A Label-Free Proteomic and Complementary Metabolomic Analysis of Leaves of the Resurrection Plant Xerophyta schlechteri during Dehydration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Growth Conditions and Dehydration Treatments
2.2. Label-Free Proteomic Analyses and Workflow
2.2.1. Protein Extraction, Sample Solubilization and Quantification
2.2.2. On-Bead Hydrophilic Interaction Liquid Chromatography (HILIC) and Trypsin Digestion
2.2.3. Liquid Chromatography–Mass Spectrometry (LC-MS)
2.2.4. Database Identification of Peptides and Proteins
2.3. Gene Ontology Analysis Pipeline
2.3.1. Protein Annotation and GO-Term Retrieval
2.3.2. Mercator and MapMan Analysis and Workflow
2.4. Metabolomics Workflow
2.4.1. Gas Chromatography–Mass Spectrometry (GC-MS) Analysis
2.4.2. LC/MS Metabolite Profiling of Phytohormones
2.4.3. Data Processing
3. Results
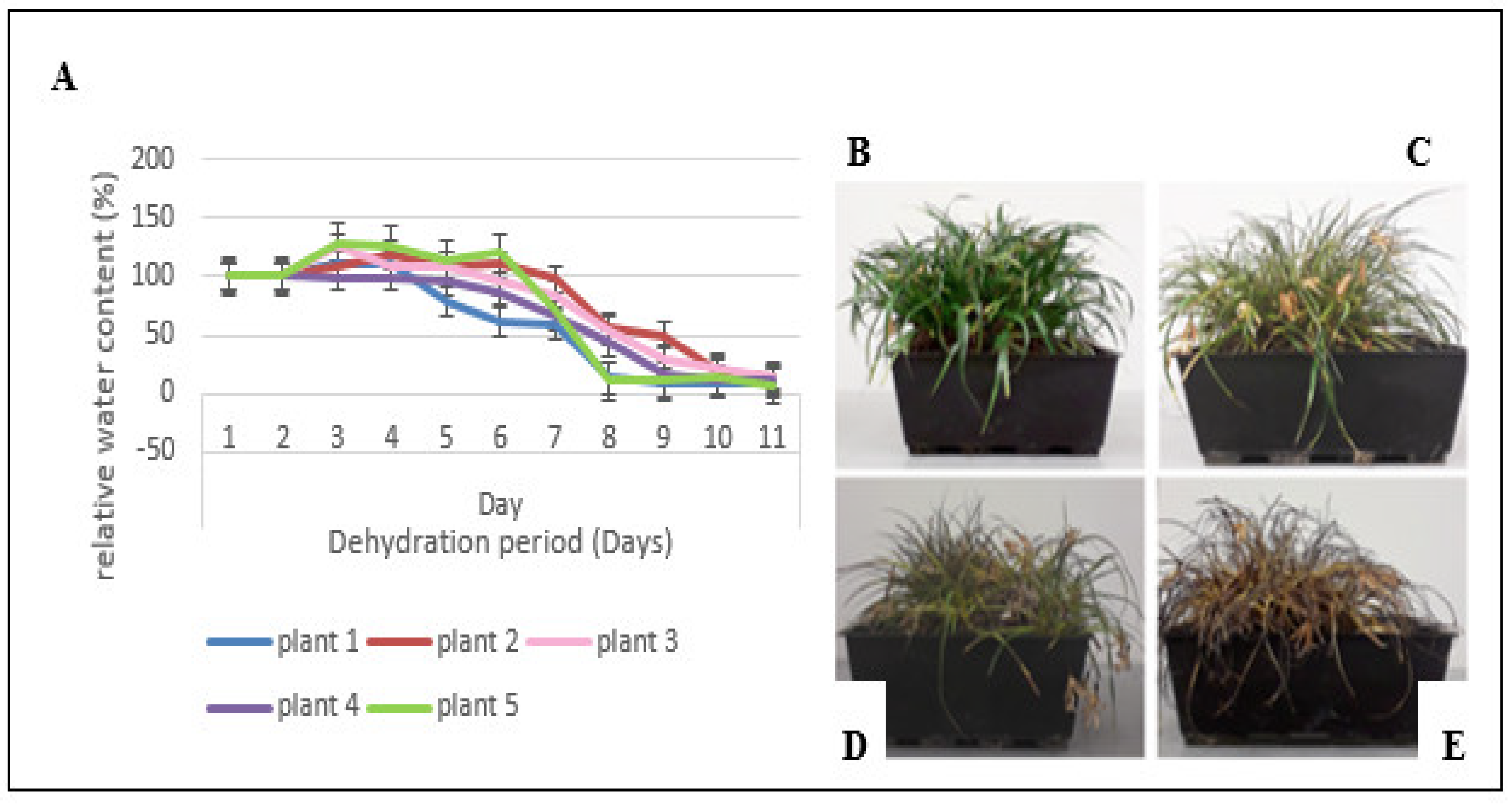
3.1. Physiological Characterization
3.2. Proteomic Analysis
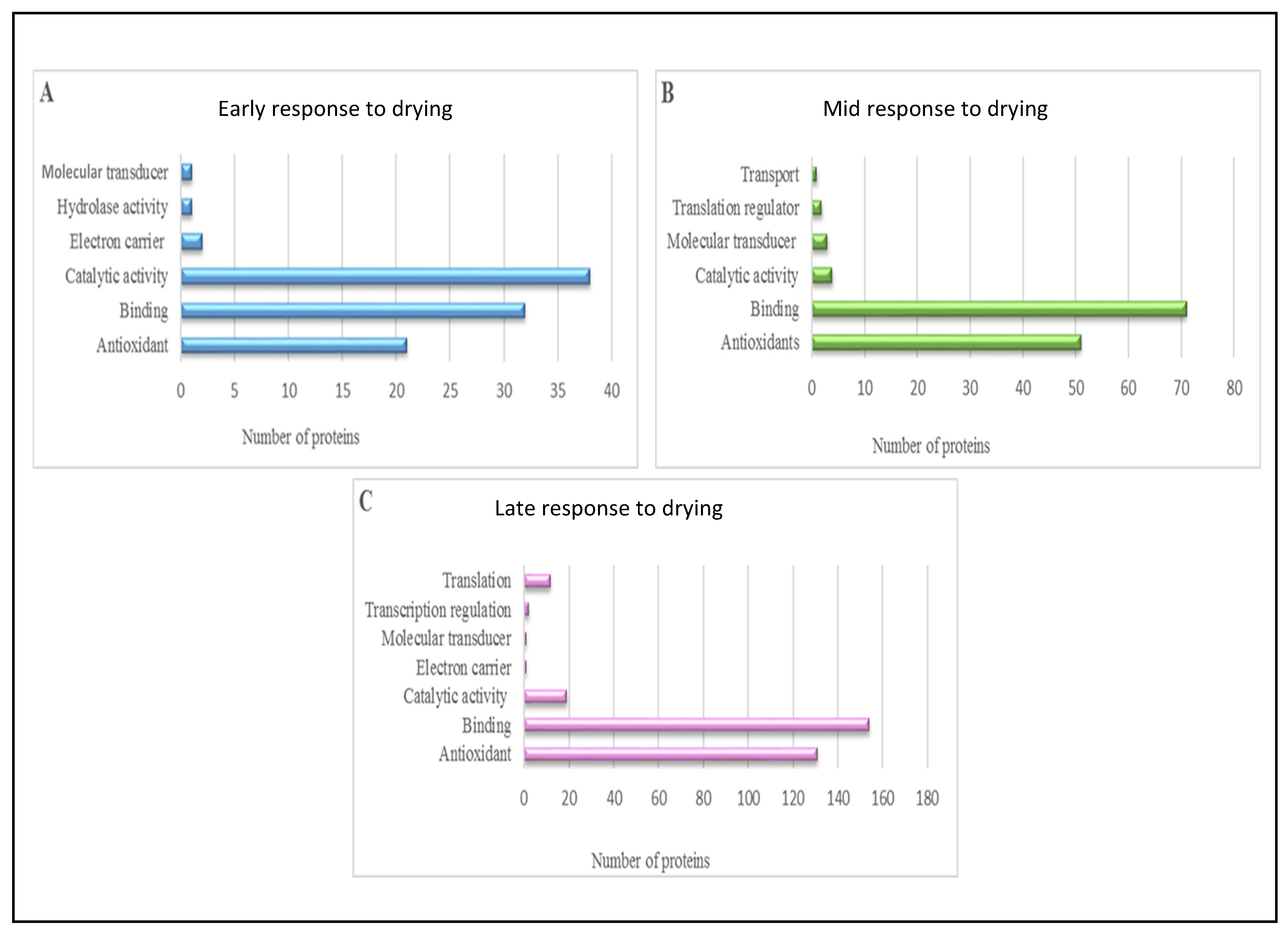
3.2.1. Blast2GO Analyses
3.2.2. MapMan Analyses
3.3. Metabolomic Analysis
Validating and Correlating a Subset of Proteomic Data with Metabolomics Data
4. Discussion
4.1. Physiological Characterization
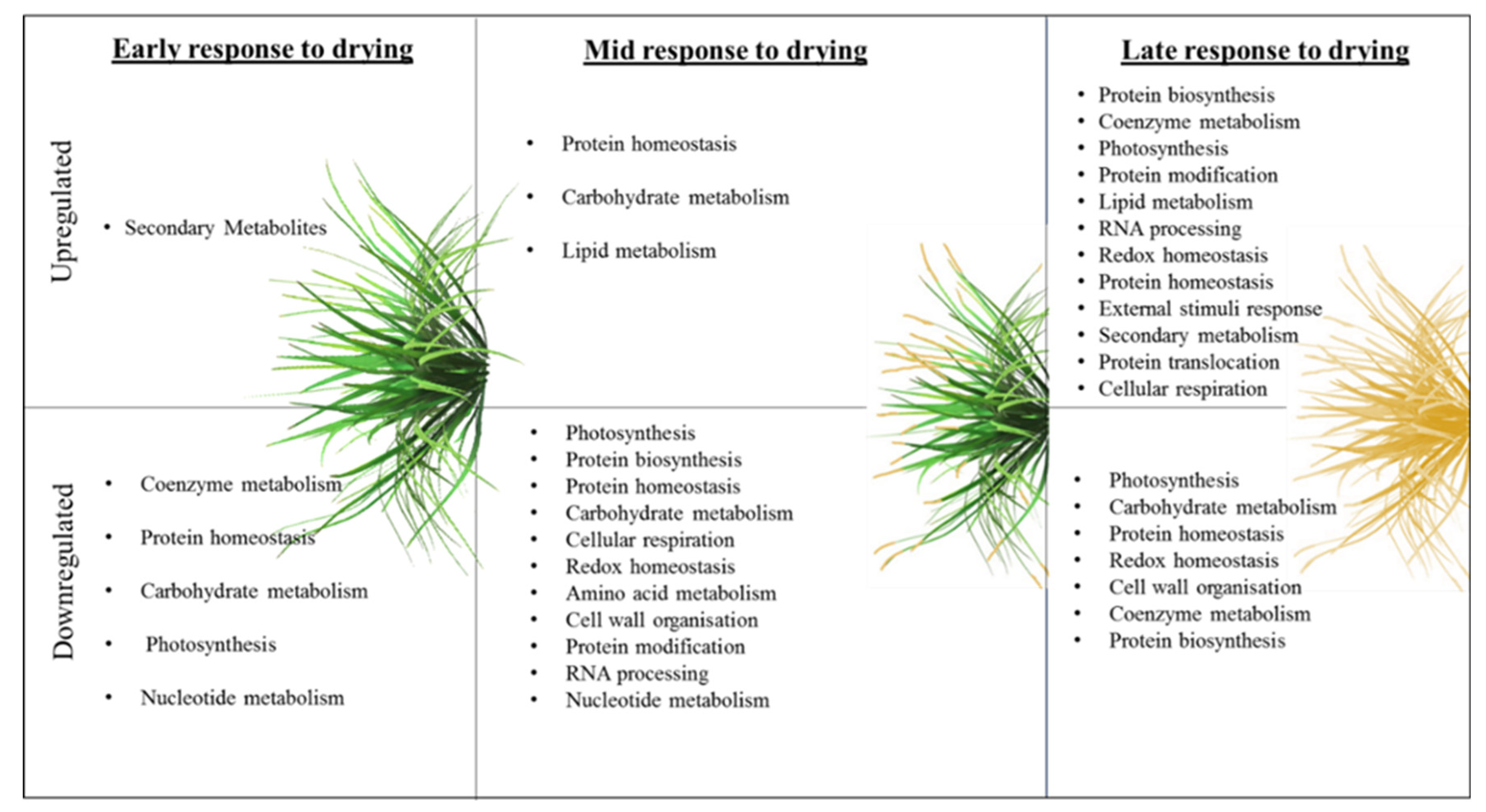
4.2. Insights Gleaned from Proteomic and Metabolomic Analyses
4.2.1. Early Response to Drying
4.2.2. Mid-Response to Drying
4.2.3. Late Response to Drying
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Marks, R.A.; Farrant, J.M.; Nicholas McLetchie, D.; VanBuren, R. Unexplored dimensions of variability in vegetative desiccation tolerance. Am. J. Bot. 2021, 108, 346–358. [Google Scholar] [CrossRef]
- Bewley, J.D. Physiological aspects of desiccation tolerance. Annu. Rev. Plant Physiol. 1979, 30, 195–238. [Google Scholar] [CrossRef]
- Dinakar, C.; Bartels, D. Desiccation tolerance in resurrection plants: New insights from transcriptome, proteome and metabolome analysis. Front. Plant Sci. 2013, 4, 482. [Google Scholar] [CrossRef] [Green Version]
- Dinakar, C.; Djilianov, D.; Bartels, D. Photosynthesis in desiccation tolerant plants: Energy metabolism and antioxidative stress defense. Plant Sci. 2012, 182, 29–41. [Google Scholar] [CrossRef]
- Farrant, J.M.; Brandt, W.; Lindsey, G.G. An Overview of Mechanisms of Desiccation Tolerance in Selected Angiosperm Resurrection Plants. Plant Stress 2007, 1, 72–84. [Google Scholar]
- Gechev, T.S.; Dinakar, C.; Benina, M.; Toneva, V.; Bartels, D. Molecular mechanisms of desiccation tolerance in resurrection plants. Cell. Mol. Life Sci. 2012, 69, 3175–3186. [Google Scholar] [CrossRef]
- Lyall, R.; Gechev, T. Multi-Omics Insights into the Evolution of Angiosperm Resurrection Plants. Annu. Plant Rev. Online 2020, 3, 77–110. [Google Scholar]
- Moore, J.P.; Le, N.T.; Brandt, W.F.; Driouich, A.; Farrant, J.M. Towards a systems-based understanding of plant desiccation tolerance. Trends Plant Sci. 2009, 14, 110–117. [Google Scholar] [CrossRef]
- Oliver, M.J.; Farrant, J.M.; Hilhorst, H.W.M.; Mundree, S.; Williams, B.; Bewley, J.D. Desiccation Tolerance: Avoiding Cellular Damage During Drying and Rehydration. Annu. Rev. Plant Biol. 2020, 71, 435–460. [Google Scholar] [CrossRef] [Green Version]
- Vicré, M.; Farrant, J.M.; Driouich, A. Insights into the cellular mechanisms of desiccation tolerance among angiosperm resurrection plant species. Plant Cell Environ. 2004, 27, 1329–1340. [Google Scholar] [CrossRef]
- Zhang, Q.; Bartels, D. Molecular responses to dehydration and desiccation in desiccation-tolerant angiosperm plants. J. Exp. Bot. 2018, 69, 3211–3222. [Google Scholar] [CrossRef]
- Mundree, S.G.; Baker, B.; Mowla, S.; Peters, S.; Marais, S.; Vander Willigen, C.; Govender, K.; Maredza, A.; Muyanga, S.; Farrant, J.M. Physiological and molecular insights into drought tolerance. Afr. J. Biotechnol. 2002, 1, 28–38. [Google Scholar]
- Farrant, J.M.; Cooper, K.; Hilgart, A.; Abdalla, K.O.; Bentley, J.; Thomson, J.A.; Dace, H.J.; Peton, N.; Mundree, S.G.; Rafudeen, M.S. A molecular physiological review of vegetative desiccation tolerance in the resurrection plant Xerophyta viscosa (Baker). Planta 2015, 242, 407–426. [Google Scholar] [CrossRef] [Green Version]
- Mundree, S.G.; Farrant, J.M. Some physiological and molecular insights into the mechanisms of desiccation tolerance in the resurrection plant Xerophyta viscosa Baker. In Plant Tolerance to Abiotic Stresses in Agriculture: Role of Genetic Engineering; Springer: Berlin/Heidelberg, Germany, 2000; pp. 201–222. [Google Scholar]
- Sherwin, H.W.; Farrant, J.M. Protection mechanisms against excess light in the resurrection plants Craterostigma wilmsii and Xerophyta viscosa. Plant Growth Regul. 1998, 24, 203–210. [Google Scholar] [CrossRef]
- Costa, M.D.; Artur, M.A.; Maia, J.; Jonkheer, E.; Derks, M.F.; Nijveen, H.; Williams, B.; Mundree, S.G.; Jimenez-Gomez, J.M.; Hesselink, T.; et al. A footprint of desiccation tolerance in the genome of Xerophyta viscosa. Nat. Plants 2017, 3, 17038. [Google Scholar] [CrossRef] [Green Version]
- Illing, N.; Denby, K.J.; Collett, H.; Shen, A.; Farrant, J.M. The signature of seeds in resurrection plants: A molecular and physiological comparison of desiccation tolerance in seeds and vegetative tissues. Integr. Comp. Biol. 2005, 45, 771–787. [Google Scholar] [CrossRef] [PubMed]
- Radermacher, A.L.; du Toit, S.F.; Farrant, J.M. Desiccation-driven senescence in the resurrection plant Xerophyta schlechteri (Baker) NL Menezes: Comparison of anatomical, ultrastructural, and metabolic responses between senescent and non-senescent tissues. Front. Plant Sci. 2019, 10, 1396. [Google Scholar] [CrossRef]
- Artur, M.A.S.; Rienstra, J.; Dennis, T.J.; Farrant, J.M.; Ligterink, W.; Hilhorst, H. Structural Plasticity of Intrinsically Disordered LEA Proteins from Xerophyta schlechteri Provides Protection In Vitro and In Vivo. Front. Plant Sci. 2019, 10, 1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmore, J.M.; Liu, J.; Smith, B.; Phinney, B.; Coaker, G. Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol. Cell. Proteom. 2012, 11, M111.014555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larrainzar, E.; Wienkoop, S.; Weckwerth, W.; Ladrera, R.; Arrese-Igor, C.; Gonzalez, E.M. Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiol. 2007, 144, 1495–1507. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Feng, J.; Campbell, K.B.; Scheffler, B.E.; Garrett, W.M.; Thibivilliers, S.; Stacey, G.; Naiman, D.Q.; Tucker, M.L.; Pastor-Corrales, M.A.; et al. Quantitative proteomic analysis of bean plants infected by a virulent and avirulent obligate rust fungus. Mol. Cell. Proteom. 2009, 8, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Poliakov, A.; Russell, C.W.; Ponnala, L.; Hoops, H.J.; Sun, Q.; Douglas, A.E.; van Wijk, K.J. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol. Cell. Proteom. 2011, 10, M110.007039. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, S.P.; Ventura, J.A.; Aguilar, C.; Nakayasu, E.S.; Choi, H.; Sobreira, T.J.; Nohara, L.L.; Wermelinger, L.S.; Almeida, I.C.; Zingali, R.B.; et al. Label-free quantitative proteomics reveals differentially regulated proteins in the latex of sticky diseased Carica papaya L. plants. J. Proteom. 2012, 75, 3191–3198. [Google Scholar] [CrossRef] [Green Version]
- Du Toit, S.F.; Bentley, J.; Farrant, J.M. NaDES formation in vegetative desiccation tolerance: Prospects and challenges. Eutectic Solvents Stress Plants 2021, 97, 225. [Google Scholar]
- Farrant, J.M.; Hilhorst, H.W.M. Crops for dry environments. Curr. Opin. Biotechnol. 2021, in press. [Google Scholar]
- Barr, H.; Weatherley, P. A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Pevzner, P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014, 5, 5277. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.Q.; Dasari, S.; Chambers, M.C.; Litton, M.D.; Sobecki, S.M.; Zimmerman, L.J.; Halvey, P.J.; Schilling, B.; Drake, P.M.; Gibson, B.W.; et al. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 2009, 8, 3872–3881. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A Fast and Simple Web Server for Genome Scale Functional Annotation of Plant Sequence Data; Technical Report No. 0140-7791; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Szymanska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, J.P.; Nguema-Ona, E.E.; Vicré-Gibouin, M.; Sørensen, I.; Willats, W.G.; Driouich, A.; Farrant, J.M. Arabinose-rich polymers as an evolutionary strategy to plasticize resurrection plant cell walls against desiccation. Planta 2013, 237, 739–754. [Google Scholar] [CrossRef]
- Tshabuse, F.; Farrant, J.M.; Humbert, L.; Moura, D.; Rainteau, D.; Espinasse, C.; Idrissi, A.; Merlier, F.; Acket, S.; Rafudeen, M.S.; et al. Glycerolipid analysis during desiccation and recovery of the resurrection plant Xerophyta humilis (Bak) Dur and Schinz. Plant Cell Environ. 2018, 41, 533–547. [Google Scholar] [CrossRef]
- Radermacher, A.L.; Williams, B.; Iranzadeh, A.; Dace, H.; Mundree, S.; Hilhorst, H.W.; Farrant, J.M. Desiccation-driven senescence and its repression in Xerophyta schlechteri are regulated at extremely low water contents. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ingle, R.A.; Schmidt, U.G.; Farrant, J.M.; Thomson, J.A.; Mundree, S.G. Proteomic analysis of leaf proteins during dehydration of the resurrection plant Xerophyta viscosa. Plant Cell Environ. 2007, 30, 435–446. [Google Scholar] [CrossRef]
- Xu, X.; Legay, S.; Sergeant, K.; Zorzan, S.; Leclercq, C.C.; Charton, S.; Giarola, V.; Liu, X.; Challabathula, D.; Renaut, J. Molecular insights into plant desiccation tolerance: Transcriptomics, proteomics and targeted metabolite profiling in Craterostigma plantagineum. Plant J. 2021, 107, 377–398. [Google Scholar] [CrossRef] [PubMed]
- Farrant, J.M.; Moore, J.P.; Hilhorst, H.W. Unifying Insights into the Desiccation Tolerance Mechanisms of Resurrection Plants and Seeds. Front. Plant Sci. 2020, 11, 1089. [Google Scholar] [CrossRef]
- Oliver, M.J.; Guo, L.; Alexander, D.C.; Ryals, J.A.; Wone, B.W.; Cushman, J.C. A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell 2011, 23, 1231–1248. [Google Scholar] [CrossRef] [Green Version]
- Gechev, T.S.; Benina, M.; Obata, T.; Tohge, T.; Sujeeth, N.; Minkov, I.; Hille, J.; Temanni, M.R.; Marriott, A.S.; Bergstrom, E.; et al. Molecular mechanisms of desiccation tolerance in the resurrection glacial relic Haberlea rhodopensis. Cell. Mol. Life Sci. 2013, 70, 689–709. [Google Scholar] [CrossRef]
- Hara, M.; Furukawa, J.; Sato, A.; Mizoguchi, T.; Miura, K. Abiotic stress and role of salicylic acid in plants. In Abiotic Stress Responses Plants; Ahmad, P., Prasad, M., Eds.; Springer: New York, NY, USA, 2012; pp. 235–251. [Google Scholar]
- Wasternack, C.; Strnad, M. Jasmonates are signals in the biosynthesis of secondary metabolites—Pathways, transcription factors and applied aspects—A brief review. New Biotechnol. 2019, 48, 1–11. [Google Scholar] [CrossRef]
- Yobi, A.; Schlauch, K.A.; Tillett, R.L.; Yim, W.C.; Espinoza, C.; Wone, B.W.; Cushman, J.C.; Oliver, M.J. Sporobolus stapfianus: Insights into desiccation tolerance in the resurrection grasses from linking transcriptomics to metabolomics. BMC Plant Biol. 2017, 17, 67. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Song, X.; Bartels, D. Enzymes and Metabolites in Carbohydrate Metabolism of Desiccation Tolerant Plants. Proteomes 2016, 4, 40. [Google Scholar] [CrossRef] [Green Version]
- Dar, N.A.; Amin, I.; Wani, W.; Wani, S.A.; Shikari, A.B.; Wani, S.H.; Masoodi, K.Z. Abscisic acid: A key regulator of abiotic stress tolerance in plants. Plant Gene 2017, 11, 106–111. [Google Scholar] [CrossRef]
- Ghoumari, A.M.; Ibanez, C.; El-Etr, M.; Leclerc, P.; Eychenne, B.; O’Malley, B.W.; Baulieu, E.E.; Schumacher, M. Progesterone and its metabolites increase myelin basic protein expression in organotypic slice cultures of rat cerebellum. J. Neurochem. 2003, 86, 848–859. [Google Scholar] [CrossRef]
- Kitano, H. Systems biology: A brief overview. Science 2002, 295, 1662–1664. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.P.; Farrant, J.M.; Lindsey, G.G.; Brandt, W.F. The South African and Namibian populations of the resurrection plant Myrothamnus flabellifolius are genetically distinct and display variation in their galloylquinic acid composition. J. Chem. Ecol. 2005, 31, 2823–2834. [Google Scholar] [CrossRef] [PubMed]
- Boeckx, T.; Winters, A.L.; Webb, K.J.; Kingston-Smith, A.H. Polyphenol oxidase in leaves: Is there any significance to the chloroplastic localization? J. Exp. Bot. 2015, 66, 3571–3579. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Parthasarathy, A.; Savka, M.A.; Hudson, A.O. The Synthesis and Role of beta-Alanine in Plants. Front. Plant Sci 2019, 10, 921. [Google Scholar] [CrossRef] [Green Version]
- Kranner, I.; Birtic, S.; Anderson, K.M.; Pritchard, H.W. Glutathione half-cell reduction potential: A universal stress marker and modulator of programmed cell death? Free Radic. Biol. Med. 2006, 40, 2155–2165. [Google Scholar] [CrossRef]
- Lea, P.J.; Miflin, B.J. Alternative route for nitrogen assimilation in higher plants. Nature 1974, 251, 614–616. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, M.; Abiko, T.; Yamaya, T. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J. Exp. Bot. 2007, 58, 2319–2327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Oster, U.; Kruse, E.; Rudiger, W.; Grimm, B. Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 1999, 120, 695–704. [Google Scholar] [CrossRef] [Green Version]
- Collett, H.; Butowt, R.; Smith, J.; Farrant, J.; Illing, N. Photosynthetic genes are differentially transcribed during the dehydration-rehydration cycle in the resurrection plant, Xerophyta humilis. J. Exp. Bot. 2003, 54, 2593–2595. [Google Scholar] [CrossRef] [PubMed]
- Garrone, A.; Archipowa, N.; Zipfel, P.F.; Hermann, G.; Dietzek, B. Plant Protochlorophyllide Oxidoreductases A and B: Catalytic Efficiency and Initial Reaction Steps. J. Biol. Chem. 2015, 290, 28530–28539. [Google Scholar] [CrossRef] [Green Version]
- Kruger, N.J.; von Schaewen, A. The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant Biol. 2003, 6, 236–246. [Google Scholar] [CrossRef]
- Xiong, Y.; DeFraia, C.; Williams, D.; Zhang, X.; Mou, Z. Deficiency in a cytosolic ribose-5-phosphate isomerase causes chloroplast dysfunction, late flowering and premature cell death in Arabidopsis. Physiol. Plant 2009, 137, 249–263. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Strabala, T.J.; Wagner, A. Potential transgenic routes to increase tree biomass. Plant Sci. 2013, 212, 72–101. [Google Scholar] [CrossRef] [Green Version]
- Ermakova, M.; Lopez-Calcagno, P.E.; Raines, C.A.; Furbank, R.T.; von Caemmerer, S. Overexpression of the Rieske FeS protein of the Cytochrome b6f complex increases C4 photosynthesis. Commun. Biol. 2019, 2, 314. [Google Scholar] [CrossRef] [Green Version]
- Ishihara, S.; Hirata, A.; Nogami, S.; Beauvais, A.; Latge, J.P.; Ohya, Y. Homologous subunits of 1,3-beta-glucan synthase are important for spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 2007, 6, 143–156. [Google Scholar] [CrossRef] [Green Version]
- Jarosz, D.F.; Lindquist, S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 2010, 330, 1820–1824. [Google Scholar] [CrossRef] [Green Version]
- Walford, S.; Thomson, J.; Farrant, J.; Mundree, S. The HSP90 family of chaperones: A look at the structure, function and mode of action. Curr. Top. Plant Biol. 2003, 4, 1–25. [Google Scholar]
- Huang, W.; Pi, L.; Liang, W.; Xu, B.; Wang, H.; Cai, R.; Huang, H. The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 2006, 18, 2479–2492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, M.A.; Gilding, E.K.; Shafee, T.; Harris, K.S.; Kaas, Q.; Poon, S.; Yap, K.; Jia, H.; Guarino, R.; Chan, L.Y.; et al. Molecular basis for the production of cyclic peptides by plant asparaginyl endopeptidases. Nat. Commun. 2018, 9, 2411. [Google Scholar] [CrossRef]
- Diaz-Villanueva, J.F.; Diaz-Molina, R.; Garcia-Gonzalez, V. Protein Folding and Mechanisms of Proteostasis. Int. J. Mol. Sci. 2015, 16, 17193–17230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdalla, K.O.; Rafudeen, M.S. Analysis of the nuclear proteome of the resurrection plant Xerophyta viscosa in response to dehydration stress using iTRAQ with 2DLC and tandem mass spectrometry. J. Proteom. 2012, 75, 2361–2374. [Google Scholar] [CrossRef]
- Morano, K.A.; Thiele, D.J. Heat shock factor function and regulation in response to cellular stress, growth, and differentiation signals. Gene Expr. 1999, 7, 271–282. [Google Scholar]
- Lin, C.T.; Xu, T.; Xing, S.L.; Zhao, L.; Sun, R.Z.; Liu, Y.; Moore, J.P.; Deng, X. Weighted Gene Co-expression Network Analysis (WGCNA) Reveals the Hub Role of Protein Ubiquitination in the Acquisition of Desiccation Tolerance in Boea hygrometrica. Plant Cell Physiol. 2019, 60, 2707–2719. [Google Scholar] [CrossRef] [PubMed]
- Coleman, H.D.; Ellis, D.D.; Gilbert, M.; Mansfield, S.D. Up-regulation of sucrose synthase and UDP-glucose pyrophosphorylase impacts plant growth and metabolism. Plant Biotechnol. J. 2006, 4, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A. Glucose activation and metabolism through UDP-glucose pyrophosphorylase in plants. Phytochemistry 1994, 37, 1507–1515. [Google Scholar] [CrossRef]
- Meng, M.; Geisler, M.; Johansson, H.; Harholt, J.; Scheller, H.V.; Mellerowicz, E.J.; Kleczkowski, L.A. UDP-glucose pyrophosphorylase is not rate limiting, but is essential in Arabidopsis. Plant Cell Physiol. 2009, 50, 998–1011. [Google Scholar] [CrossRef] [Green Version]
- Farrant, J.M.; Cooper, K.; Dace, H.J.; Bentley, J.; Hilgart, A.; Shabala, S. Desiccation tolerance. In Plant Stress Physiology; CAB International: Wallingford, UK, 2017; pp. 217–252. [Google Scholar]
- Kosugi, M.; Miyake, H.; Yamakawa, H.; Shibata, Y.; Miyazawa, A.; Sugimura, T.; Satoh, K.; Itoh, S.; Kashino, Y. Arabitol provided by lichenous fungi enhances ability to dissipate excess light energy in a symbiotic green alga under desiccation. Plant Cell Physiol. 2013, 54, 1316–1325. [Google Scholar] [CrossRef] [Green Version]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to Environmental Stresses. Plant Cell 1995, 7, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Narvaez-Vasquez, J.; Florin-Christensen, J.; Ryan, C.A. Positional specificity of a phospholipase A activity induced by wounding, systemin, and oligosaccharide elicitors in tomato leaves. Plant Cell 1999, 11, 2249–2260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudek, H.; Datta, S.R.; Franke, T.F.; Birnbaum, M.J.; Yao, R.; Cooper, G.M.; Segal, R.A.; Kaplan, D.R.; Greenberg, M.E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 1997, 275, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.; Chiapello, M.; Montero, H.; Gehrig, P.; Grossmann, J.; O’Holleran, K.; Hartken, D.; Walters, F.; Yang, S.-Y.; Hillmer, S. A rice Serine/Threonine receptor-like kinase regulates arbuscular mycorrhizal symbiosis at the peri-arbuscular membrane. Nat. Commun. 2018, 9, 4677. [Google Scholar] [CrossRef] [Green Version]
- Bartels, D.; Schneider, K.; Terstappen, G.; Piatkowski, D.; Salamini, F. Molecular cloning of abscisic acid-modulated genes which are induced during desiccation of the resurrection plant Craterostigma plantagineum. Planta 1990, 181, 27–34. [Google Scholar] [CrossRef]
- Gaff, D.F.; Bartels, D.; Gaff, J.L. Changes in gene expression during drying in a desiccation-tolerant grass Sporobolus stapfianus and a desiccation-sensitive grass Sporobolus pyramidalis. Funct. Plant Biol. 1997, 24, 617–622. [Google Scholar] [CrossRef]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadok, W.; Schoppach, R. Potential involvement of root auxins in drought tolerance by modulating nocturnal and daytime water use in wheat. Ann. Bot. 2019, 124, 969–978. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabier, H.; Tabb, D.L.; Farrant, J.M.; Rafudeen, M.S. A Label-Free Proteomic and Complementary Metabolomic Analysis of Leaves of the Resurrection Plant Xerophyta schlechteri during Dehydration. Life 2021, 11, 1242. https://doi.org/10.3390/life11111242
Gabier H, Tabb DL, Farrant JM, Rafudeen MS. A Label-Free Proteomic and Complementary Metabolomic Analysis of Leaves of the Resurrection Plant Xerophyta schlechteri during Dehydration. Life. 2021; 11(11):1242. https://doi.org/10.3390/life11111242
Chicago/Turabian StyleGabier, Hawwa, David L. Tabb, Jill M. Farrant, and Mohamed Suhail Rafudeen. 2021. "A Label-Free Proteomic and Complementary Metabolomic Analysis of Leaves of the Resurrection Plant Xerophyta schlechteri during Dehydration" Life 11, no. 11: 1242. https://doi.org/10.3390/life11111242
APA StyleGabier, H., Tabb, D. L., Farrant, J. M., & Rafudeen, M. S. (2021). A Label-Free Proteomic and Complementary Metabolomic Analysis of Leaves of the Resurrection Plant Xerophyta schlechteri during Dehydration. Life, 11(11), 1242. https://doi.org/10.3390/life11111242





