(De)Activation (Ir)Reversibly or Degradation: Dynamics of Post-Translational Protein Modifications in Plants
Abstract
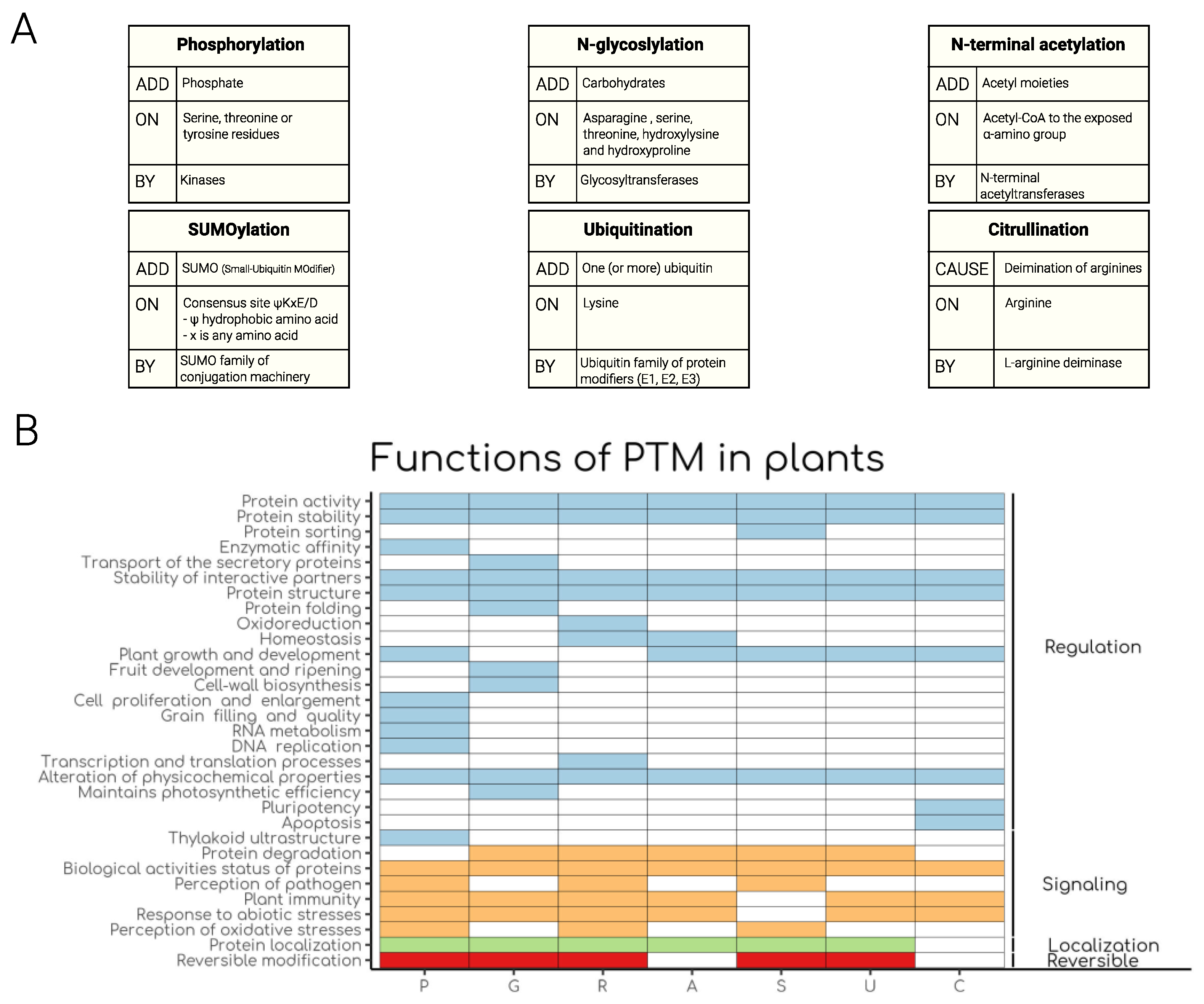
1. Introduction
2. Phosphorylation: Regulation of Plant Signalling Processes
3. Function of N-Glycosylation in Plants
4. Redox Regulation, Signaling and Functional Significance in Plants
5. Emerging Roles of N-Terminal Acetylation in Plants
6. Protein Ubiquitination in Plants
7. SUMOylation in Plant Development and Stress Responses
8. Citrullination Discovery and Potential Roles in Plants
9. Concluding Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gough, C.; Sadanandom, A. Understanding and exploiting post-translational modifications for plant disease resistance. Biomolecules 2021, 11, 1122. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Mehta, D.; Mishra, N.; Nayak, D.; Sunil, S. Role of host-mediated post-translational modifications (PTMs) in RNA virus pathogenesis. Int. J. Mol. Sci. 2021, 22, 323. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Groen, A.J.; Thomas, L.; Lilley, K.S.; Gehring, C. A quantitative phosphoproteome analysis of cGMP-dependent cellular responses in Arabidopsis thaliana. Mol. Plant. 2016, 9, 621–623. [Google Scholar] [CrossRef]
- Ryslava, H.; Doubnerova, V.; Kavan, D.; Vanek, O. Effect of posttranslational modifications on enzyme function and assembly. J. Proteom. 2013, 92, 80–109. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Triplett, L.; Chen, X.-L. Emerging roles of posttranslational modifications in plant-pathogenic fungi and bacteria. Ann. Rev. Phytopathol. 2021, 59, 99–124. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C. The increasing diversity and complexity of the RNA-binding protein repertoire in plants. Proc. R. Soc. B 2020, 287, 20201397. [Google Scholar] [CrossRef]
- Withers, J.; Dong, X. Post-translational regulation of plant immunity. Curr. Opin. Plant Biol. 2017, 38, 124–132. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Noor, J.J.; Gohain, B.; Gulabani, H.; Dnyaneshwar, I.K.; Singla, A. Post-translational modifications in regulationof pathogen surveillance and signaling in plants: The inside- (and perturbations from) outside story. IUBMB Life 2015, 67, 524–532. [Google Scholar] [CrossRef]
- Nongpiur, R.; Soni, P.; Karan, R.; Singla-Pareek, S.L.; Pareek, A. Histidine kinases in plants: Cross talk between hormone and stress responses. Plant Signal Behav. 2012, 7, 1230–1237. [Google Scholar] [CrossRef]
- Sözen, C.; Schenk, S.T.; Boudsocq, M.; Chardin, C.; Almeida-Trapp, M.; Krapp, A.; Hirt, H.; Mithöfer, A.; Colcombet, J. Wounding and Insect Feeding Trigger two Independent MAPK Pathways with Distinct Regulation and Kinetics. Plant Cell 2020, 32, 1988–2003. [Google Scholar] [CrossRef]
- Kumar, K.; Raina, S.K.; Sultan, S.M. Arabidopsis MAPK signaling pathways and their cross talks in abiotic stress response. J. Plant Biochem. Biotechnol. 2020, 29, 700–714. [Google Scholar] [CrossRef]
- Muleya, V.; Marondedze, C.; Wheeler, J.I.; Thomas, L.; Mok, Y.F.; Griffin, M.D.; Manallack, D.T.; Kwezi, L.; Lilley, K.S.; Gehring, C.; et al. Phosphorylation of the dimeric cytoplasmic domain of the phytosulfokine receptor, PSKR1. Biochem. J. 2016, 473, 3081–3098. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, C.; Motzkus, M.; Sauter, M. Phosphorylation of the phytosulfokine peptide receptor PSKR1 controls receptor activity. J. Exp. Bot. 2017, 68, 1411–1423. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Goshe, M.B.; Soderblom, E.J.; Phinney, B.S.; Kuchar, J.A.; Li, J. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 2005, 17, 1685–1703. [Google Scholar] [CrossRef]
- Kutschmar, A.; Rzewuski, G.; Stührwohldt, N.; Beemster, G.T.S.; Inzé, D.; Sauter, M. PSK-α promotes root growth in Arabidopsis. New Phytol. 2009, 181, 820–831. [Google Scholar] [CrossRef]
- Matsuzaki, Y.; Ogawa-Ohnishi, M.; Mori, A.; Matsubayashi, Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 2010, 329, 1065–1067. [Google Scholar] [CrossRef]
- Stührwohldt, N.; Dahlke, R.I.; Kutschmar, A.; Peng, X.; Sun, M.X.; Sauter, M. Phytosulfokine peptide signaling controls pollen tube growth and funicular pollen tube guidance in Arabidopsis thaliana. Physiol. Plant 2015, 153, 643–653. [Google Scholar] [CrossRef]
- Takeo, K.; Ito, T. Subcellular localization of VIP1 is regulated by phosphorylation and 14-3-3 proteins. FEBS Lett. 2017, 519, 1972–1981. [Google Scholar] [CrossRef]
- Ishida, S.; Yuasa, T.; Nakata, M.; Takahashi, Y. A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor repression of shoot growth in response to gibberellins. Plant Cell 2008, 20, 3273–3288. [Google Scholar] [CrossRef]
- Fukazawa, J.; Sakai, T.; Ishida, S.; Yamaguchi, I.; Kamiya, Y.; Takahashi, Y. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 2000, 12, 901–915. [Google Scholar] [CrossRef]
- Ishida, S.; Fukazawa, J.; Yuasa, T.; Takahashi, Y. Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator repression of shoot growth by gibberellins. Plant Cell 2004, 16, 2641–2651. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, D.; Ishida, S.; Fukazawa, J.; Takahashi, Y. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 2001, 12, 2483–2497. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Visconti, S.; Aducci, P.; Marra, M. 14-3-3 proteins in plant hormone signaling: Doing several things at once. Front Plant Sci. 2018, 13, 297. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.; Leung, S.; Cumming, C.; Shaw, M.; Albert, N.; McCallum, J.; McManus, M.T. Genotypic variation in sulphur assimilation and metabolism of onion (Allium cepa L.). II: Characterisation of ATP sulphurylase activity. Phytochem 2011, 72, 888–896. [Google Scholar] [CrossRef]
- Chevalier, D.; Morris, E.R.; Walker, J.C. 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu. Rev. Plant Biol. 2009, 60, 67–91. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH Oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef]
- Morales, J.; Kadota, Y.; Zipfel, C.; Molina, A.; Torres, M.A. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot 2016, 67, 1663–1676. [Google Scholar] [CrossRef]
- Luo, X.; Wu, W.; Liang, Y.; Xu, N.; Wang, Z.; Zou, H.; Liu, J. Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J. 2020, 39, e102856. [Google Scholar] [CrossRef]
- Yu, G.; Xian, L.; Xue, H.; Yu, W.; Rufian, J.S.; Sang, Y.; Morcillo, R.J.L.; Wang, Y.; Macho, A.P. A bacterial effector protein prevents MAPK-mediated phosphorylation of SGT1 to suppress plant immunity. PLoS Pathog. 2020, 25, e1008933. [Google Scholar] [CrossRef]
- Zhou, Q.; Meng, Q.; Tan, X.; Ding, W.; Ma, K.; Xu, Z.; Huang, X.; Gao, H. Protein phosphorylation changes during systemic acquired resistance in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 2457. [Google Scholar] [CrossRef]
- Hao, X.; Zeng, M.; Wang, J.; Zeng, Z.; Dai, J.; Xie, Z.; Yang, Y.; Tian, L.; Chen, L.; Li, D. A node-expressed transporter OsCCX2 is involved in grain cadmium accumulation of rice. Front. Plant Sci. 2018, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S.; Ma, J.F. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Li, S.F.; Huang, F.L.; Qiu, J.H.; Zhang, J.; Sheng, Z.H.; Tang, S.Q.; Wei, X.J.; Hu, P.S. The phosphoproteomic response of rice seedlings to cadmium stress. Int. J. Mol. Sci. 2017, 18, 2055. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wu, X.; Tian, F.; Chen, Q.; Luo, P.; Zhang, F.; Wan, X.; Zhong, Y.; Liu, Q.; Lin, T. Changes in proteome and protein phosphorylation reveal the protective roles of exogenous nitrogen in alleviating cadmium toxicity in poplar plants. Int. J. Mol. Sci. 2020, 21, 278. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.-J.; Sun, M.-H.; Lu, J.; Hu, D.-G.; Kang, H.; You, C.-X.; Hao, Y.-J. Phosphorylation of a malate transporter promotes malate excretion and reduces cadmium uptake in apple. J. Exp. Bot. 2020, 71, 3437–3449. [Google Scholar] [CrossRef]
- Costa, A.R.; Rodrigues, M.E.; Henriques, M.; Oliveira, R.; Azeredo, J. Glycosylation: Impact, control and improvement during therapeutic protein production. Crit. Rev. Biotechnol. 2014, 34, 281–299. [Google Scholar] [CrossRef]
- Mendez-Yañez, A.; Ramos, P.; Morales-Quintana, L. Role of glycoproteins during fruit ripening and seed development. Cells 2021, 10, 2095. [Google Scholar] [CrossRef]
- Xu, C.; Ng, D.T. Glycosylation-directed quality control of protein folding. Nat. Rev. Mol. Cell Biol. 2015, 16, 742–752. [Google Scholar] [CrossRef]
- Lombard, J. The multiple evolutionary origins of the eukaryotic N-glycosylation pathway. Biol. Direct. 2016, 11, 36. [Google Scholar] [CrossRef]
- Wang, P.; Wang, H.; Gai, J.; Tian, X.; Zhang, X.; Lv, Y.; Jian, Y. Evolution of protein N-glycosylation process in Golgi apparatus which shapes diversity of protein N-glycan structures in plants, animals and fungi. Sci. Rep. 2017, 7, 40301. [Google Scholar] [CrossRef]
- Nagashima, Y.; Schaewen, A.; Koiwa, H. Function of N-glycosylation in plants. Plant Sci. 2008, 274, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao-Hui, C.; Batoux, M.; Nekrasov, V.; Roux, M.; Chinchilla, D.; Zipfel, C.; Jones, J.D. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. USA 2009, 106, 15973–15978. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Hong, Z.; Su, W.; Li, J. A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2009, 106, 13612–13617. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.S.; Baek, J.H.; Macoy, D.M.; Chakraborty, R.; Cha, J.Y.; Hwang, D.J.; Lee, Y.H.; Lee, S.Y.; Kim, W.Y.; Kim, M.G. N-glycosylation process in both ER and Golgi plays pivotal role in plant immunity. J. Plant Biol. 2015, 58, 374–382. [Google Scholar] [CrossRef]
- Zhang, M.; Henquet, M.; Chen, Z.; Zhang, H.; Zhang, Y.; Ren, X.; van der Krol, S.; Gonneau, M.; Bosch, D.; Gong, Z.; et al. LEW3, encoding a putative α-1,2-mannosyltransferase (ALG11) in N-linked glycoprotein, plays vital roles in cell-wall biosynthesis and the abiotic stress response in Arabidopsis thaliana. Plant J. 2009, 60, 983–999. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, H.; Du, H.; Bao, Z.; Shi, Q. Sugar metabolic and N-glycosylated profiles unveil the regulatory mechanism of tomato quality under salt stress. Environ. Exp. Bot. 2020, 177, 104145. [Google Scholar] [CrossRef]
- Liu, C.; Niu, G.; Zhang, H.; Sun, Y.; Sun, S.; Yu, F.; Lu, S.; Yang, Y.; Li, J.; Hong, Z.; et al. Trimming of N-glycans by the Golgi-localized α-1, 2-mannosidases, MNS1 and MNS2, is crucial for maintaining RSW2 protein abundance during salt stress in Arabidopsis. Mol. Plant 2018, 11, 678–690. [Google Scholar] [CrossRef]
- Jiao, Q.-S.; Niu, G.-T.; Wang, F.-F.; Dong, J.-Y.; Chen, T.-S.; Zhou, C.-F.; Hong, Z. N-glycosylation regulates photosynthetic efficiency of Arabidopsis thaliana. Photosynthetica 2020, 58, 72–79. [Google Scholar] [CrossRef]
- Song, W.; Mentink, R.A.; Henquet, M.G.L.; Cordewener, J.H.G.; van Dijk, A.D.J.; Bosch, D.; America, A.H.P.; van der Krol, A.R. N-glycan occupancy of Arabidopsis N-glycoproteins. J. Proteom. 2013, 93, 343–355. [Google Scholar] [CrossRef]
- Ghosh, S.; Meli, V.S.; Kumar, A.; Thakur, A.; Chakraborty, N.; Chakraborty, S.; Datta, A. The N-glycan processing enzymes α-mannosidase and β-D-N-acetylhexosaminidase are involved in ripening-associated softening in the non-climateric fruits of capsicum. J. Exp. Bot. 2010, 62, 571–582. [Google Scholar] [CrossRef]
- Bose, S.K.; He, Y.; Howlader, P.; Wang, W.; Yin, H. The N-glycan processing enzymes beta-D-N acetylhexosaminidase are involvedin ripening-associated softening in strawberry fruit. J. Food Sci. Technol. 2020, 58, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; Van Damme, E.J. N-glycans: The making of a varied toolbox. Plant Sci. 2015, 239, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.; Seifert, G.; Doblin, M.S.; Johnson, K.L.; Ruprecht, C.; Pfrengle, F.; Bacic, A.; Estevez, J.M. Cracking the “Sugar Code”: A snapshot of N- and O-glycosylation pathways and functions in plants cells. Front. Plant Sci. 2021, 12, 640919. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Turek, I.; Parrott, B.; Thomas, L.; Jankovic, B.; Lilley, K.S.; Gehring, C. Structural and functional characteristics of cGMP-dependent methionine oxidation in Arabidopsis thaliana proteins. Cell Commun. Signal 2013, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Chibani, K.; Jacquot, J.-P.; Rouhier, N. Cysteine-based redox regulation and signaling in plants. Front. Plant Sci. 2013, 4, 105. [Google Scholar] [CrossRef]
- Tikkanen, M.; Aro, E.M. Thylakoid protein phosphorylation in dynamic regulation of photosystem II in higher plants. Biochim. Biophys. Acta 2012, 1817, 232–238. [Google Scholar] [CrossRef]
- Rochaix, J.D. Regulation and dynamics of the light-harvesting system. Annu. Rev. Plant Biol. 2014, 65, 287–309. [Google Scholar] [CrossRef]
- Michelet, L.; Zaffagnini, M.; Morisse, S.; Sparla, F.; Pérez-Pérez, M.E.; Francia, F.; Danon, A.; Marchand, C.; Fermani, S.; Trost, P.; et al. Redox regulation of the Calvin-Benson cycle: Something old, something new. Front. Plant Sci. 2013, 4, 470. [Google Scholar] [CrossRef]
- Buchanan, B.B.; Balmer, Y. Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 2005, 56, 187–220. [Google Scholar] [CrossRef]
- Pedersen, T.A.; Kirk, M.; Bassham, J.A. Inhibition of photophosphorylation and photosynthetic carbon cycle reactions by fatty acids and esters. Biochim. Biophys. Acta 1966, 112, 189–203. [Google Scholar] [CrossRef]
- Grabsztunowicz, M.; Koskela, M.M.; Mulo, P. Post-translational modifications in regulation of chloroplast function: Recent advances. Front. Plant Sci. 2017, 8, 240. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.; Durner, J.; Navarre, D.A.; Klessig, D.F. Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol. Plant Microbe Interact. 2000, 13, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Abat, J.K.; Mattoo, A.K.; Deswal, R. S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata- ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 2008, 275, 2862–2872. [Google Scholar] [CrossRef] [PubMed]
- Abat, J.K.; Deswal, R. Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: Change in S-nitrosylation of rubisco is responsible for the inactivation of its carboxylase activity. Proteomics 2009, 9, 4368–4380. [Google Scholar] [CrossRef] [PubMed]
- Kötting, O.; Kossmann, J.; Zeeman, S.C.; Lloyd, J.R. Regulation of starch metabolism: The age of enlightenment? Curr. Opin. Plant Biol. 2010, 13, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P. Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol. 2011, 155, 1566–1577. [Google Scholar] [CrossRef]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef]
- Lindermayr, C.; Sell, S.; Muller, B.; Leister, D.; Durner, J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 2010, 22, 2894–2907. [Google Scholar] [CrossRef]
- Akter, S.; Huang, J.; Waszczak, C.; Jacques, S.; Gevaert, K.; van Breusegem, F.; Messens, J. Cysteines under ROS attack in plants: A proteomics view J. Exp. Bot. 2015, 66, 2935–2944. [Google Scholar] [CrossRef]
- D’Autreaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef]
- Cheng, Z.; Wu, J.; Setterdahl, A.; Reddie, K.; Carroll, K.; Hammad, L.A.; Karty, J.A.; Bauer, C.E. Activity of the tetrapyrrole regulator CrtJ is controlled by oxidation of a redox active cysteine located in the DNA binding domain. Mol. Microbiol. 2012, 85, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Rey, P.; Tarrago, L. Physiological roles of plant methionine sulfoxide reductases in redox homeostasis and signaling. Antioxidants 2018, 7, 114. [Google Scholar] [CrossRef]
- Tossounian, M.A.; Wahni, K.; Van Molle, I.; Vertommen, D.; Rosado, L.A.; Messens, J. Redox regulated methionine oxidation of Arabidopsis thaliana glutathione transferase Phi9 induces H-site flexibility. Protein Sci. 2019, 28, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Rinalducci, S.; Murgiano, L.; Zolla, L. Redox proteomics: Basic principles and future perspectives for the detection of protein oxidation in plants. J. Exp. Bot. 2008, 59, 3781–3801. [Google Scholar] [CrossRef] [PubMed]
- Jacques, S.; Ghesquie, B.; De Bocka, P.-J.; Demola, H.; Wahni, K.; Willems, P.; Messens, J.; Van Breusegem, F.; Gevaert, K. Protein methionine sulfoxide dynamics in Arabidopsis thaliana under oxidative stress. Mol. Cell Prot. 2015, 14, 1217–1229. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Perluigi, M. Redox proteomics: A key tool for new insights into protein modification with relevance to disease. Antioxid. Redox Signal 2017, 26, 277–279. [Google Scholar] [CrossRef]
- Kwon, S.J.; Kwon, S.I.; Bae, M.S.; Cho, E.J.; Park, O.K. Role of the methionine sulfoxide reductase MsrB3 in cold acclimation in Arabidopsis. Plant Cell Physiol. 2007, 48, 1713–1723. [Google Scholar] [CrossRef]
- Stadtman, E.R.; van Remmen, H.; Richardson, A.; Wehr, N.B.; Levine, R.L. Methionine oxidation and aging. Biochim. Biophys. Acta 2005, 1703, 135–140. [Google Scholar] [CrossRef]
- Matamoros, M.A.; Becana, M. Molecular responses of legumes to abiotic stress: Post-translational modifications of proteins and redox signaling. J. Exp. Bot. 2021, 72, 5876–5892. [Google Scholar] [CrossRef]
- Kim, J.S.; Park, H.M.; Chae, S.; Lee, T.H.; Hwang, D.J.; Oh, S.D.; Park, J.S.; Song, D.G.; Pan, C.H.; Choi, D.; et al. A pepper MSRB2 gene confers drought tolerance in rice through the protection of chloroplast-targeted genes. PLoS ONE 2014, 9, e90588. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Li, C.W.; Koh, K.W.; Chuang, H.Y.; Chen, Y.R.; Lin, C.S.; Chan, M.T. MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress. J. Exp. Bot. 2014, 65, 5049–5062. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xian, M. Chemical methods to detect S-nitrosation. Curr. Opin. Chem. Biol. 2011, 15, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Leichert, L.I.; Gehrke, F.; Gudiseva, H.V.; Blackwell, T.; Ilbert, M.; Walker, A.K.; Strahler, J.R.; Andrews, P.C.; Jakob, U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 8197–8202. [Google Scholar] [CrossRef]
- Hagglund, P.; Bunkenborg, J.; Maeda, K.; Svensson, B. Identification of thioredoxin disulfide targets using a quantitative proteomics approach based on isotope-coded affinity tags. J. Proteome Res. 2008, 7, 5270–5276. [Google Scholar] [CrossRef]
- Aksnes, H.; Hole, K.; Arnesen, T. Molecular, cellular, and physiological significance of N-terminal acetylation. Int. Rev. Cell Mol. Biol. 2015, 316, 267–305. [Google Scholar]
- Arnesen, T.; Van Damme, P.; Polevoda, B.; Helsens, K.; Evjenth, R.; Colaert, N.; Varhaug, J.E.; Vandekerckhove, J.; Lillehaug, J.R.; Sherman, F.; et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA 2009, 106, 8157–8162. [Google Scholar] [CrossRef]
- Bienvenut, W.V.; Sumpton, D.; Martinez Am Lilla, S.; Espagne, C.; Meinnel, T.; Giglione, C. Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-α-acetylation features. Mol. Cell Prot. 2012, 11, M111.015131. [Google Scholar] [CrossRef]
- Falb, M.; Aivaliotis, M.; Garcia-Rizo, C.; Bisle, B.; Tebbe, A.; Klein, C.; Konstantinidis, K.; Siedler, F.; Pfeiffer, F.; Oesterhelt, D. Archael N-terminal protein maturation commonly involves N-terminal acetylation: A large-scale proteomics survey. J. Mol. Biol. 2006, 362, 915–924. [Google Scholar] [CrossRef]
- Starheim, K.K. Protein N-terminal acetyltransferases: When the start matters1. Trends Biochem. Sci. 2012, 2012, 152–161. [Google Scholar] [CrossRef]
- Pesaresi, P.; Gardner, N.A.; Masiero, S.; Dietzmann, A.; Eichacker, L.; Wickner, R.; Salamini, F.; Leister, D. Cytoplasmic N-terminal protein acetylation is required for efficient photosynthesis in Arabidopsis. Plant Cell 2003, 15, 1817–1832. [Google Scholar] [CrossRef] [PubMed]
- Giglione, C. N-terminal protein modifications: Bringing back into play the ribosome. Biochemie 2015, 114, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Linster, E.; Wirtz, M. N-terminal acetylation: An essential protein modification emerges as an important regulator of stress responses. J. Exp. Bot. 2018, 69, 4555–4568. [Google Scholar] [CrossRef]
- Dissmeyer, N.; Rivas, S.; Graciet, E. Life and death of proteins after protease cleavage: Protein degradation by the N-end rule pathway. New Phytol. 2018, 218, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Aksnes, H.; Drazic, A.; Marie, M.; Arnesen, T. First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem. Sci. 2016, 41, 746–760. [Google Scholar] [CrossRef]
- Gibbs, D.J. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol. 2014, 24, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Huang, Y.; Li, L.; Gannon, P.; Linster, E.; Huber, M.; Kapos, P.; Bienvenut, W.; Polevoda, B.; Meinnel, T.; et al. Two N-terminal acetyltransferases antagonistically regulate the stability of a nod-like receptor in Arabidopsis. Plant Cell 2015, 27, 1547–1562. [Google Scholar] [CrossRef]
- Silva, R.D.; Martinho, R.G. Developmental roles of protein N-terminal acetylation. Proteomics 2015, 15, 2402–2409. [Google Scholar] [CrossRef]
- Linster, E.; Stephan, I.; Bienvenut, W.V. Downregulation of N-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nat. Commun. 2015, 6, 7640. [Google Scholar] [CrossRef]
- Ferrandez-Ayela, A.; Micol-Ponce, R.; Sanchez-Garcia, A.B.; Alonso-Peral, M.M.; Micol, J.L.; Ponce, M.R. Mutation of an Arabidopsis NatB N-alpha-termianl acetylation complex component causes pleiotropic developmental defects. PLoS ONE 2013, 8, e80697. [Google Scholar]
- Huber, M.; Bienvenut, W.V.; Linster, E.; Stephan, I.; Armbruster, L.; Sticht, C.; Layer, D.C.; Lapouge, K.; Meinnel, T.; Sinning, I.; et al. NatB-mediated N-terminal acetylation affects growth and biotic stress responses. Plant Physiol. 2020, 182, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-Q.; Zou, Y.-J.; Li, X.-F.; Wu, L.; Guo, G.-Q. Stabilization of ACOs by NatB mediated-N-terminal acetylation is required for ethylene homeostasis. BMC Plant Biol. 2021, 21, 320. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Sharma, B.; Joshi, D.; Yadav, P.K.; Gupta, A.K.; Bhatt, T.K. Role of ubiquitin-mediated degradation system in plant biology. Front. Plant Sci. 2016, 7, 806. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, M.; Sarcevic, B. Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine. Cell Div. 2010, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Vierstra, R.D. The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 2011, 62, 299–334. [Google Scholar] [CrossRef]
- Doroodian, P.; Hua, Z. The ubiquitin switch in plant stress response. Plants 2021, 10, 246. [Google Scholar] [CrossRef]
- Abd-Hamid, N.A.; Ahmad-Fauzi, M.I.; Zainal, Z.; Ismail, I. Diverse and dynamic roles of F-box proteins in plant biology. Planta 2020, 251, 68. [Google Scholar] [CrossRef]
- Ban, Z.; Estelle, M. CUL3 E3 ligases in plant development and environmental response. Nat. Plants 2021, 7, 6–16. [Google Scholar] [CrossRef]
- Fonseca, S.; Rubio, V. Arabidopsis CRL4 complexes: Surveying chromatin states and gene expression. Front. Plant Sci. 2019, 10, 1095. [Google Scholar] [CrossRef]
- Barberon, M.; Zelazny, E.; Robert, S.; Conéjéro, G.; Curie, C.; Friml, J.; Vert, G. Monoubiquitin-dependent endocytosis of the IRON-REGULATED TRANSPORTER 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 2011, 108, E450–E458. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Li, X.; Wang, Z.; Ding, M.; Sun, Y.; Dong, F.; Chen, F.; Liu, L.; Doughty, J.; Li, Y.; et al. Histone H2B monoubiquitination mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 is involved in anther development by regulating tapetum degradation-related genes in rice. Plant Physiol. 2015, 168, 1389–1405. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Claus, L.A.N.; Leslie, M.E.; Kai, T.; Zhiping, W.; Jun, L.; Xiao, Y.; Bo, L.; Jinggeng, Z.; Daniel , V.S.; et al. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 2020, 581, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Grubb, L.E.; Derbyshire, P.; Dunning, K.E.; Zipfel, C.; Menke, F.L.H.; Monaghan, J. Large-scale identification of ubiquitination sites on membrane-associated proteins in Arabidopsis thaliana seedlings. Plant Physiol. 2021, 185, 1483–1488. [Google Scholar] [CrossRef] [PubMed]
- Orr, J.N.; Waugh, R.; Colas, I. Ubiquitination in plant meiosis: Recent advances and high throughput methods. Front. Plant Sci. 2021, 12, 667314. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Yang, M.; Wang, P.; Zhao, Y.; Ma, C. Interplay between the ubiquitin proteasome system and ubiquitin-mediated autophagy in plants. Cells 2016, 9, 2219. [Google Scholar] [CrossRef]
- Nagels Durand, A.; Pauwels, L.; Goossens, A. The ubiquitin system and jasmonate signaling. Plants 2016, 5, 6. [Google Scholar] [CrossRef]
- Adams, E.H.G.; Spoel, S.H. The ubiquitin-proteasome system as a transcriptional regulator of plant immunity. J. Exp. Bot. 2018, 69, 4529–4537. [Google Scholar] [CrossRef]
- Yu, F.; Wu, Y.; Xie, Q. Ubiquitin-proteasome system in ABA signaling: From perception to action. Mol. Plant 2016, 9, 21–33. [Google Scholar] [CrossRef]
- Chico, J.M.; Chini, A.; Fonseca, S.; Solano, R. JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 2008, 11, 486–494. [Google Scholar] [CrossRef]
- Chico, J.M.; Lechner, E.; Fernandez-Barbero, G.; Canibano, E.; García-Casado, G.; Franco-Zorrilla, J.M.; Hammann, P.; Zamarreño, A.M.; García-Mina, J.M.; Rubio, V.; et al. CUL3BPM E3 ubiquitin ligases regulate MYC2, MYC3, and MYC4 stability and JA responses. Proc. Natl. Acad. Sci. USA 2020, 117, 6205–6215. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467.e1415. [Google Scholar] [CrossRef]
- Skelly, M.J.; Furniss, J.J.; Grey, H.L.; Wong, K.W.; Spoel, S.H. Dynamic ubiquitination determines transcriptional activity of the plant immune coactivator NPR1. eLife 2019, 8, e47005. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zeng, L. Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity. Plant Commun. 2020, 1, 100041. [Google Scholar] [CrossRef] [PubMed]
- Denay, G.; Chahtane, H.; Tichtinsky, G.; Parcy, F. A flower is born: An update on Arabidopsis floral meristem formation. Curr. Opin. Plant Biol. 2017, 35, 15–22. [Google Scholar] [CrossRef]
- Chae, E.; Tan, Q.K.-G.; Hill, T.A.; Irish, V.F. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 2008, 135, 1235–1245. [Google Scholar] [CrossRef]
- Chahtane, H.; Zhang, B.; Norberg, M.; LeMasson, M.; Thevenon, E.; Bako, L.; Benlloch, R.; Holmlund, M.; Parcy, F.; Nilsson, O.; et al. LEAFY activity is post-transcriptionally regulated by BLADE ON PETIOLE2 and CULLIN3 in Arabidopsis. New Phytol. 2018, 220, 579–592. [Google Scholar] [CrossRef]
- Zhang, B.; Holmlund, M.; Lorrain, S.; Norberg, M.; Bako, L.; Fankhauser, C.; Nilsson, O. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife 2017, 6, e26759. [Google Scholar] [CrossRef]
- Ling, Q.; Huang, W.; Baldwin, A.; Jarvis, P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 2012, 338, 655–659. [Google Scholar] [CrossRef]
- Ling, Q.; Broad, W.; Trosch, R.; Topel, M.; Sert, T.D.; Lymperopoulos, P.; Baldwin, A.; Jarvis, P.R. Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 2019, 363, eaav4467. [Google Scholar] [CrossRef] [PubMed]
- Brunoud, G.; Wells, D.M.; Oliva, M.; Larrieu, A.; Mirabet, V.; Beeckman, T.; Kepinski, S.; Traas, J.; Bennett, M.J.; Vernoux, T. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 2012, 482, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Martin-Arevalillo, R.; Vernoux, T. Shining light on plant hormones with genetically encoded biosensors. Biol. Chem. 2019, 400, 477–486. [Google Scholar] [CrossRef]
- Yang, X.; Wen, Z.; Zhang, D.; Li, Z.; Li, D.; Nagalakshmi, U.; Dinesh-Kumar, S.P.; Zhang, Y. Proximity labeling: An emerging tool for probing in planta molecular interactions. Plant Commun. 2021, 2, 100137. [Google Scholar] [CrossRef] [PubMed]
- Melchior, F. SUMO: Ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 2003, 28, 612–618. [Google Scholar] [CrossRef]
- Hickey, C.M.; Wilson, N.R.; Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012, 13, 755–766. [Google Scholar] [CrossRef]
- Lois, L.M.; Lima, C.D. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005, 24, 439–451. [Google Scholar] [CrossRef]
- Reiter, K.H.; Ramachandran, A.; Xia, X.; Boucher, L.E.; Bosch, J.; Matunis, M.J. Characterization and structural insights into selective E1-E2 interactions in the human and Plasmodium falciparum SUMO conjugation systems. J. Biol. Chem. 2016, 291, 3860–3870. [Google Scholar] [CrossRef]
- Liu, B.; Lois, L.M.; Reverter, D. Structural insights into SUMO E1–E2 interactions in Arabidopsis uncovers a distinctive platform for securing SUMO conjugation specificity across evolution. Biochem. J. 2019, 476, 2127–2139. [Google Scholar] [CrossRef]
- Liu, B.; Lois, L.M.; Reverter, D. Structural analysis and evolution of specificity of the SUMO UFD E1-E2 interactions. Sci. Rep. 2017, 7, 41998. [Google Scholar] [CrossRef]
- Bernier-Villamor, V.; Sampson, D.A.; Matunis, M.J.; Lima, C.D. Structural Basis for E2-mediated SUMO Conjugation Revealed by a Complex between Ubiquitin-Conjugating Enzyme Ubc9 and RanGAP1. Cell 2002, 108, 345–356. Available online: https://www.ncbi.nlm.nih.gov/pubmed/11853669 (accessed on 17 February 2022).
- Augustine, R.C.; Vierstra, R.D. SUMOylation: Re-wiring the plant nucleus during stress and development. Curr. Opin. Plant Biol. 2018, 45, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, I.A.; D’Souza, R.C.J.; Yang, B.; Verlaan-de Vries, M.; Mann, M.; Vertegaal, A.C.O. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014, 21, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Tomanov, K.; Zeschmann, A.; Hermkes, R.; Eifler, K.; Ziba, I.; Grieco, M.; Novatchkova, M.; Hofmann, K.; Hesse, H.; Bachmair, A. Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 2014, 26, 4547–4560. [Google Scholar] [CrossRef] [PubMed]
- Geoffroy, M.C.; Hay, R.T. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 2009, 10, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007, 8, 550–555. [Google Scholar] [CrossRef]
- Elrouby, N. Regulation of plant cellular and organismal development by SUMO. Adv. Exp. Med. Biol. 2017, 963, 227–247. [Google Scholar] [CrossRef]
- Saracco, S.A.; Miller, M.J.; Kurepa, J.; Vierstra, R.D. Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007, 145, 119–134. [Google Scholar] [CrossRef]
- Miura, K.; Lee, J.; Jin, J.B.; Yoo, C.Y.; Miura, T.; Hasegawa, P.M. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 5418–5423. [Google Scholar] [CrossRef]
- Lois, L.M.; Lima, C.D.; Chua, N.-H. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 2003, 15, 1347–1359. [Google Scholar] [CrossRef]
- Lee, J.; Nam, J.; Park, H.C.; Na, G.; Miura, K.; Jin, J.B.; Yoo, C.Y.; Baek, D.; Kim, D.H.; Jeong, J.C.; et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007, 49, 79–90. [Google Scholar] [CrossRef]
- Orosa-Puente, B.; Leftley, N.; von Wangenheim, D.; Banda, J.; Srivastava, A.K.; Hill, K.; Truskina, J.; Bhosale, R.; Morris, E.; Srivastava, M.; et al. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 2018, 362, 1407–1410. [Google Scholar] [CrossRef] [PubMed]
- Benlloch, R.; Lois, L.M. Sumoylation in plants: Mechanistic insights and its role in drought stress. J. Exp. Bot. 2018, 69, 4539–4554. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Fuertes, D.; Perez-Gil, J.; Lois, L.M. SUMOylation in phytopathogen interactions: Balancing invasion and resistance. Front. Cell Dev. Biol. 2021, 9, 703795. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Miquel, L.; Seguí, J.; Lois, L.M. Distinctive properties of Arabidopsis SUMO paralogues support the in vivo predominant role of AtSUMO1/2 isoforms. Biochem. J. 2011, 436, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Van den Burg, H.A.; Kini, R.K.; Schuurink, R.C.; Takken, F.L. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 2010, 22, 1998–2016. [Google Scholar] [CrossRef]
- Saleh, A.; Withers, J.; Mohan, R.; Marqués, J.; Gu, Y.; Yan, S.; Zavaliev, R.; Nomoto, M.; Tada, Y.; Dong, X. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 2015, 18, 169–182. [Google Scholar] [CrossRef]
- Castaño-Miquel, L.; Seguí, J.; Manrique, S.; Teixeira, I.; Carretero-Paulet, L.; Atencio, F.; Lois, L.M. Diversification of SUMO-activating enzyme in Arabidopsis: Implications in SUMO conjugation. Mol. Plant 2013, 6, 1646–1660. [Google Scholar] [CrossRef][Green Version]
- Castro, P.H.; Bachmair, A.; Bejarano, E.R.; Coupland, G.; Lois, L.M.; Sadanandom, A.; Van Den Burg, H.A.; Vierstra, R.D.; Azevedo, H. Revised nomenclature and functional overview of the ULP gene family of plant deSUMOylating proteases. J. Exp. Bot. 2018, 69, 4505–4509. [Google Scholar] [CrossRef]
- Roy, D.; Sadanandom, A. SUMO mediated regulation of transcription factors as a mechanism for transducing environmental cues into cellular signaling in plants. Cell. Mol. Life Sci. 2021, 78, 2641–2664. [Google Scholar] [CrossRef]
- Miller, M.J.; Barrett-Wilt, G.A.; Hua, Z.; Vierstra, R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 16512–16517. [Google Scholar] [CrossRef]
- Nukarinen, E.; Tomanov, K.; Ziba, I.; Weckwerth, W.; Bachmair, A. Protein sumoylation and phosphorylation intersect in Arabidopsis signaling. Plant J. 2017, 91, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Miquel, L.; Mas, A.; Teixeira, I.; Seguí, J.; Perearnau, A.; Thampi, B.N.; Schapire, A.L.; Rodrigo, N.; La Verde, G.; Manrique, S.; et al. SUMOylation inhibition mediated by disruption of SUMO E1-E2 interactions confers plant susceptibility to necrotrophic fungal pathogens. Mol. Plant 2017, 10, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Morrell, R.; Sadanandom, A. Dealing with stress: A review of plant SUMO proteases. Front. Plant Sci. 2019, 10, 1122. [Google Scholar] [CrossRef] [PubMed]
- Rosa, M.T.; Abreu, I.A. Exploring the regulatory levels of SUMOylation to increase crop productivity. Curr. Opin. Plant Biol. 2019, 49, 43–51. [Google Scholar] [CrossRef]
- Tarcsa, E.; Marekov, L.N.; Mei, G.; Melino, G.; Lee, S.C.; Steinert, P.M. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J. Biol. Chem. 1996, 271, 30709–30716. [Google Scholar] [CrossRef]
- Smith, D.G.; Young, E.G. The combined amino acids in several species of marine algae. J. Biol. Chem. 1955, 217, 845–853. [Google Scholar] [CrossRef]
- Linsky, T.; Fast, W. Mechanistic similarity and diversity among the guanidine-modifying members of the pentein superfamily. Biochim. Biophys. Acta 2010, 1804, 1943–1953. [Google Scholar] [CrossRef][Green Version]
- Rogers, G.E.; Simmonds, D.H. Content of citrulline and other amino-acids in a protein of hair follicles. Nature 1958, 182, 186–187. [Google Scholar] [CrossRef]
- Christophorou, M.A.; Castelo-Branco, G.; Halley-Stott, R.P.; Oliveira, C.S.; Loos, R.; Radzisheuskaya, A.; Mowen, K.A.; Bertone, P.; Silva, J.C.; Zernicka-Goetz, M.; et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 2014, 507, 104–108. [Google Scholar] [CrossRef]
- Yuzhalin, A.E. Citrullination in Cancer. Cancer Res. 2019, 79, 1274–1284. [Google Scholar] [CrossRef]
- Van Venrooij, W.J.; Pruijn, G.J. Citrullination: A small change for a protein with great consequences for rheumatoid arthritis. Arthritis Res. 2000, 2, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Klareskog, L.; Lundberg, K.; Malmstrom, V. Autoimmunity in rheumatoid arthritis: Citrulline immunity and beyond. Adv. Immunol. 2013, 118, 129–158. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Elia, G.; Thomas, L.; Wong, A.; Gehring, C. Citrullination of proteins as a specific response mechanism in plants. Front. Plant Sci. 2021, 12, 377. [Google Scholar] [CrossRef] [PubMed]
- Joshi, V.; Fernie, A.R. Citrulline metabolism in plants. Amino Acids 2017, 49, 1543–1559. [Google Scholar] [CrossRef]
- Hashigushi, A.; Komatsu, S. Posttranslational modifications and plant–environment interaction. In Methods in Enzymology; Shukla, A.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; Volume 586, pp. 97–113. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muleya, V.; Lois, L.M.; Chahtane, H.; Thomas, L.; Chiapello, M.; Marondedze, C. (De)Activation (Ir)Reversibly or Degradation: Dynamics of Post-Translational Protein Modifications in Plants. Life 2022, 12, 324. https://doi.org/10.3390/life12020324
Muleya V, Lois LM, Chahtane H, Thomas L, Chiapello M, Marondedze C. (De)Activation (Ir)Reversibly or Degradation: Dynamics of Post-Translational Protein Modifications in Plants. Life. 2022; 12(2):324. https://doi.org/10.3390/life12020324
Chicago/Turabian StyleMuleya, Victor, L. Maria Lois, Hicham Chahtane, Ludivine Thomas, Marco Chiapello, and Claudius Marondedze. 2022. "(De)Activation (Ir)Reversibly or Degradation: Dynamics of Post-Translational Protein Modifications in Plants" Life 12, no. 2: 324. https://doi.org/10.3390/life12020324
APA StyleMuleya, V., Lois, L. M., Chahtane, H., Thomas, L., Chiapello, M., & Marondedze, C. (2022). (De)Activation (Ir)Reversibly or Degradation: Dynamics of Post-Translational Protein Modifications in Plants. Life, 12(2), 324. https://doi.org/10.3390/life12020324






