Abstract
Streptococcus suis (S. suis) is an important swine pathogen and an emerging zoonotic agent worldwide. Serotype 9 is the most prevalent serotype in several European countries but it is relatively rare in China. In this study, through the investigation of the serotypes of 279 S. suis strains isolated from China from 2015 to 2017, it was found that serotype 9 is the second most prevalent serotype (43 out of 279), behind serotype 2 (83 out of 279). Next, the 43 serotype 9 isolates were sequenced and compared with those from the Netherland (28) and the U.K. (eight). For the purpose of comparison, the strain D12 (GCA_000231905), which has completed genome sequences, was also incorporated. Phylogenetic tree analysis showed that the strains from China and the U.K. were heterogeneous. In contrast, all but one from the Netherland belonged to the same clade. The dominant clades of Chinese strains (33) and strains from the Netherland (27) were very similar. Both of them may have originated from the same strain about 70 years ago. Then, the distributions of virulence-associated genes and antibiotic resistance genes among different clades and sources were analyzed. By comparison, strains from the Netherland carried more virulence-associated genes and those from the U.K. had more antibiotic resistance genes. Additionally, some virulence-associated genes (salK and salR) and antibiotic resistance genes (lincomycin and spectinomycin) existed only in several Chinese strains. In conclusion, our data displayed the population characteristics and differences of S. suis serotype 9 between China and Europe, suggesting that they have taken different evolutionary paths.
1. Introduction
Streptococcus suis, an important emerging zoonotic pathogen, causes significant economic losses in the swine industry and severe systemic infections in humans [,]. The first human infection case occurred in Denmark in 1968. Since then, there have been over 1600 S. suis infections in humans worldwide [,]. In particular, human S. suis cases have dramatically increased over the past two decades []. Previously, it was thought that S. suis caused only sporadic human infection cases. However, two epidemics in China in 1998 and 2005 provoked considerable public health concerns worldwide []. In recent years, S. suis has been established as the leading and second cause of adult meningitis in Vietnam and in Thailand, respectively, and the third most common cause of community-acquired bacterial meningitis in Hong Kong []. The growing threat of S. suis to humans highlights the critical need to better understand the prevalent characterization of S. suis for the promotion of public health [].
Based on the differences in capsular polysaccharide antigens, a total of 33 serotypes were described for S. suis []. Serotype 2 is the most prevalent and virulent worldwide, followed by serotype 9 []. However, in several European countries, including Germany, Spain, the Netherland, and Belgium, serotype 9 is more prevalent than other serotypes []. It is noteworthy that these serotype distribution characteristics are not constant. For example, serotype 2 was the most identified isolate in the Netherland before 1996. In Spain, a greater increase in serotype 9 was observed, from 4.4% or lower in 1991–1995 to 54–65% in 1998–2002 []. These findings suggest that continuous serotyping is necessary for an accurate understanding of the epidemic regularity of S. suis [].
Genomic research based on next-generation sequencing is an important strategy for studying pathogenic bacteria, and is widely used to investigate pathogenesis and drug resistance mechanisms []. The S. suis population has rich genetic diversity, even among strains belonging to the same serotype. For example, the serotype 2 strains from Europe and Asia are considerably different in genotype from those of North America []. Importantly, their virulence phenotypes also show significant differences, suggesting that the virulence potential of S. suis is genetically related []. At present, the importance of serotype 9 strains is growing, especially in several European countries and China. However, the genomic research on S. suis has mainly focused on serotype 2 strains, and limited data are available for serotype 9 strains [].
Here, when investigating the epidemic characteristics of S. suis in China, it was found that, compared with more than a decade ago, the prevalence of serotype 9 strains has increased significantly in recent years. Considering that similar phenomena have already taken place in several European countries, it was of interest to evaluate whether this increase in serotype 9 strains in China is attributable to the import of European strains or the evolution of Chinese strains. To answer this question, we analyzed strains isolated from China, the Netherland, and the U.K. in this study. Through comparative genomic analysis, we not only described the evolutionary characteristics of serotype 9 strains isolated from China, but also showed their virulence and drug resistance potential.
2. Materials and Methods
2.1. Bacterial Isolation and Identification
A total of 279 S. suis strains isolated from diseased pigs were provided by the diagnostic center of Keqian Biology, Wuhan, China. These strains were isolated from 17 provinces or regions in China from 2015 to 2017 (Anhui, Chongqing, Fujian, Guangdong, Guangxi, Hebei, Henan, Hubei, Hunan, Jiangsu, Jiangxi, Liaoning, Shaanxi, Shandong, Shanxi, Sichuan, and Zhejiang), involving seven isolation sources (lung, arthrosis, brain, spleen, kidneys, liver, and heart). Streptococcus suis was identified using a species-specific PCR assay targeting the gdh gene [].
2.2. Serotyping of S. suis
Streptococcus suis serotyping was performed using a coagglutination test, with commercial specific sera against serotypes 1–31 and 33 (Statens Serum Institut, Copenhagen, Denmark) [].
2.3. Genome Sequencing Analysis
Based on the serotyping results, 43 serotype 9 isolates were chosen for genome sequencing analysis. The serotype 9 strains were cultured overnight at 37 °C in tryptic soy broth (TSB) (Difco, Detroit, MI, USA) with 10% bovine serum. Genomic DNA was extracted with the Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA), and then randomly fragmented by a Covaris instrument to an average size of 350 nucleotides. Next, the sequence libraries were sequenced using an IlluminaHiSeq PE150 instrument. To ensure the accuracy and reliability of subsequent information analysis, raw data were filtered to obtain effective data (clean data). Finally, the sequences were assembled using SOAPdenovo [].
2.4. Phylogenetic Analysis
For the purpose of comparison, 37 additionally available serotype 9 genomes were obtained from public databases and used in this study, including 28 isolates from the Netherland, eight isolates from the U.K., and D12 from China. By BlastP and cluster analysis, 205 single-copy orthologous genes were obtained and aligned by MUSCLE. Subsequently, a phylogenetic tree was constructed in MEGA (1000 bootstrap replications) [].
2.5. Comparative Analysis of Virulence Potential
To compare the virulence potentials of the 43 isolated serotype 9 strains from this study and 37 additional serotype 9 strains, 37 virulence-associated genes of S. suis were chosen, including adcR, ccpA, ciaH, ciaR, cps2C, cps2E, cps2F, dltA, dppIV, endoD, epf, fbpS, feoB, fur, gapdh, glnA, gtfA, hp0197, IgA1, luxS, manN, mrp, neuB, nox, pgdA, purA, purD, salK, salR, sly, srtA, ssnA, sspA, SSU0308, treR, troA, and virA []. Using BlastP analysis (E-value < 1 × 10−5), the distribution of virulence-associated genes in each strain was obtained. Finally, the statistical results were visualized using the ggplot2 drawing package in R.
2.6. Comparative Analysis of Drug Resistance Potential
To identify potential antibiotic resistance genes in the genomic sequences of these serotype 9 strains, the protein sequence alignment of antibiotic resistance genes in the Antibiotic Resistance Genes database (ARDB, http://ardb.cbcb.umd.edu/, accessed on 26 November 2021) and serotype 9 strain genomic sequences was conducted using BlastP (E-value < 1 × 10−5) []. Finally, the statistical results of various antibiotic resistance genes were visualized using the ggplot2 drawing package in R.
2.7. Nucleotide Sequence Accession Number
Reads of newly sequenced strains obtained in this study were deposited in GenBank under the following accession numbers: PRJNA765159.
3. Results
3.1. Serotypes of S. suis Isolates
Of 279 S. suis isolates, serotype 2 (29.7%) was most prevalent, followed by serotypes 9 (15.4%), 3 (12.5%), 8 (8.6%), 7 (6.5%), and 1/2 (2.9%). Further, 38 isolates were nontypable and serotypes 1, 4, 5, 6, 11, 13, 14, 15, 21, 23, 28, and 29 (0.6–1.8%) rarely appeared. In addition, serotypes 10, 12, 16, 17, 18, 19, 20, 22, 24, 25, 26, 27, 30, 31, and 33 were not isolated in this study (Figure 1). The detailed background information of the 279 S. suis isolates is provided in Supplementary Table S1.
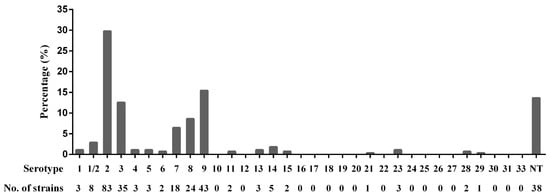
Figure 1.
Serotype distribution of 279 S. suis isolates. NT: nontypable.
3.2. Evolution Analysis
To assess the evolutionary relationships between the 43 isolated serotype 9 strains from this study and the 37 additional serotype 9 strains, a phylogenetic analysis was performed based on the concatenated DNA sequence obtained by joining 205 single-copy core gene sequences. The divergence time was estimated using a relaxed molecular clock []. A Bayesian tree revealed that the 43 serotype 9 strains isolated in this study were divided into one dominant clade containing 33 strains and five smaller clades. The 28 isolated serotype 9 strains from the Netherland were divided into one dominant clade and one small clade containing only one strain, the eight isolated serotype 9 strains from the U.K. were equally divided into four clades, and the reference strain D12 occupied one independent clade (Figure 2). By analyzing the divergence time of different clades, it was found that the dominant clades of Chinese strains and the strains from the Netherland were very similar and may have originated from the same strain about 70 years ago.
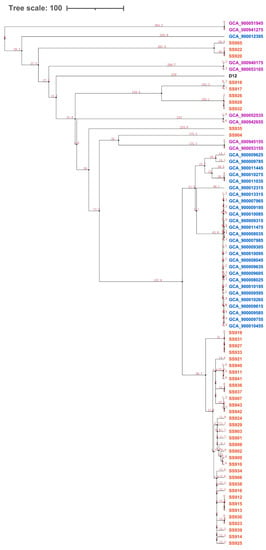
Figure 2.
Phylogenetic relationship and evolutionary time scale based on 205 single-copy orthologous genes identified in 80 S. suis genomes. Blue, Netherlandish strains; purple, British strains; red, Chinese strains; black, reference strain.
3.3. Distribution of Virulence-Associated Genes among Serotype 9 Strains
To evaluate the virulence potential of above serotype 9 strains from different sources, the presence of 37 S. suis virulence-associated genes was studied in the 80 serotype 9 genomes. A total of 24 virulence-associated genes were present in all 80 tested serotype 9 genomes (ccpA, ciaH, ciaR, cps2C, cps2F, dppIV, fbpS, feoB, fur, gtfA, hp0197, IgA1, luxS, manN, nox, pgdA, purA, purD, srtA, ssnA, sspA, SSU0308, treR, and troA) and 13 were partially present (adcR, cps2E, dltA, endoD, epf, gapdh, glnA, mrp, neuB, salK, salR, sly, and virA). Statistical analysis showed that the distributions of some virulence-associated genes were significantly different between serotype 9 strains from three sources. More strains from the Netherland contained sly, and strains from the Netherland contained more copies of ciaH, ciaR, and manN. More strains from the Netherland and the U.K. contained mrp and more copies of nox. Strains from the Netherland and China contained more copies of ccpA. Strains from China and the U.K. contained more copies of virA, and salK and salR existed only in Chinese strains (Figure 3). These results suggest that serotype 9 strains from the Netherland possess more virulence potential compared to those from China and the U.K.

Figure 3.
The distribution of 37 S. suis virulence-associated genes in the 80 serotype 9 genomes. Abscissa number represents the number of gene, the amount of each gene in strain was expressed with the length of red bar. Blue, Netherlandish strains; purple, British strains; red, Chinese strains; black, reference strain.
3.4. Difference in Drug Resistance Potential among Tested Strains
To evaluate the drug resistance potential between Chinese strains and those from the Netherland and the U.K., all serotype 9 strain genomic sequences (including 43 Chinese strains, 28 strains from the Netherland, eight strains from the U.K., and D12) were analyzed using BlastP in ARDB. A total of 29 types of antibiotic resistance genes (ARGs) were tested, which involved almost all common antibiotic types [], including genes for resistance against amikacin, bacitracin, butirosin, chloramphenicol, ciprofloxacin, dibekacin, gentamincin_b, isepamicin, kanamycin, kasugamycin, lincomycin, lincosamide, lividomycin, macrolide, neomycin, netilmicin, norfloxacin, paromomycin, penicillin, ribostamycin, sisomicin, spectomycin, sulfonamide, teicoplanin, tetracycline, tigecycline, tobramycin, trimethoprim, and vancomycin. A total of ten were present in all 80 tested serotype 9 genomes (bacitracin, ciprofloxacin, lincosamide, macrolide, norfloxacin, penicillin, teicoplanin, tetracycline, trimethoprim, and vancomycin), eleven were partially present (amikacin, chloramphenicol, dibekacin, isepamicin, kanamycin, lincomycin, netilmicin, sisomicin, spectomycin, tigecycline, and tobramycin), and eight were absent (butirosin, gentamincin_b, kasugamycin, lividomycin, neomycin, paromomycin, ribostamycin, and sulfonamide). Statistical analysis showed that more strains from the U.K. contained resistance genes against chloramphenicol, kanamycin, and tigecycline, as well as more copies of genes associated with resistance against bacitracin, ciprofloxacin, norfloxacin, penicillin, and tetracycline. Strains from the U.K. and the Netherland contained more copies of genes associated with resistance against lincosamide, macrolide, and vancomycin. Strains from the Netherland contained more copies of genes associated with resistance against teicoplanin, and resistance genes against lincomycin and spectomycin existed only in Chinese strains (Figure 4). These results suggest that serotype 9 strains from the U.K. possess more drug resistance potential compared to those from China and the Netherland.

Figure 4.
The distribution of 29 types of antibiotic resistance genes in the 80 serotype 9 genomes. Abscissa number represents the number of gene, the amount of each gene in strain was expressed with the length of red bar. Blue, Netherlandish strains; purple, British strains; red, Chinese strains; black, reference strain.
4. Discussion
In recent years, the growing prevalence of human S. suis disease in Southeast and East Asia has caused growing concern about this pathogen among the scientific community []. This highlights the importance of investigating the epidemiological characteristics of S. suis. However, there are limited related studies, especially regarding the epidemiological characteristics of S. suis in China, where two large-scale outbreaks occurred in 1998 and 2005. Here, we genotypically and phenotypically analyzed 279 S. suis strains isolated from 17 provinces of China, providing valuable reference information for clinical treatment strategies.
In China, between 2003 and 2007, serotype 2 (43.2%, 176/407) was the most prevalent serotype, followed by serotype 3 (14.7%, 60/407), and the isolation rate of serotype 9 was only 1.2% (5 in 407) []. However, our data show that the serotype distribution of S. suis in China has changed significantly over the past 10 years. Although serotype 2 remained the most prevalent from 2015 to 2017, the isolation rate fell to 29.7% (80/279). Serotype 9 (15.4%, 43 out of 279) exceeded serotype 3, becoming the second most prevalent serotype, which is the most frequent serotype in many swine-producing European countries, including Germany, Spain, the Netherland, and Belgium []. Moreover, the serotype distributions of S. suis in China also differed from those of several other Asian countries; for example, the most prevalent serotypes in Korea were serotype 3 (15.8%) and serotype 2 (15.0%), whereas serotype 9 only comprised 3.8% []. Because serotype 9 displaced serotype 2 and became the most prevalent serotype in several European countries, our data imply that the same danger may also exist in China. In the absence of an effective vaccine against serotype 9 in China, serotype 9 could cause enormous economic losses, as seen in the affected European countries. Therefore, in China, an effective vaccine against serotype 9 should be developed or imported from abroad as soon as possible, and the serotype 9 epidemic trend must be carefully monitored.
Considering the finding that S. suis serotype 9 is the most frequent serotype in many swine-producing European countries, it is necessary to investigate whether the increase in serotype 9 strains in China is attributed to the import of European strains or to the evolution of local Chinese strains. To answer this question, we assessed the evolutionary relationships between 43 serotype 9 strains from China and 36 from the Netherland and the U.K. The phylogenetic tree indicated that the last divergence between Chinese serotype 9 strains and those from the Netherland and the U.K. was 70 years ago. Considering the distribution differences of virulence-associated genes and antibiotic resistance genes, we inferred that the increase in serotype 9 strains in China may be attributed to the evolution of local Chinese strains. In addition, regarding the reason why the epidemic features of serotype 9 strains in China occurred similarly to several European countries, we hypothesize that it may be because the breeding patterns and the environment of the pig industry in China are becoming similar to those of several European countries, creating a similar evolutionary pressure for serotype 9 strains in China as in European countries.
The traditional virulence markers mrp, epf, and sly are frequently used to assess the virulence potential of S. suis [], and are mainly associated with serotype 2 strains []. Previous studies indicated that S. suis serotype 9 isolates from different regions displayed different genotypes of mrp and sly. For example, the presence rates of mrp and sly in Spanish strains were significantly higher than in Canadian strains []. A similar feature between strains from China and the Netherland was also found in this study. The presence rates of mrp and sly in strains from the Netherland were 100% (28 in 28) and 96.4% (27 in 28), respectively. Conversely, the presence rates of mrp and sly were only 20.9% (9 in 43) and 9.3% (4 in 43), respectively, in Chinese strains. In addition, almost all serotype 9 strains were epf−, even with sporadic positive isolates; they also lacked mrp and sly []. However, in the present study, one serotype 9 isolate from China (SS904) was identified as mrp+epf+sly+. Because the mrp+epf+sly+ strain has strong virulence potential [], although this virulence genotype is not a dominant clade at present, it must be maintained adequate vigilance and concern.
The overuse and misuse of antibiotics has accelerated the emergence of antibiotic-resistant bacteria and ARGs, increasing the therapeutic difficulty of treating human and animal pathogens []. At present, antimicrobial resistance is recognized as a serious global public health problem that is one of the most important challenges of contemporary medicine []. In this study, all serotype 9 strains contained at least ten types of ARGs, including resistance against bacitracin, ciprofloxacin, lincosamide, macrolide, norfloxacin, penicillin, teicoplanin, tetracycline, trimethoprim, and vancomycin. Similarly, high detection rates of ARGs associated with resistance to tetracycline, lincosamides, and macrolides were reported in Canadian serotype 9 strains [], in agreement with previous resistance data of S. suis strains from China []. Furthermore, some studies have found that the resistance rates of S. suis to trimethoprim, ciprofloxacin, and norfloxacin have significantly increased in recent years []. Therefore, based on the high detection rates of related ARGs, the potential problems of resistance against bacitracin, penicillin, teicoplanin, and vancomycin in S. suis are highly important, although these rates of resistance remain relatively low at present.
In conclusion, our data describe the serotype epidemic characteristics of S. suis in China in recent years and show that serotype 9 significantly increased and became the second most prevalent serotype. Furthermore, we assessed the evolutionary relationships between Chinese serotype 9 strains and some European strains, and then compared their virulence potential and drug resistance potential. These results suggested that the increase in serotype 9 strains in China could be attributed to the evolution of Chinese local strains, but not the import of strains from some European countries where serotype 9 is the most frequent serotype of S. suis. These findings increase our understanding about the evolution of S. suis.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/life11121324/s1. Supplementary Table S1. The information of 279 tested isolates.
Author Contributions
Q.Z. and M.J. conceived and designed the research; the experiments were performed mainly by H.Y. and J.H.; X.H. and M.H. assisted the execution of experiments; H.Y., J.H., Q.Z. and M.J. analyzed the data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Major special projects of technological innovation in Hubei Province (2020ABA016).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: PRJNA765159.
Acknowledgments
We acknowledge the diagnostic center of Keqian Biology, Wuhan, China, for providing the suspected S. suis isolates.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ye, C.; Zheng, H.; Zhang, J.; Jing, H.; Wang, L.; Xiong, Y.; Wang, W.; Zhou, Z.; Sun, Q.; Luo, X.; et al. Clinical, Experimental, and Genomic Differences between Intermediately Pathogenic, Highly Pathogenic, and Epidemic Streptococcus suis. J. Infect. Dis. 2009, 199, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, J.; Yu, J.; Xu, Z.; Liu, L.; Song, Y.; Sun, X.; Zhang, A.; Jin, M. Hp1330 contributes to Streptococcus suis virulence by inducing toll-like receptor 2- and erk1/2-dependent pro-inflammatory responses and influencing in vivo s. Suis loads. Front. Immunol. 2017, 8, 869. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, G.; Li, S.; Wang, M.; Song, J.; Wang, J.; Tang, J.; Li, M.; Hu, F. Role of a Type IV–Like Secretion System of Streptococcus suis 2 in the Development of Streptococcal Toxic Shock Syndrome. J. Infect. Dis. 2011, 204, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lin, L.; Fu, L.; Han, L.; Zhang, A. Neutrophil extracellular Taps play an important role in clearance of Streptococcus suis in vivo. Microbiol. Immunol. 2016, 60, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-I.; Jeon, A.B.; Jung, B.-Y.; Byun, J.-W.; Gottschalk, M.; Kim, A.; Kim, J.W.; Kim, H.-Y. Capsular serotypes, virulence-associated genes and antimicrobial susceptibility of Streptococcus suis isolates from pigs in Korea. J. Vet. Med. Sci. 2017, 79, 780–787. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Yang, Y.; Yan, S.; Liu, J.; Xu, Z.; Yu, J.; Song, Y.; Zhang, A.; Jin, M. A novel pro-inflammatory protein of Streptococcus suis 2 induces the toll-like receptor 2-dependent expression of pro-inflammatory cytokines in raw 264.7 macrophages via activation of erk1/2 pathway. Front. Microbiol. 2015, 6, 178. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Zhu, Y.; Ma, Y.; Ma, J.; Zhang, Y.; Yuan, L.; Pan, Z.; Wu, Z.; Yao, H. Multilocus sequence typing and virulence genotyping of Streptococcus suis serotype 9 isolates revealed high genetic and virulence diversity. FEMS Microbiol. Lett. 2017, 364, 364. [Google Scholar] [CrossRef]
- Dekker, N.; Bouma, A.; Daemen, I.; Vernooij, H.; Van Leengoed, L.; Wagenaar, J.A.; Stegeman, A. Effect of Simultaneous Exposure of Pigs to Streptococcus suis Serotypes 2 and 9 on Their Colonization and Transmission, and on Mortality. Pathogens 2017, 6, 46. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Du, P.; Qiu, X.; Kerdsin, A.; Roy, D.; Bai, X.; Xu, J.; Vela, A.I.; Gottschalk, M. Genomic comparisons of Streptococcus suis serotype 9 strains recovered from diseased pigs in Spain and Canada. Vet. Res. 2018, 49, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okwumabua, O.; O’Connor, M.; Shull, E. A polymerase chain reaction (pcr) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol. Lett. 2003, 218, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Li, R.; Zhang, A.; He, H.; Hua, Y.; Xia, J.; Cai, X.; Chen, H.; Jin, M. Characterization of Streptococcus suis isolates from the diseased pigs in China between 2003 and 2007. Vet. Microbiol. 2009, 137, 196–201. [Google Scholar] [CrossRef]
- Pan, F.; Zhang, H.; Dong, X.; Ye, W.; He, P.; Zhang, S.; Zhu, J.X.; Zhong, N. Comparative genomic analysis of multidrug-resistant Streptococcus pneumoniae isolates. Infect. Drug Resist. 2018, 11, 659–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fittipaldi, N.; Segura, M.; Grenier, D.; Gottschalk, M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012, 7, 259–279. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Yu, P.; Ge, P.; Jiang, Y.; Xu, R.; Chen, R.; Liu, X. Identification of antibiotic resistance genes in the multidrug-resistant Acinetobacter baumannii strain, MDR-SHH02, using whole-genome sequencing. Int. J. Mol. Med. 2016, 39, 364–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis Virulence Factors: Are They All Really Critical? Trends Microbiol. 2017, 25, 585–599. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, W.; Li, X.; Lu, C.; Chen, J.; Fan, W.; Huang, B. Characterization of Streptococcus suis Isolates from Slaughter Swine. Curr. Microbiol. 2013, 66, 344–349. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.-G.; Singer, A.; Zhu, Y.-G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef] [Green Version]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, P.; Wang, Y.; Fu, L.; Liu, L.; Xu, D.; Hou, Y.; Li, Y.; Fu, M.; Wang, X.; et al. Capsular serotypes, antimicrobial susceptibility, and the presence of transferable oxazolidinone resistance genes in Streptococcus suis isolated from healthy pigs in China. Vet. Microbiol. 2020, 247, 108750. [Google Scholar] [CrossRef]
- Soares, T.C.S.; Paes, A.C.; Megid, J.; Ribolla, P.; Paduan, K.D.S.; Gottschalk, M. Antimicrobial susceptibility of Streptococcus suis isolated from clinically healthy swine in Brazil. Can. J. Vet. Res. Rev. Can. Rech. Vet. 2014, 78, 145–149. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).