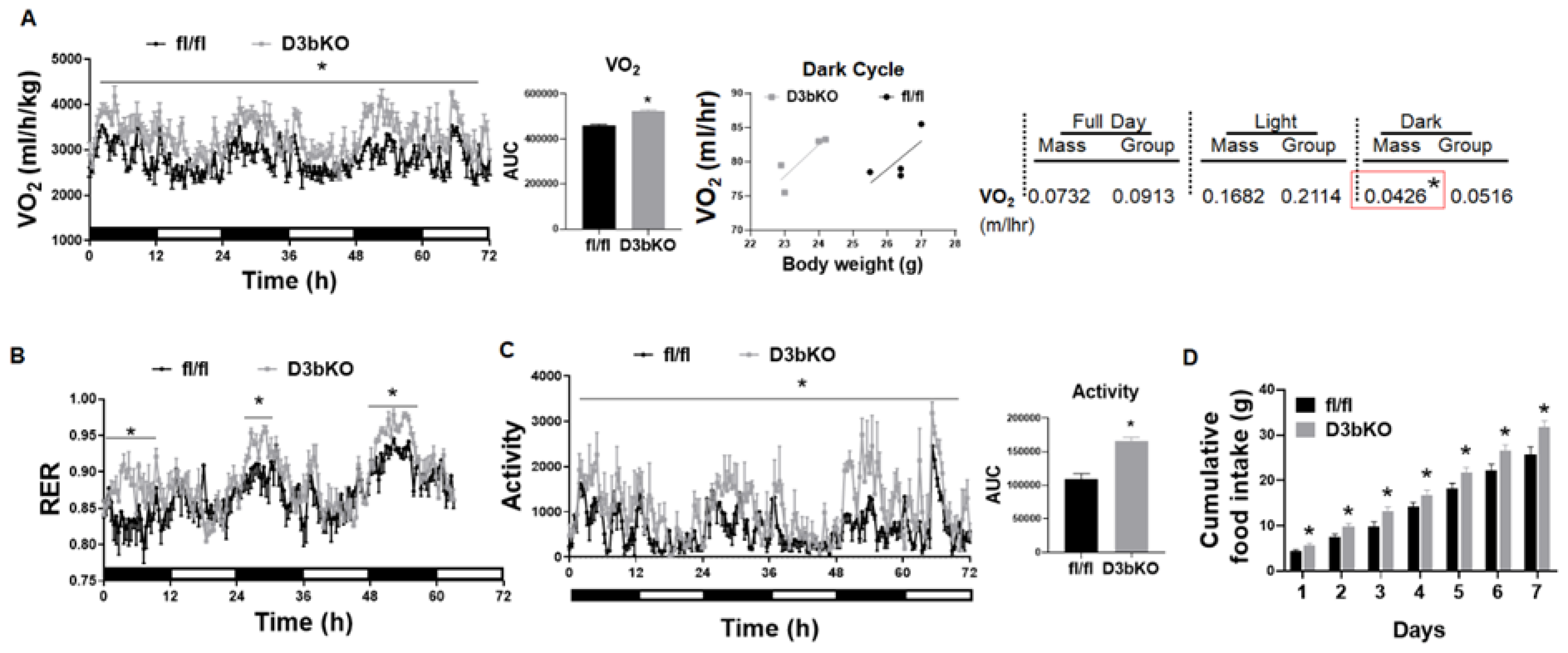
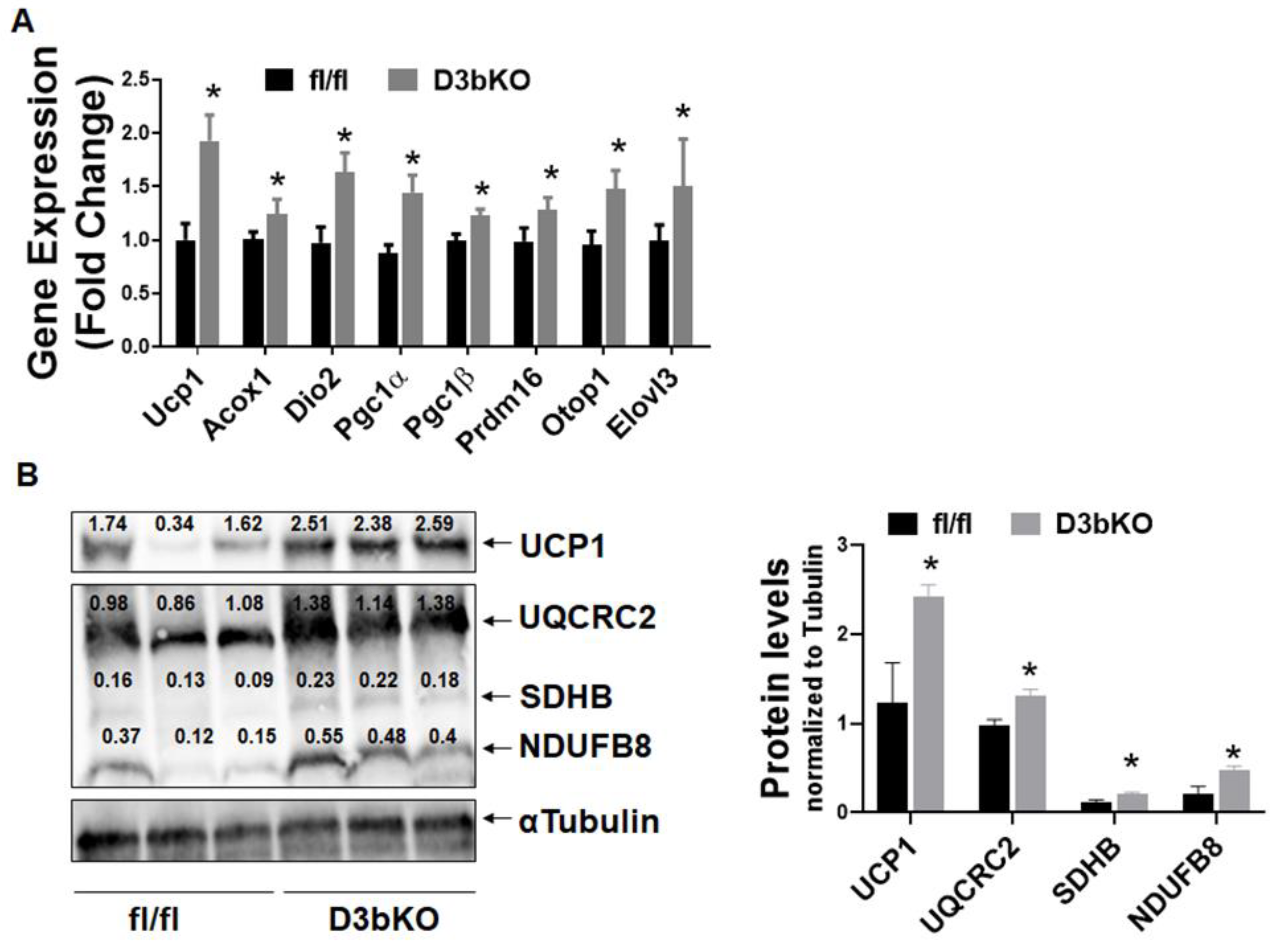
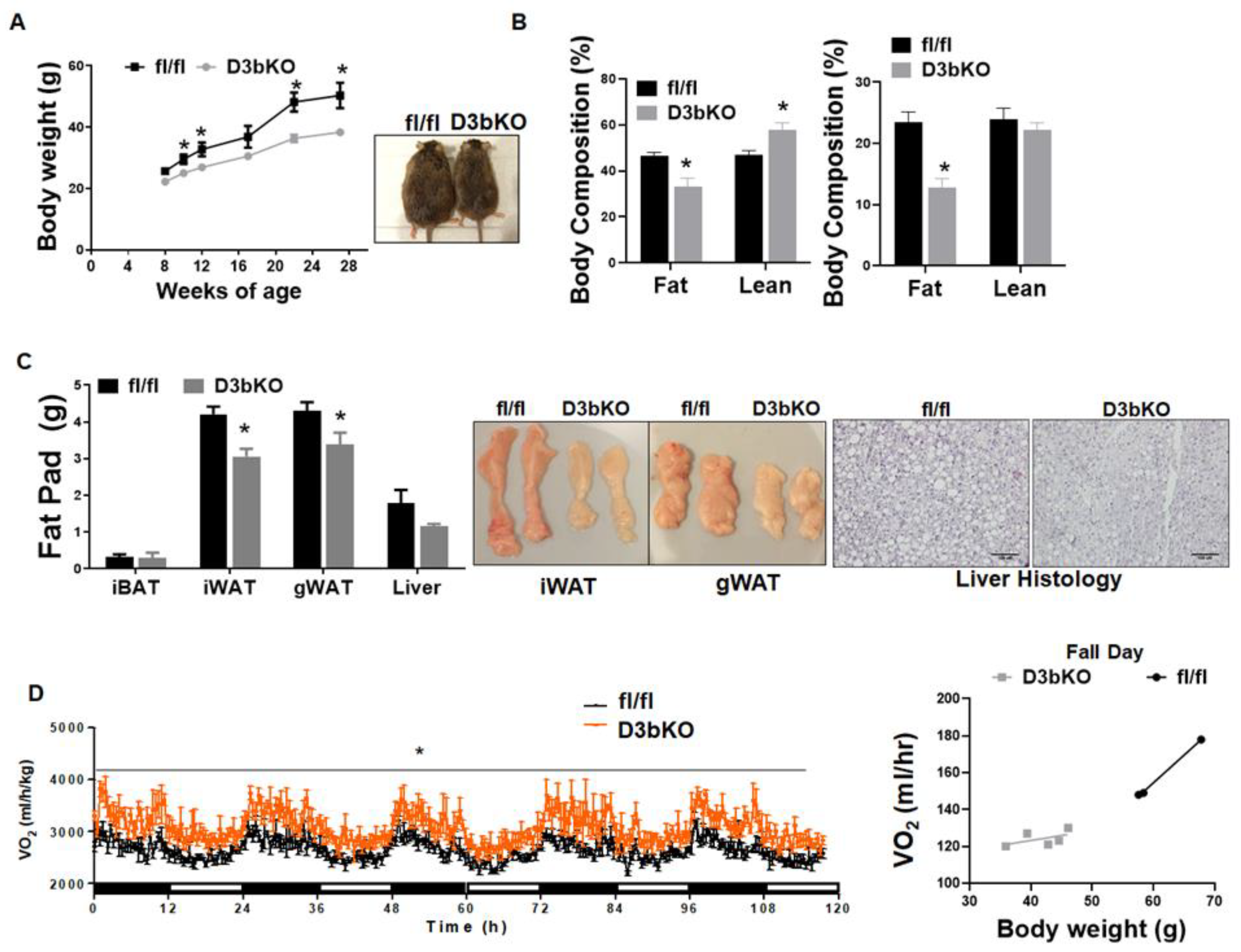
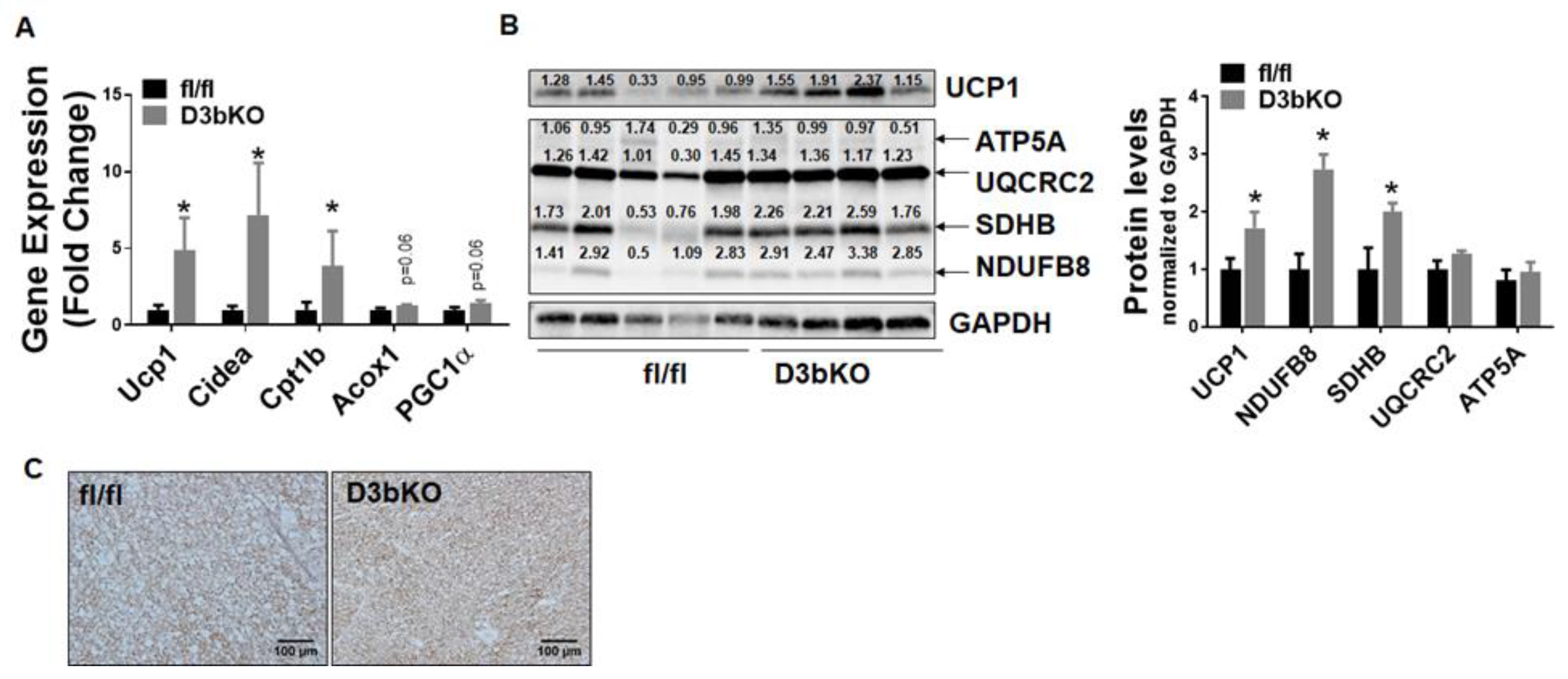
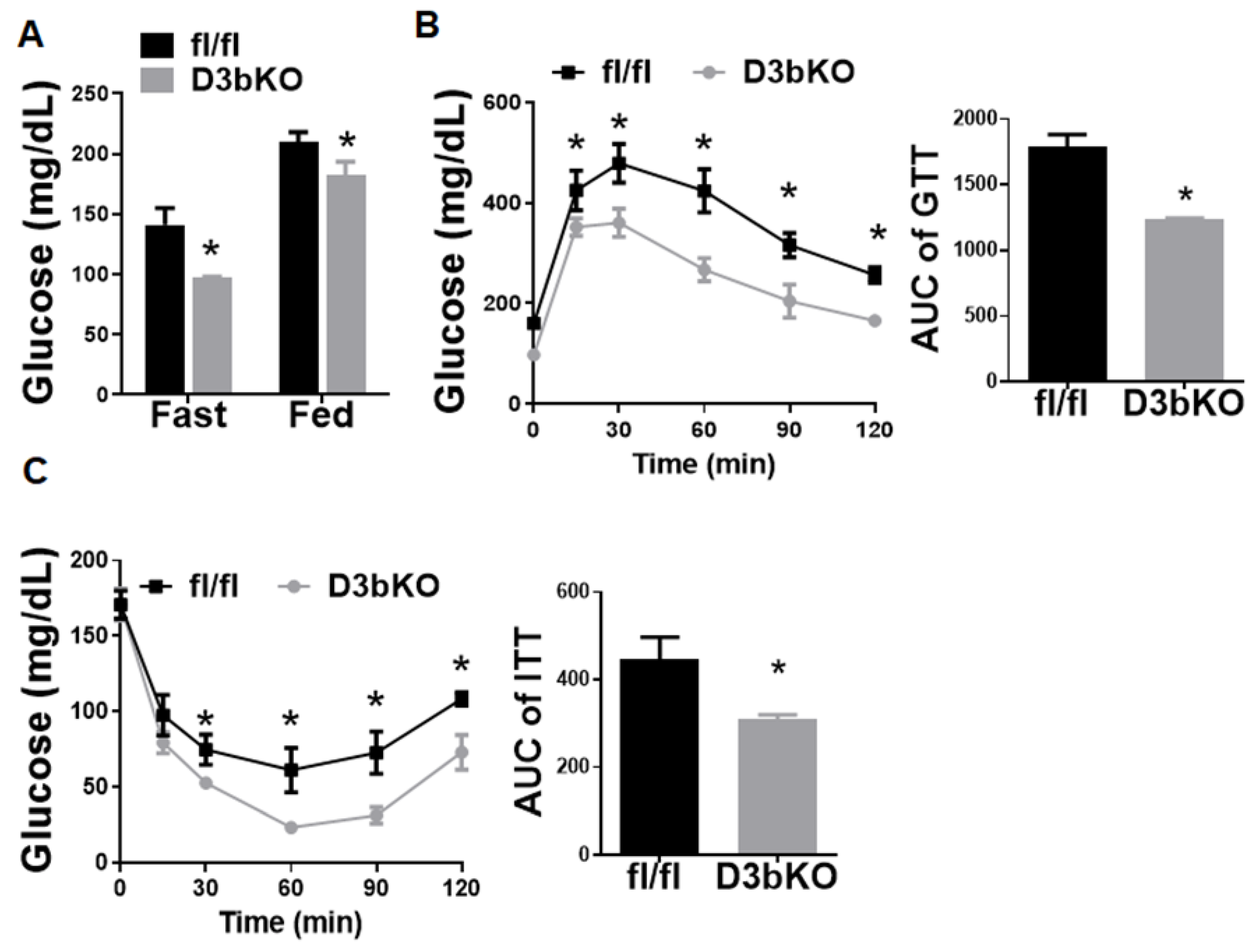
3.1. Dnmt3b Deficiency in Brown Fat Decreases Adiposity in Female Mice Fed a Chow Diet
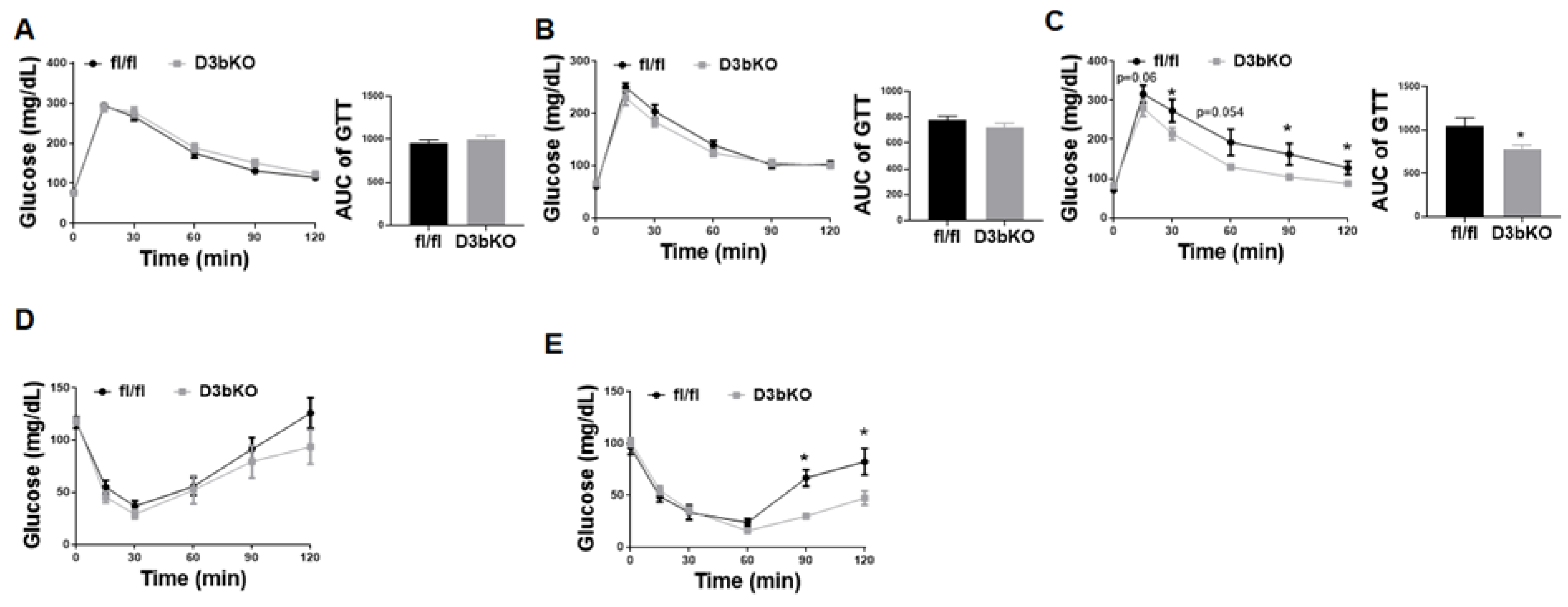
3.2. Dnmt3b Deficiency in Brown Fat Ameliorates Diet-Induced Obesity in Female Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hill, J.O.; Wyatt, H.R.; Peters, J.C. Energy balance and obesity. Circulation 2012, 126, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Lowell, B.B.; Spiegelman, B.M. Towards a molecular understanding of adaptive thermogenesis. Nat. Cell Biol. 2000, 404, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Donahoo, W.; Levine, A.J.; Melanson, E.L. Variability in energy expenditure and its components. Curr. Opin. Clin. Nutr. Metab. Care 2004, 7, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Cannon, B.; Nedergaard, J. The biochemistry of an inefficient tissue: Brown adipose tissue. Essays Biochem. 1985, 20, 110–164. [Google Scholar] [PubMed]
- Nicholls, D.G.; Locke, R.M. Thermogenic mechanisms in brown fat. Physiol. Rev. 1984, 64, 1–64. [Google Scholar] [CrossRef]
- Kazak, L.; Chouchani, E.T.; Jedrychowski, M.P.; Erickson, B.; Shinoda, K.; Cohen, P.; Vetrivelan, R.; Lu, G.Z.; Laznik-Bogoslavski, D.; Hasenfuss, S.C.; et al. A Creatine-Driven Substrate Cycle Enhances Energy Expenditure and Thermogenesis in Beige Fat. Cell 2015, 163, 643–655. [Google Scholar] [CrossRef]
- Ikeda, K.; Kang, Q.; Yoneshiro, T.; Camporez, J.P.; Maki, H.; Homma, M.; Shinoda, K.; Chen, Y.; Lu, X.; Maretich, P.; et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat. Med. 2017, 23, 1454–1465. [Google Scholar] [CrossRef]
- Ishibashi, J.; Seale, P. Medicine. Beige can be slimming. Science 2010, 328, 1113–1114. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, N.; Walden, T.B.; Shabalina, I.G.; Timmons, J.A.; Cannon, B.; Nedergaard, J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 2010, 285, 7153–7164. [Google Scholar]
- Wu, J.; Bostrom, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef]
- Brito, M.N.; Brito, N.A.; Baro, D.J.; Song, C.K.; Bartness, T.J. Differential Activation of the Sympathetic Innervation of Adipose Tissues by Melanocortin Receptor Stimulation. Endocrinology 2007, 148, 5339–5347. [Google Scholar] [CrossRef] [PubMed]
- Brito, N.A.; Brito, M.N.; Bartness, T.J. Differential sympathetic drive to adipose tissues after food deprivation, cold exposure or glucoprivation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R1445–R1452. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Levy, J.D.; Zhang, Y.; Frontini, A.; Kolodin, D.P.; Svensson, K.J.; Lo, J.C.; Zeng, X.; Ye, L.; Khandekar, M.J.; et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 2014, 156, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, H.M.; Golozoubova, V.; Cannon, B.; Nedergaard, J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009, 9, 203–209. [Google Scholar] [CrossRef]
- Seale, P.; Conroe, H.M.; Estall, J.; Kajimura, S.; Frontini, A.; Ishibashi, J.; Cohen, P.; Cinti, S.; Spiegelman, B.M. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Investig. 2011, 121, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- van Marken Lichtenbelt, W.D.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.; Kemerink, G.J.; Bouvy, N.D.; Schrauwen, P.; Teule, G.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009, 360, 1500–1508. [Google Scholar] [CrossRef]
- Virtanen, K.A.; Lidell, M.E.; Orava, J.; Heglind, M.; Westergren, R.; Niemi, T.; Taittonen, M.; Laine, J.; Savisto, N.J.; Enerback, S.; et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009, 360, 1518–1525. [Google Scholar] [CrossRef]
- Edwards, T.M.; Myers, J.P. Environmental exposures and gene regulation in disease etiology. Environ. Health Perspect. 2007, 115, 1264–1270. [Google Scholar] [CrossRef]
- Skinner, M.K.; Manikkam, M.; Guerrero-Bosagna, C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol. Metab. 2010, 21, 214–222. [Google Scholar] [CrossRef]
- Luczak, M.W.; Jagodzinski, P.P. The role of DNA methylation in cancer development. Folia Histochem. Cytobiol. 2006, 44, 143–154. [Google Scholar] [PubMed]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Jing, J.; Li, F.; Zha, L.; Yang, X.; Wu, R.; Wang, S.; Xue, B.; Shi, H. The histone methyltransferase Suv39h regulates 3T3-L1 adipogenesis. Adipocyte 2020, 9, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, R.; Shan, W.; Yu, L.; Xue, B.; Shi, H. DNA Methylation Biphasically Regulates 3T3-L1 Preadipocyte Differentiation. Mol. Endocrinol. 2016, 30, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Wu, R.; Yang, X.; Kou, S.; MacDougald, O.A.; Yu, L.; Shi, H.; Xue, B. Inhibiting DNA methylation switches adipogenesis to osteoblastogenesis by activating Wnt10a. Sci. Rep. 2016, 6, 25283. [Google Scholar] [CrossRef]
- Li, F.; Wu, R.; Cui, X.; Zha, L.; Yu, L.; Shi, H.; Xue, B. Histone Deacetylase 1 (HDAC1) Negatively Regulates Thermogenic Program in Brown Adipocytes via Coordinated Regulation of Histone H3 Lysine 27 (H3K27) Deacetylation and Methylation. J. Biol. Chem. 2016, 291, 4523–4536. [Google Scholar] [CrossRef]
- Zha, L.; Li, F.; Wu, R.; Artinian, L.; Rehder, V.; Yu, L.; Liang, H.; Xue, B.; Shi, H. The Histone Demethylase UTX Promotes Brown Adipocyte Thermogenic Program Via Coordinated Regulation of H3K27 Demethylation and Acetylation. J. Biol. Chem. 2015, 290, 25151–25163. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Jing, J.; Movahed, M.; Cui, X.; Cao, Q.; Wu, R.; Chen, Z.; Yu, L.; Pan, Y.; Shi, H.; et al. Epigenetic Interaction between UTX and DNMT1 Regulates Diet-Induced Myogenic Remodeling in Brown Fat. bioRxiv 2020, 2, 1–22. [Google Scholar] [CrossRef]
- Dodge, J.E.; Okano, M.; Dick, F.; Tsujimoto, N.; Chen, T.; Wang, S.; Ueda, Y.; Dyson, N.; Li, E. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 2005, 280, 17986–17991. [Google Scholar] [CrossRef] [PubMed]
- Mina, A.I.; LeClair, R.A.; LeClair, K.B.; Cohen, D.E.; Lantier, L.; Banks, A.S. CalR: A Web-Based Analysis Tool for Indirect Calorimetry Experiments. Cell Metab. 2018, 28, 656–666.e1. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cao, Q.; Yu, L.; Shi, H.; Xue, B.; Shi, H. Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity. JCI Insight 2016, 1, e87748. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cao, Q.; Cui, X.; Jing, J.; Li, F.; Shi, H.; Xue, B.; Shi, H. Dnmt3b Deficiency in Myf5+-Brown Fat Precursor Cells Promotes Obesity in Female Mice. Biomolecules 2021, 11, 1087. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.L.; Barr, C.L.; Ryu, V.; Cao, Q.; Xue, B.; Bartness, T.J. Separate and shared sympathetic outflow to white and brown fat coordinately regulates thermoregulation and beige adipocyte recruitment. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 312, R132–R145. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Jing, J.; Cui, X.; Shi, H.; Xue, B. Sympathetic nerve innervation is required for beigeing in white fat. Physiol. Rep. 2019, 7, e14031. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.D.; Klingenspor, M.; Tschop, M.H. Revisiting energy expenditure: How to correct mouse metabolic rate for body mass. Nat. Metab. 2021, 3, 1134–1136. [Google Scholar] [CrossRef]
- Campion, J.; Milagro, F.I.; Martinez, J.A. Individuality and epigenetics in obesity. Obes. Rev. 2009, 10, 383–392. [Google Scholar] [CrossRef]
- Holness, M.J.; Sugden, M.C. Epigenetic regulation of metabolism in children born small for gestational age. Curr. Opin. Clin. Nutr. Metab. Care 2006, 9, 482–488. [Google Scholar] [CrossRef]
- Ling, C.; Groop, L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes 2009, 58, 2718–2725. [Google Scholar] [CrossRef] [PubMed]
- Maier, S.; Olek, A. Diabetes: A candidate disease for efficient DNA methylation profiling. J. Nutr. 2002, 132, 2440S–2443S. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Szarc vel Szic, K.; Ndlovu, M.N.; Haegeman, G.; Vanden Berghe, W. Nature or nurture: Let food be your epigenetic medicine in chronic inflammatory disorders. Biochem. Pharmacol. 2010, 80, 1816–1832. [Google Scholar] [CrossRef] [PubMed]
- Noer, A.; Boquest, A.C.; Collas, P. Dynamics of adipogenic promoter DNA methylation during clonal culture of human adipose stem cells to senescence. BMC Cell Biol. 2007, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Barres, R.; Yan, J.; Egan, B.; Treebak, J.T.; Rasmussen, M.; Fritz, T.; Caidahl, K.; Krook, A.; O’Gorman, D.J.; Zierath, J.R. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012, 15, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Barres, R.; Osler, M.E.; Yan, J.; Rune, A.; Fritz, T.; Caidahl, K.; Krook, A.; Zierath, J.R. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009, 10, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Milagro, F.I.; Campion, J.; Garcia-Diaz, D.F.; Goyenechea, E.; Paternain, L.; Martinez, J.A. High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J. Physiol. Biochem. 2009, 65, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, E.C.; Garretson, J.T.; Wu, R.; Shi, H.; Xue, B. Neuronal Dnmt1 Deficiency Attenuates Diet-Induced Obesity in Mice. Endocrinology 2018, 159, 145–162. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Wang, X.; Jia, L.; Mondal, A.K.; Diallo, A.; Hawkins, G.A.; Das, S.K.; Parks, J.S.; Yu, L.; Shi, H.; et al. Inhibiting DNA Methylation by 5-Aza-2′-deoxycytidine ameliorates atherosclerosis through suppressing macrophage inflammation. Endocrinology 2014, 155, 4925–4938. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, X.; Liu, D.; Yu, L.; Xue, B.; Shi, H. Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Mol. Endocrinol. 2014, 28, 565–574. [Google Scholar] [CrossRef]
- Yang, Q.; Liang, X.; Sun, X.; Zhang, L.; Fu, X.; Rogers, C.J.; Berim, A.; Zhang, S.; Wang, S.; Wang, B.; et al. AMPK/alpha-Ketoglutarate Axis Dynamically Mediates DNA Demethylation in the Prdm16 Promoter and Brown Adipogenesis. Cell Metab. 2016, 24, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Michael, L.F.; Wu, Z.; Cheatham, R.B.; Puigserver, P.; Adelmant, G.; Lehman, J.J.; Kelly, D.P.; Spiegelman, B.M. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. USA 2001, 98, 3820–3825. [Google Scholar] [CrossRef]
- Shore, A.; Karamitri, A.; Kemp, P.; Speakman, J.R.; Lomax, M.A. Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue. Diabetologia 2010, 53, 1164–1173. [Google Scholar] [CrossRef]
- Gallagher, D.; Heymsfield, S.B.; Heo, M.; Jebb, S.A.; Murgatroyd, P.R.; Sakamoto, Y. Healthy percentage body fat ranges: An approach for developing guidelines based on body mass index. Am. J. Clin. Nutr. 2000, 72, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Palmer, B.F.; Clegg, D.J. The sexual dimorphism of obesity. Mol. Cell. Endocrinol. 2015, 402, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.L.; Bessesen, D.H.; Stotz, S.; Peelor, F.F.; Miller, B.F.; Horton, T.J. Adrenergic control of lipolysis in women compared with men. J. Appl. Physiol. 2014, 117, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, R.; Teixeira, D.; Calhau, C. Estrogen signaling in metabolic inflammation. Mediators Inflamm. 2014, 2014, 615917. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Cui, X.; Jing, J.; Wang, S.; Shi, H.; Xue, B.; Shi, H. Brown Fat Dnmt3b Deficiency Ameliorates Obesity in Female Mice. Life 2021, 11, 1325. https://doi.org/10.3390/life11121325
Li F, Cui X, Jing J, Wang S, Shi H, Xue B, Shi H. Brown Fat Dnmt3b Deficiency Ameliorates Obesity in Female Mice. Life. 2021; 11(12):1325. https://doi.org/10.3390/life11121325
Chicago/Turabian StyleLi, Fenfen, Xin Cui, Jia Jing, Shirong Wang, Huidong Shi, Bingzhong Xue, and Hang Shi. 2021. "Brown Fat Dnmt3b Deficiency Ameliorates Obesity in Female Mice" Life 11, no. 12: 1325. https://doi.org/10.3390/life11121325
APA StyleLi, F., Cui, X., Jing, J., Wang, S., Shi, H., Xue, B., & Shi, H. (2021). Brown Fat Dnmt3b Deficiency Ameliorates Obesity in Female Mice. Life, 11(12), 1325. https://doi.org/10.3390/life11121325





