A Soft Voting Ensemble-Based Model for the Early Prediction of Idiopathic Pulmonary Fibrosis (IPF) Disease Severity in Lungs Disease Patients
Abstract
:1. Introduction
- ◦
- Applying different state-of-art machine learning algorithms on IPF data;
- ◦
- Investigating the performance of all applied machine learning algorithms;
- ◦
- Developing a soft voting ensemble model for the prediction of severity of IPF disease in patients with lung disease;
- ◦
- Evaluating the performance of the proposed soft-voting ensemble-based model; and
- ◦
- To the best of our knowledge, we are among the pioneers for applying machine learning and proposing a soft-voting ensemble approach on an IPF dataset with improved accuracy.
2. Related Works
3. Research Materials and Methods
3.1. Data Source
3.2. Data Preprocessing
3.3. Data Sampling
3.4. Feature Selection
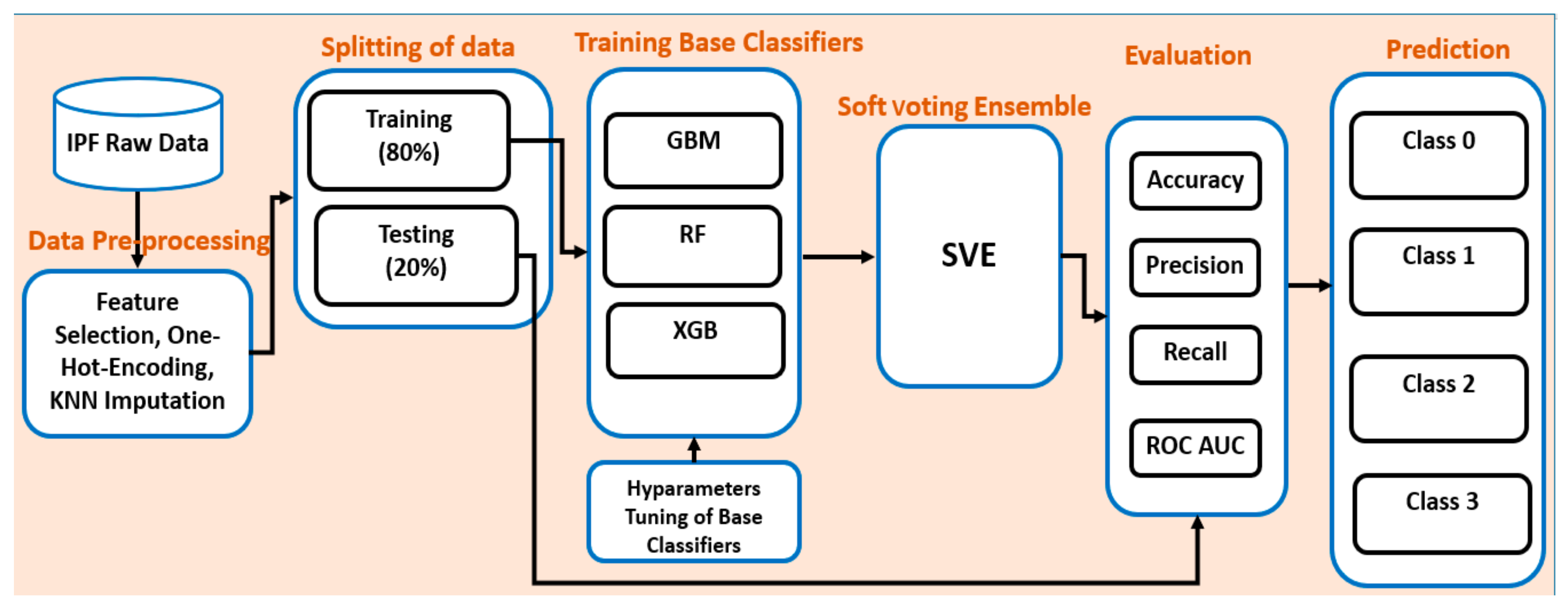
4. Proposed Ensemble Model
| Algorithm 1 To Develop a Soft Voting Ensemble Model for the Prediction of IPF Disease Severity | |
| 1 | Let’s the whole IPF dataset consists of i instances and X features. |
| 2 | The class variable is Y so labels_Yi = [4] |
| 3 | Function F: X→ labels_Yi |
| 4 | Procedure KNN Imputation (IPF_dataset) |
| 5 | Procedure Split_data (IPF_dataset) |
| 6 | Training_data, Testing_data = split (IPF_dataset) |
| 7 | Procedure datasampling (IPF_dataset) |
| 8 | return (IPF_dataset) |
| 10 | C1 = GBM (Training_dataset, Testing_data) |
| 12 | C2 = RF (Training_dataset, Testing_data) |
| 13 | C3 = XGB (Training_dataset, Testing_data) |
| 14 | Procedure ensemble_model (Training_dataset, Testing_data) |
| 15 | soft_voting_ensemble = concatenate (C1, C2, C3) |
| 16 | soft_voting_ensemble.fit (Training_dataset, Testing_data) |
| 17 | Predictions = soft_voting_ensemble.predict (Testing_data) |
4.1. Why Soft Voting Ensemble Is Better and Conspicuous among Other Classifiers
4.2. Overall Workflow of the Proposed Ensemble Model
4.3. Implementation Environment
5. Experimental Results and Discussion
5.1. Experimental Results
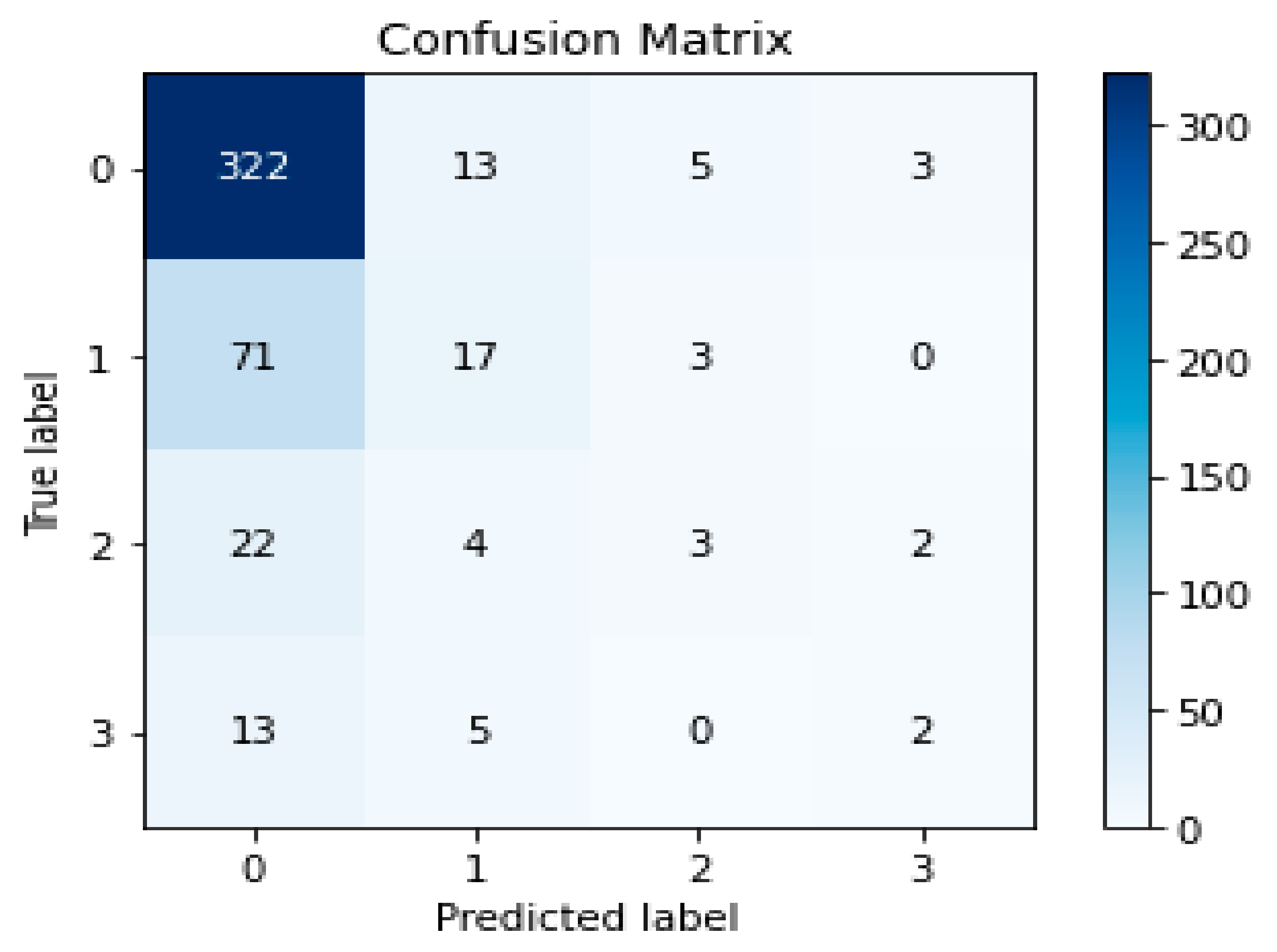
5.2. Evaluation
5.3. Discussion
5.4. Limitations
5.5. Internal and External Thread to the Validity
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Raghu, G.; Collard, H.R.; Egan, J.J.; Martinez, F.J.; Behr, J.; Brown, K.K.; Colby, T.V.; Cordier, J.F.; Flaherty, K.R.; Lasky, J.A.; et al. An official ATS/ERS/JRS/ALAT statement: Idiopathic pulmonary fibrosis: Evidence-based guidelines for diagnosis and management. Am. J. Respir. Crit. Care Med. 2011, 183, 788–824. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G.; Freudenberger, T.D.; Yang, S.; Curtis, J.R.; Spada, C.; Hayes, J.; Sillery, J.K.; Pope, C.E., 2nd; Pellegrini, C.A. High prevalence of abnormal acid gastro-oesophageal reflux in idiopathic pulmonary fibrosis. Eur. Respir. J. 2006, 27, 136–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idiopathic Pulmonary Fibrosis (IPF). Available online: https://www.webmd.com/lung/what-is-idiopathic-pulmonary-fibrosis (accessed on 5 April 2021).
- Kim, S.Y.; Diggans, J.; Pankratz, D.; Huang, J.; Pagan, M.; Sindy, N.; Tom, E.; Anderson, J.; Choi, Y.; Lynch, D.A.; et al. Classification of usual interstitial pneumonia in patients with interstitial lung disease: Assessment of a machine learning approach using high-dimensional transcriptional data. Lancet Respir. Med. 2015, 3, 473–482. [Google Scholar] [CrossRef]
- Wolters, P.J.; Blackwell, T.S.; Eickelberg, O.; Loyd, J.; Kaminski, N.; Jenkins, G.; Maher, T.M.; Molina, M.M.; Noble, P.W.; Raghu, G.; et al. Time for a change: Is idiopathic pulmonary fibrosis still idiopathic and only fibrotic? Lancet Respir. Med. 2018, 6, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Selman, M.; Pardo, A. Idiopathic pulmonary fibrosis: An epithelial/fibroblastic cross-talk disorder. Respir. Res. 2001, 3, 1–8. [Google Scholar]
- Song, J.W.; Hong, S.-B.; Lim, C.-M.; Koh, Y.; Kim, D.S. Acute exacerbation of idiopathic pulmonary fibrosis: Incidence, risk factors and outcome. Eur. Respir. J. 2011, 37, 356–363. [Google Scholar] [CrossRef]
- Martinez, F.J.; Collard, H.R.; Pardo, A.; Raghu, G.; Richeldi, L.; Selman, M.; Swigris, J.J.; Taniguchi, H.; Wells, A.U. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Primers 2017, 3, 17074. [Google Scholar] [CrossRef]
- Mekov, E.; Miravitlles, M.; Petkov, R. Artificial intelligence and machine learning in respiratory medicine. Expert Rev. Respir. Med. 2020, 14, 559–564. [Google Scholar] [CrossRef]
- Walsh, S.L.F.; Humphries, S.M.; Wells, A.U.; Brown, K.K. Imaging research in fibrotic lung disease; applying deep learning to unsolved problems. Lancet Respir. Med. 2020, 8, 1144–1153. [Google Scholar] [CrossRef]
- Walsh, S.L.F.; Calandriello, L.; Silva, M.; Sverzellati, N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: A case-cohort study. Lancet Respir. Med. 2018, 6, 837–845. [Google Scholar] [CrossRef]
- Schwartz, D.; Helmers, R.A.; Galvin, J.R.; Van Fossen, D.S.; Frees, K.L.; Dayton, C.S.; Burmeister, L.F.; Hunninghake, G.W. Determinants of survival in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 1994, 149, 450–454. [Google Scholar] [CrossRef]
- Raghu, G.; Weycker, D.; Edelsberg, J.; Bradford, W.Z.; Oster, G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2006, 174, 810–816. [Google Scholar] [CrossRef]
- Olson, A.L.; Swigris, J.J. Idiopathic pulmonary fibrosis: Diagnosis and epidemiology. Clin. Chest Med. 2012, 33, 41–50. [Google Scholar] [CrossRef]
- King, T.E., Jr.; Tooze, J.A.; Schwarz, M.I.; Brown, K.R.; Cherniack, R.M. Predicting survival in idiopathic pulmonary fibrosis: Scoring system and survival model. Am. J. Respir. Crit. Care Med. 2001, 164, 1171–1181. [Google Scholar] [CrossRef]
- Ryerson, C.J.; Hartman, T.; Elicker, B.M.; Ley, B.; Lee, J.S.; Abbritti, M.; Jones, K.D.; King, T.E.; Ryu, J.; Collard, H.R. Clinical features and outcomes in combined pulmonary fibrosis and emphysema in idiopathic pulmonary fibrosis. Chest 2013, 144, 234–240. [Google Scholar] [CrossRef]
- Fell, C.D.; Martinez, F.J.; Liu, L.X.; Murray, S.; Han, M.K.; Kazerooni, E.A.; Gross, B.H.; Myers, J.; Travis, W.D.; Colby, T.V.; et al. Clinical predictors of a diagnosis of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2010, 181, 832–837. [Google Scholar] [CrossRef]
- Shi, Y.; Wong, W.K.; Goldin, J.G.; Brown, M.S.; Kim, G.H.J. Prediction of progression in idiopathic pulmonary fibrosis using CT scans at baseline: A quantum particle swarm optimization-Random Forest approach. Artif. Intell. Med. 2019, 100, 101709. [Google Scholar] [CrossRef]
- Christe, A.; Peters, A.A.; Drakopoulos, D.; Heverhagen, J.; Geiser, T.; Stathopoulou, T.; Christodoulidis, S.; Anthimopoulos, M.; Mougiakakou, S.G.; Ebner, L. Computer-aided diagnosis of pulmonary fibrosis using deep learning and CT images. Investig. Radiol. 2019, 54, 627. [Google Scholar] [CrossRef] [Green Version]
- Hussain, A.; Choi, H.-E.; Kim, H.-J.; Aich, S.; Saqlain, M.; Kim, H.-C. Forecast the Exacerbation in Patients of Chronic Obstructive Pulmonary Disease with Clinical Indicators Using Machine Learning Techniques. Diagnostics 2021, 11, 829. [Google Scholar] [CrossRef]
- Park, S.C.; Tan, J.; Wang, X.; Lederman, D.; Leader, J.K.; Kim, S.H.; Zheng, B. Computer-aided detection of early interstitial lung diseases using low-dose CT images. Phys. Med. Biol. 2011, 56, 1139. [Google Scholar] [CrossRef]
- Zelaya, C.V.G. Towards explaining the effects of data preprocessing on machine learning. In Proceedings of the 2019 IEEE 35th International Conference on Data Engineering (ICDE), Macao, China, 8–11 April 2019. [Google Scholar]
- García, S.; Luengo, J.; Herrera, F. Data Preprocessing in Data Mining; Springer: Berlin, Germany, 2015; Volume 72. [Google Scholar]
- Newgard, C.D.; Lewis, R.J. Missing data: How to best account for what is not known. JAMA 2015, 314, 940–941. [Google Scholar] [CrossRef]
- Zhang, S. Nearest neighbor selection for iteratively kNN imputation. J. Syst. Softw. 2012, 85, 2541–2552. [Google Scholar] [CrossRef]
- Ghaemi, M.; Feizi-Derakhshi, M.-R. Feature selection using forest optimization algorithm. Pattern Recognit. 2016, 60, 121–129. [Google Scholar] [CrossRef]
- Han, W.; Huang, Z.; Li, S.; Jia, Y. Distribution-sensitive unbalanced data oversampling method for medical diagnosis. J. Med Syst. 2019, 43, 39. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Ren, Y.; Zhang, L.; Suganthan, P. Ensemble classification and regression-recent developments, applications and future directions. IEEE Comput. Intell. Mag. 2016, 11, 41–53. [Google Scholar] [CrossRef]

| Features | Feature Details | Feature Type | |
|---|---|---|---|
| 1 | Sex | Male = 1, female = 0 | Categorical |
| 2 | Ht | Height | Continuous |
| 3 | Wt | Weight | Continuous |
| 4 | BMI | Body mass index | Continuous |
| 5 | Age | Age of patient | Continuous |
| 6 | Job | Housewife = 1, Office worker = 2, commerce = 3, construction site = 4 | Categorical |
| 7 | Toxic_chem | Toxic chemical yes = 1, No = 0 | Categorical |
| 8 | Toxic_wooddust | Toxic wood dust yes = 1, No = 0 | Categorical |
| 9 | Toxic_mineraldust | Yes = 1, No = 0 | Categorical |
| 10 | Smoking | Yes = 1, No = 0 | Categorical |
| 11 | DxAge | Diagnose age | Continuous |
| 12 | BAL_Done | Bronchoalveolar lavage, yes = 1, No = 0 | Categorical |
| 13 | ABG_Done | Arterial blood gases, yes = 1, No = 0 | Categorical |
| 14 | PFT_FEVm | Pulmonary function test forced expiratory volume measure | Continuous |
| 15 | PFT_FEVpc | Pulmonary function test forced expiratory predicted value | Continuous |
| 16 | PFT_FVCm | Pulmonary function test forced vital capacity | Continuous |
| 17 | PFT_FVCpc | Pulmonary function test forced vital capacity predicted value | Continuous |
| 18 | PFT_FFpc | Pulmonary function test free fluid | Continuous |
| 19 | PFT_DLCOm | Diffusing capacity of lungs for carbon monoxide measure | Continuous |
| 20 | PFT_DLCOpc | Diffusing capacity of lungs for carbon monoxide predicted value | Continuous |
| 21 | CT_GGO | Ground-glass opacity, Yes = 1, No = 0 | Categorical |
| 22 | RheumaYN | Rhema, Yes = 1, No = 0 | Categorical |
| 23 | AntiCCPYN | Anti-CCP, Yes = 1, No = 0 | Categorical |
| 24 | NT-Pro BNP | NT-Pro BNP, Yes = 1, No = 0 | Categorical |
| 25 | EchoYN | Echo, Yes = 1, No = 0 | Categorical |
| 26 | RxHomeYN | Home oxygen treatment, Yes = 1, No = 0 | Categorical |
| Classifiers | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| GBM | 0.6907 | 0.6300 | 0.6900 | 0.6400 |
| RF | 0.6948 | 0.6300 | 0.6900 | 0.6600 |
| XGB | 0.6989 | 0.6300 | 0.7000 | 0.6400 |
| SVE | 0.7100 | 0.6400 | 0.7100 | 0.6600 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, S.; Hussain, A.; Aich, S.; Park, M.S.; Chung, M.P.; Jeong, S.H.; Song, J.W.; Lee, J.H.; Kim, H.C. A Soft Voting Ensemble-Based Model for the Early Prediction of Idiopathic Pulmonary Fibrosis (IPF) Disease Severity in Lungs Disease Patients. Life 2021, 11, 1092. https://doi.org/10.3390/life11101092
Ali S, Hussain A, Aich S, Park MS, Chung MP, Jeong SH, Song JW, Lee JH, Kim HC. A Soft Voting Ensemble-Based Model for the Early Prediction of Idiopathic Pulmonary Fibrosis (IPF) Disease Severity in Lungs Disease Patients. Life. 2021; 11(10):1092. https://doi.org/10.3390/life11101092
Chicago/Turabian StyleAli, Sikandar, Ali Hussain, Satyabrata Aich, Moo Suk Park, Man Pyo Chung, Sung Hwan Jeong, Jin Woo Song, Jae Ha Lee, and Hee Cheol Kim. 2021. "A Soft Voting Ensemble-Based Model for the Early Prediction of Idiopathic Pulmonary Fibrosis (IPF) Disease Severity in Lungs Disease Patients" Life 11, no. 10: 1092. https://doi.org/10.3390/life11101092
APA StyleAli, S., Hussain, A., Aich, S., Park, M. S., Chung, M. P., Jeong, S. H., Song, J. W., Lee, J. H., & Kim, H. C. (2021). A Soft Voting Ensemble-Based Model for the Early Prediction of Idiopathic Pulmonary Fibrosis (IPF) Disease Severity in Lungs Disease Patients. Life, 11(10), 1092. https://doi.org/10.3390/life11101092









