A Free-Energy Landscape Analysis of Calmodulin Obtained from an NMR Data-Utilized Multi-Scale Divide-and-Conquer Molecular Dynamics Simulation
Abstract
:1. Introduction
2. Materials and Methods
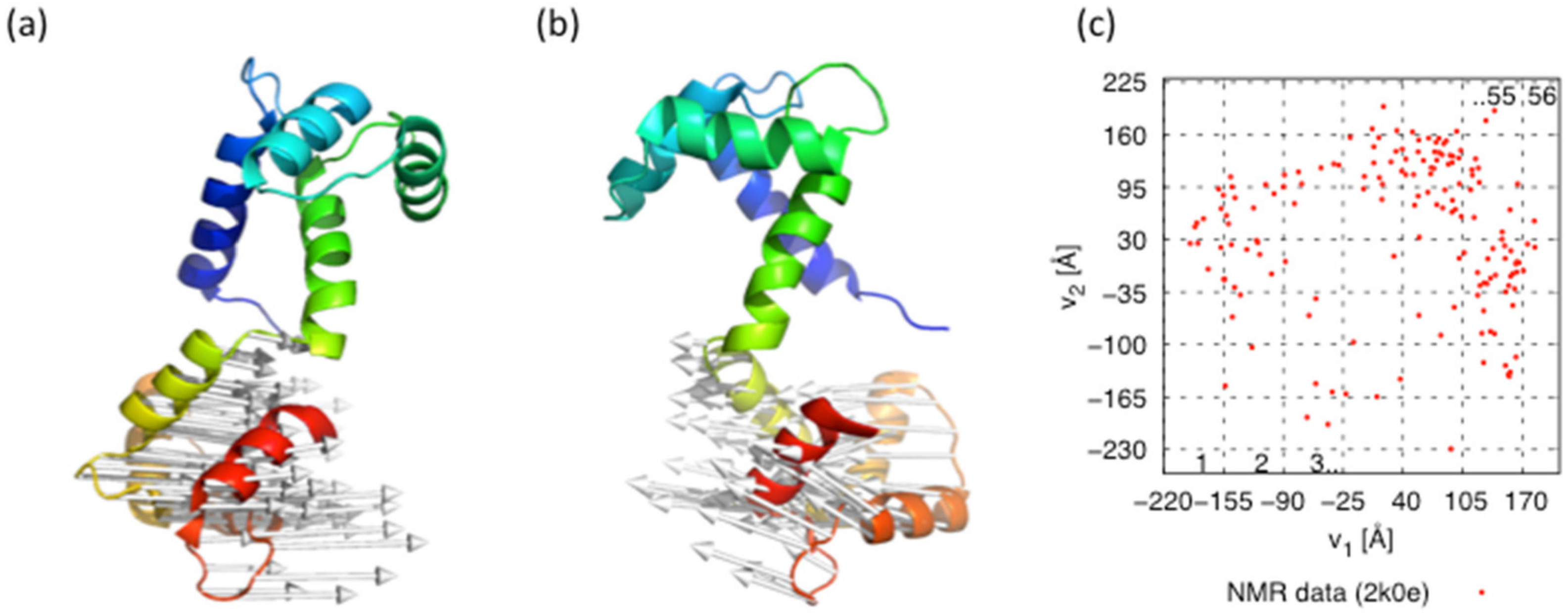
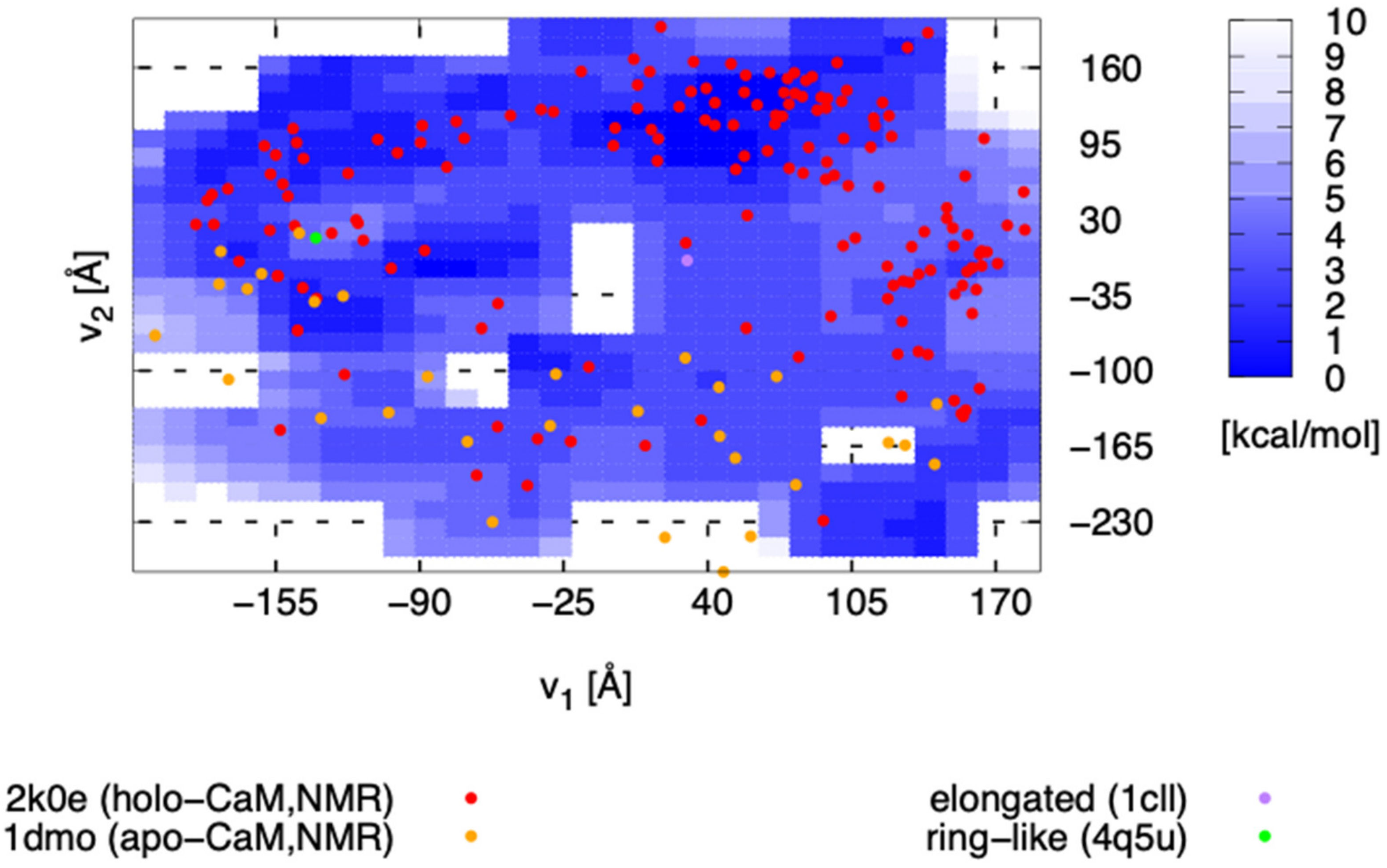
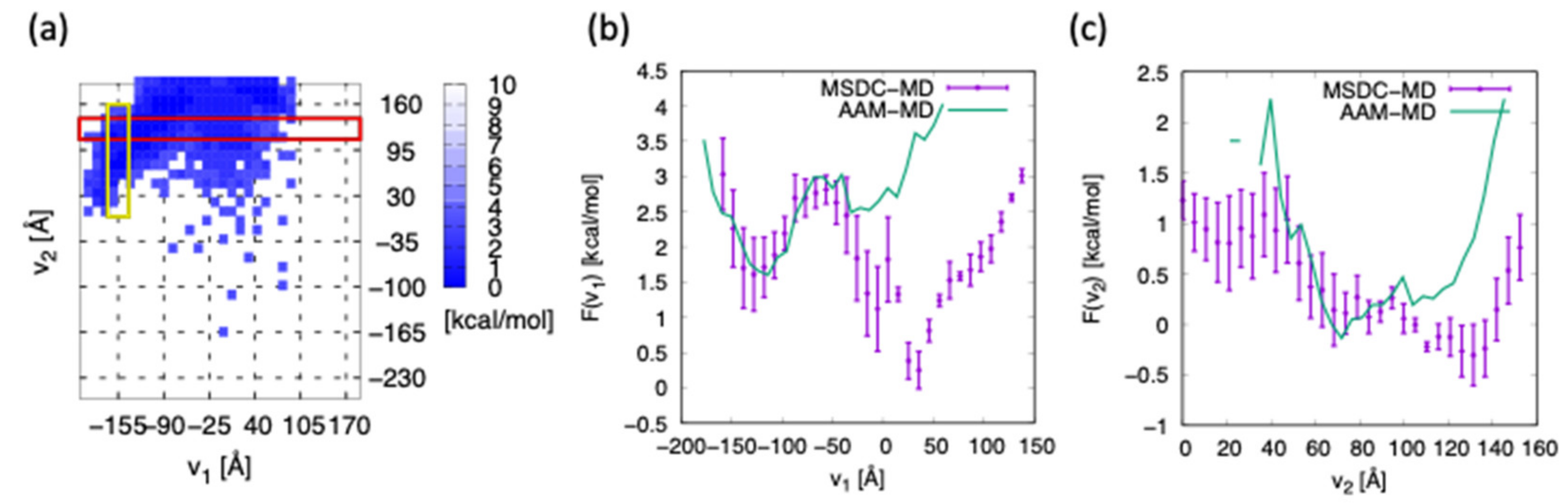
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Putney, J.W. Calcium signaling. Cell 2005, 80, 1–540. [Google Scholar]
- Berridge, M.J.; Bootman, M.D.; Lipp, P. Calcium—A life and death signal. Nature 1998, 395, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Chin, D.; Means, A.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef]
- Gifford, J.; Walsh, M.; Vogel, H. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef]
- Klee, C.B.; Vanaman, T.C. Calmodulin. Adv. Protein Chem. 1982, 35, 213–321. [Google Scholar] [PubMed]
- Kakiuchi, S.; Yamazaki, R. Calcium dependent phosphodiesterase activity and its activating factor (PAF) from brain. Studies on cyclic 3′,5′-nucleotide phosphodiesterase (III). Biochem. Biophys. Res. Commun. 1970, 41, 1104–1110. [Google Scholar] [CrossRef]
- Cheung, W.Y. Cyclic 3′,5′-nucleotide phosphodiesterase. Demonstration of an activator. Biochem. Biophys. Res. Commun. 1970, 38, 533–538. [Google Scholar] [CrossRef]
- Teo, T.S.; Wang, T.H.; Wang, J.H. Purification and properties of the protein activator of bovine heart cyclic adenosine 3′,5′-monophosphate phosphodiesterase. J. Biol. Chem. 1973, 248, 588–595. [Google Scholar] [CrossRef]
- Teo, T.S.; Wang, J.H. Mechanism of activation of a cyclic adenosine 3′:5′ monophosphate phosphodiesterase from bovine heart by calcium ions. Identification of the protein activator as a Ca2+ binding protein. J. Biol. Chem. 1973, 248, 5950–5955. [Google Scholar] [CrossRef]
- Kumar, V.; Chichili, V.; Zhong, L. Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin. Sci. Rep. 2013, 3, 1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tansey, M.G.; Luby-Phelps, K.; Kamm, K.E.; Stull, J.T. Ca2+-dependent phosphorylation of myosin light chain kinase decreases the Ca2+ sensitivity of light chain phosphorylation within smooth muscle cells. J. Biol. Chem. 1994, 269, 9912–9920. [Google Scholar] [CrossRef]
- Means, A.R.; Chafouleas, J.G. Calmodulin is involved in the regulation of cell proliferation. Cell Biol. Int. Rep. 1983, 7, 481–482. [Google Scholar] [CrossRef] [Green Version]
- Kawai, T.; Nomura, F.; Katsuaki, H.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Shizuo, A. Death-associated protein kinase 2 is a new calcium/calmodulin-dependent protein kinase that signals apoptosis through its catalytic activity. Oncogene 1999, 18, 3471–3480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fladmark, K.E.; Fladmark, K.E.; Brustugun, O.T.; Mellgren, G.; Krakstad, C.; Bøe, R.; Vintermyr, O.K.; Schulman, H.; Døskeland, S.O. Ca2+/calmodulin-dependent protein kinase II is required for microcystin-induced apoptosis. J. Biol. Chem. 2002, 277, 2804–2811. [Google Scholar] [CrossRef] [Green Version]
- Racioppi, L.; Means, A.R. Calcium/calmodulin-dependent kinase IV in immune and inflammatory responses: Novel routes for an ancient traveller. Trends Immunol. 2008, 29, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Curtis, J.; Finkbeiner, S. Sending signals from the synapse to the nucleus: Possible roles for CaMK, Ras/ERK, and SAPK pathways in the regulation of synaptic plasticity and neuronal growth. J. Neurosci. Res. 1999, 58, 88–95. [Google Scholar] [CrossRef]
- Prichard, L.; Deloulme, J.C.; Storm, D.R. Interactions between neurogranin and calmodulin in vivo. J. Biol. Chem. 1999, 274, 7689–7694. [Google Scholar] [CrossRef] [Green Version]
- Soderling, T.R. CaM-kinases: Modulators of synaptic plasticity. Curr. Opin. Neurobiol. 2000, 10, 375–380. [Google Scholar] [CrossRef]
- Ikura, M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem. Sci. 1996, 21, 14–17. [Google Scholar] [CrossRef]
- Grabarek, Z. Structural Basis for Diversity of the EF-hand Calcium-binding Proteins. J. Mol. Biol. 2006, 359, 509–525. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, W.; Kirberger, M.; Lee, H.W.; Ayalasomayajula, G.; Yang, J.J. Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins Struct. Funct. Genet. 2006, 65, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Lewit-Bentley, A.; Réty, S. EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 2000, 10, 637–643. [Google Scholar] [CrossRef]
- Kawasaki, H.; Nakayama, S.; Kretsinger, R.H. Classification and evolution of EF-hand proteins. BioMetals 1998, 11, 277–295. [Google Scholar] [CrossRef]
- Ravasi, T.; Hsu, K.; Goyette, J.; Schroder, K.; Yang, Z.; Rahimi, F.; Miranda, L.P.; Alewood, P.F.; Hume, D.A.; Geczy, C. Probing the S100 protein family through genomic and functional analysis. Genomics 2004, 84, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Mcphalen, C.A.; Strynadka, N.C.J.; James, M.N.G. Calcium-Binding Sites in Proteins: A Structural Perspective. Adv. Protein Chem. 1991, 42, 77–82. [Google Scholar] [PubMed]
- Nelson, M.R.; Chazin, W.J. Structures of EF-hand Ca2+-binding proteins: Diversity in the organization, packing and response to Ca2+ binding. Biometals 1998, 11, 297–318. [Google Scholar] [CrossRef]
- Strynadka, N.C.J.; James, M.N.G. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu. Rev. Biochem. 1989, 58, 951–999. [Google Scholar] [CrossRef]
- Chattopadhyaya, R.; Meador, W.E.; Means, A.R.; Quiocho, F.A. Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol. 1992, 228, 1177–1192. [Google Scholar] [CrossRef]
- Babu, Y.S.; Bugg, C.E.; Cook, W.J. Structure of calmodulin refined at 2.2 Å resolution. J. Mol. Biol. 1988, 204, 191–204. [Google Scholar] [CrossRef]
- Taylor, D.A.; Sack, J.S.; Maune, J.F.; Beckingham, K.; Quiocho, F.A. Structure of a recombinant calmodulin from Drosophila melanogaster refined at 2.2 Å resolution. J. Biol. Chem. 1991, 266, 21375–21380. [Google Scholar] [CrossRef]
- Rao, S.T.; Wu, S.; Satyshur, K.A.; Sundaralingam, M.; Ling, K.Y.; Kung, C. Structure of Paramecium tetraurelia calmodulin at 1.8 Å resolution. Protein Sci. 1993, 2, 436–447. [Google Scholar] [CrossRef] [Green Version]
- Ban, C.; Ramakrishnan, B.; Ling, K.Y.; Ching, K.; Sundaralingam, M. Structure of the recombinant Paramecium tetraurelia calmodulin at 1.68 Angstroms resolutin. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Rupp, B.; Marshak, D.R.; Parkin, S. Crystallization and preliminary X-ray analysis of two new crystal forms of calmodulin. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996, 52, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A.; Brunger, A.T. The 1.0 Å crystal structure of Ca2+-bound calmodulin: An analysis of disorder and implications for functionally relevant plasticity. J. Mol. Biol. 2000, 301, 1237–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, C.H.; Bai, J.; Sun, D.Y.; Cui, D.F.; Chang, W.R.; Liang, D.C. Structure of potato calmodulin PCM6: The first report of the three-dimensional structure of a plant calmodulin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 1214–1219. [Google Scholar] [CrossRef]
- Lin, J.; van den Bedem, H.; Brunger, A.T.; Wilson, M.A. Atomic resolution experimental phase information reveals extensive disorder and bound 2-methyl-2,4-pentanediol in Ca2+-calmodulin. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Mazumder, M.; Gupta, N.; Chattopadhyay, S.; Gourinath, S. Crystal structure of Arabidopsis thaliana calmodulin7 and insight into its mode of DNA binding. FEBS Lett. 2016, 590, 3029–3039. [Google Scholar] [CrossRef]
- Zhang, M.; Tanaka, T.; Ikura, M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 1995, 2, 758–767. [Google Scholar] [CrossRef]
- Ishida, H.; Nakashima, K.; Kumaki, Y.; Nakata, M.; Hikichi, K.; Yazawa, M. The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property. Biochemistry 2002, 41, 15536–15542. [Google Scholar] [CrossRef]
- Rellos, P.; Pike, A.C.W.; Niesen, F.H.; Salah, E.; Lee, W.H.; von Delft, F.; Knapp, S. Structure of the CaMKIIδ/calmodulin complex reveals the molecular mechanism of CamKII kinase activation. PLoS Biol. 2010, 8, e1000426. [Google Scholar] [CrossRef] [Green Version]
- Gifford, J.L.; Ishida, H.; Vogel, H.J. Fast methionine-based solution structure determination of calcium-calmodulin complexes. J. Biomol. NMR 2011, 50, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Meador, W.E.; Means, A.R.; Quiocho, F.A. Target enzyme recognition by calmodulin: 2.4 Å structure of a calmodulin-peptide complex. Science 1992, 257, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Gsponer, J.; Christodoulou, J.; Cavalli, A.; Bui, J.M.; Richter, B.; Dobson, C.M.; Vendruscolo, M. A Coupled Equilibrium Shift Mechanism in Calmodulin-Mediated Signal Transduction. Structure 2008, 16, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Grishaev, A.; Anthis, N.J.; Clore, G.M. Contrast-matched small-angle x-ray scattering from a heavy-atom-labeled protein in structure determination: Application to a lead-substituted calmodulin-peptide complex. J. Am. Chem. Soc. 2012, 134, 14686–14689. [Google Scholar] [CrossRef] [Green Version]
- Ye, Q.; Wang, H.; Zheng, J.; Wei, Q.; Jia, Z. The complex structure of calmodulin bound to a calcineurin peptide. Proteins Struct. Funct. Genet. 2008, 73, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Majava, V.; Kursula, P. Domain swapping and different oligomeric states for the complex between calmodulin and the calmodulin-binding domain of calcineurin A. PLoS ONE 2009, 4, e5402. [Google Scholar] [CrossRef] [Green Version]
- Fallon, J.L.; Baker, M.R.; Xiong, L.; Loy, R.E.; Yang, G.; Dirksen, R.T.; Hamilton, S.L.; Quiocho, F.A. Crystal structure of dimeric cardiac L-type calcium channel regulatory domains bridged by Ca2+·calmodulins. Proc. Natl. Acad. Sci. USA 2009, 106, 5135–5140. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, T.B.; Guo, H.F.; Cook, E.C.; Holbrook, E.; Rumi-Masante, J.; Lester, T.E.; Colbert, C.L.; Vander, K.; Craig, W.; Creamer, T.P. Stoichiometry of the Calcineurin Regulatory Domain-Calmodulin Complex. Biochemistry 2014, 53, 5779–5790. [Google Scholar] [CrossRef]
- De Diego, I.; Kuper, J.; Bakalova, N.; Kursula, P.; Wilmanns, M. Molecular basis of the death-associated protein kinase-calcium/calmodulin regulator complex. Sci. Signal. 2010, 3, ra6. [Google Scholar] [CrossRef]
- Bertini, I.; Kursula, P.; Luchinat, C.; Parigi, G.; Vahokoski, J.; Wilmanns, M.; Yuan, J. Accurate solution structures of proteins from X-ray data and a minimal set of NMR Data: Calmodulin-peptide complexes as examples. J. Am. Chem. Soc. 2009, 131, 5134–5144. [Google Scholar] [CrossRef]
- Shen, Y.; Lee, Y.S.; Soelaiman, S.; Bergson, P.; Lu, D.; Chen, A.; Beckingham, K.; Grabarek, Z.; Mrksich, M.; Tang, W.J. Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins. EMBO J. 2002, 21, 6721–6732. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Guo, Q.; Zhukovskaya, N.L.; Drum, C.L.; Bohm, A.; Tang, W.J. Structure of anthrax edema factor-calmodulin-adenosine 5′-(α, β-methylene)-triphosphate complex reveals an alternative mode of ATP binding to the catalytic site. Biochem. Biophys. Res. Commun. 2004, 317, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Shen, Y.; Zhukovskaya, N.L.; Florián, J.; Tang, W.J. Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate. J. Biol. Chem. 2004, 279, 29427–29435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Zhukovskaya, N.L.; Guo, Q.; Florián, J.; Tang, W.J. Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor. EMBO J. 2005, 24, 929–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, C.; Misra, I.; Iyanagi, T.; Kim, J.J.P. Regulation of interdomain interactions by calmodulin in inducible nitric-oxide synthase. J. Biol. Chem. 2009, 284, 30708–30717. [Google Scholar] [CrossRef] [Green Version]
- Piazza, M.; Futrega, K.; Spratt, D.E.; Dieckmann, T.; Guillemette, J.G. Structure and dynamics of calmodulin (CaM) bound to nitric oxide synthase peptides: Effects of a phosphomimetic CaM mutation. Biochemistry 2012, 51, 3651–3661. [Google Scholar] [CrossRef]
- Piazza, M.; Taiakina, V.; Guillemette, S.R.; Guillemette, J.G.; Dieckmann, T. Solution structure of calmodulin bound to the target peptide of endothelial nitric oxide synthase phosphorylated at Thr495. Biochemistry 2014, 53, 1241–1249. [Google Scholar] [CrossRef] [Green Version]
- Piazza, M.; Dieckmann, T.; Guillemette, J.G. Structural Studies of a Complex between Endothelial Nitric Oxide Synthase and Calmodulin at Physiological Calcium Concentration. Biochemistry 2016, 55, 5962–5971. [Google Scholar] [CrossRef]
- Tidow, H.; Nissen, P. Structural diversity of calmodulin binding to its target sites. FEBS J. 2013, 280, 5551–5565. [Google Scholar] [CrossRef]
- Kern, D.; Zuiderweg, E.R.P. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003, 13, 748–757. [Google Scholar] [CrossRef]
- Swain, J.F.; Gierasch, L.M. The changing landscape of protein allostery. Curr. Opin. Struct. Biol. 2006, 16, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Bohr, C.; Hasselbalch, K.; Krogh, A. Ueber einen in biologischer Beziehung wichtigen Einfluss, den die Kohlensäurespannung des Blutes auf dessen Sauerstoffbindung übt. Skand. Arch. Physiol. 1904, 16, 402–412. [Google Scholar] [CrossRef]
- Liu, J.; Nussinov, R. Allostery: An Overview of Its History, Concepts, Methods, and Applications. PLoS Comput. Biol. 2016, 12, e1004966. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, H. A Structural Comparison of ‘Real’ and ‘Model’ Calmodulin Clarified Allosteric Interactions Regulating Domain Motion. J. Biomol. Struct. Dyn. 2018, 37, 1567–1581. [Google Scholar] [CrossRef] [PubMed]
- Komeiji, Y.; Ueno, Y.; Uebayasi, M. Molecular dynamics simulations revealed Ca2+-dependent conformational change of Calmodulin. FEBS Lett. 2002, 521, 133–139. [Google Scholar] [CrossRef] [Green Version]
- Yamada, Y.; Matsuo, T.; Iwamoto, H.; Yagi, N. A compact intermediate state of calmodulin in the process of target binding. Biochemistry 2012, 51, 3963–3970. [Google Scholar] [CrossRef]
- Torrie, G.M.; Valleau, J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977, 23, 187–199. [Google Scholar] [CrossRef]
- Pai-Chi, L.; Miyashita, N.; Im, W.; Ishido, S.; Sugita, Y. Multidimensional umbrella sampling and replica-exchange molecular dynamics simulations for structure prediction of transmembrane helix dimers. J. Comput. Chem. 2013, 35, 300–308. [Google Scholar]
- Leitgeb, M.; Schröder, C.; Boresch, S. Alchemical free energy calculations and multiple conformational substates. J. Chem. Phys. 2005, 122, 084109. [Google Scholar] [CrossRef]
- Wojtas-Niziurski, W.; Meng, Y.; Roux, B.; Berneche, S. Self-Learning Adaptive Umbrella Sampling Method for the Determination of Free Energy Landscapes in Multiple Dimensions. J. Chem. Theory. Comput. 2013, 9, 1885–1895. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Shao, Q.; Xu, Z.; Liu, Y.; Yang, Z.; Cossins, B.P.; Jiang, H.; Chen, K.; Shi, J.; Zhu, W. Exploring transition pathway and free-energy profile of large-scale protein conformational change by combining normal mode analysis and umbrella sampling molecular dynamics. J. Phys. Chem. B 2014, 118, 134–143. [Google Scholar] [CrossRef]
- Shimoyama, H.; Yonezawa, Y. Atomistic detailed free-energy landscape of intrinsically disordered protein studied by multi-scale divide-and-conquer molecular dynamics simulation. J. Comput. Chem. 2021, 42, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Kurkcuoglu, Z.; Bahar, I.; Doruker, P. ClustENM: ENM-Based Sampling of Essential Conformational Space at Full Atomic Resolution. J. Chem. Theory Comput. 2016, 12, 4549–4562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabsch, W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. Sect. A Cryst. Phys. Diffr. Theor. Gen. Crystallogr. 1976, 32, 922–923. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van Der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open-source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase Quantum Mechanical Calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Shimoyama, H.; Shitaka-Takeda, M. Residue-residue interactions regulating the Ca2+-induced EF-hand conformation changes in calmodulin. J. Biochem. 2017, 162, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Cheatham, T.E.; Miller, J.L.; Fox, T.; Darden, T.A.; Kollman, P.A. Molecular Dynamics Simulations on Solvated Biomolecular Systems: The Particle Mesh Ewald Method Leads to Stable Trajectories of DNA, RNA, and Proteins. J. Am. Chem. Soc. 1995, 117, 4193–4194. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Van Gunsteren, W.F.; Berendsen, H.J.C. Computer Simulation of Molecular Dynamics: Methodology, Applications, and Perspectives in Chemistry. Angew. Chemie Int. Ed. 1990, 29, 992–1023. [Google Scholar] [CrossRef]
- DeLano, W. PyMOL. 2020. Available online: http://www.pymol.org/pymol (accessed on 30 September 2021).
- R Development Core Team 3.0.1. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Volume 201, pp. 1–16. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimoyama, H.; Shigeta, Y. A Free-Energy Landscape Analysis of Calmodulin Obtained from an NMR Data-Utilized Multi-Scale Divide-and-Conquer Molecular Dynamics Simulation. Life 2021, 11, 1241. https://doi.org/10.3390/life11111241
Shimoyama H, Shigeta Y. A Free-Energy Landscape Analysis of Calmodulin Obtained from an NMR Data-Utilized Multi-Scale Divide-and-Conquer Molecular Dynamics Simulation. Life. 2021; 11(11):1241. https://doi.org/10.3390/life11111241
Chicago/Turabian StyleShimoyama, Hiromitsu, and Yasuteru Shigeta. 2021. "A Free-Energy Landscape Analysis of Calmodulin Obtained from an NMR Data-Utilized Multi-Scale Divide-and-Conquer Molecular Dynamics Simulation" Life 11, no. 11: 1241. https://doi.org/10.3390/life11111241
APA StyleShimoyama, H., & Shigeta, Y. (2021). A Free-Energy Landscape Analysis of Calmodulin Obtained from an NMR Data-Utilized Multi-Scale Divide-and-Conquer Molecular Dynamics Simulation. Life, 11(11), 1241. https://doi.org/10.3390/life11111241






