Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and Tissue Preparation
2.2. RNA Extraction
2.3. RNA Labeling and Array Hybridization
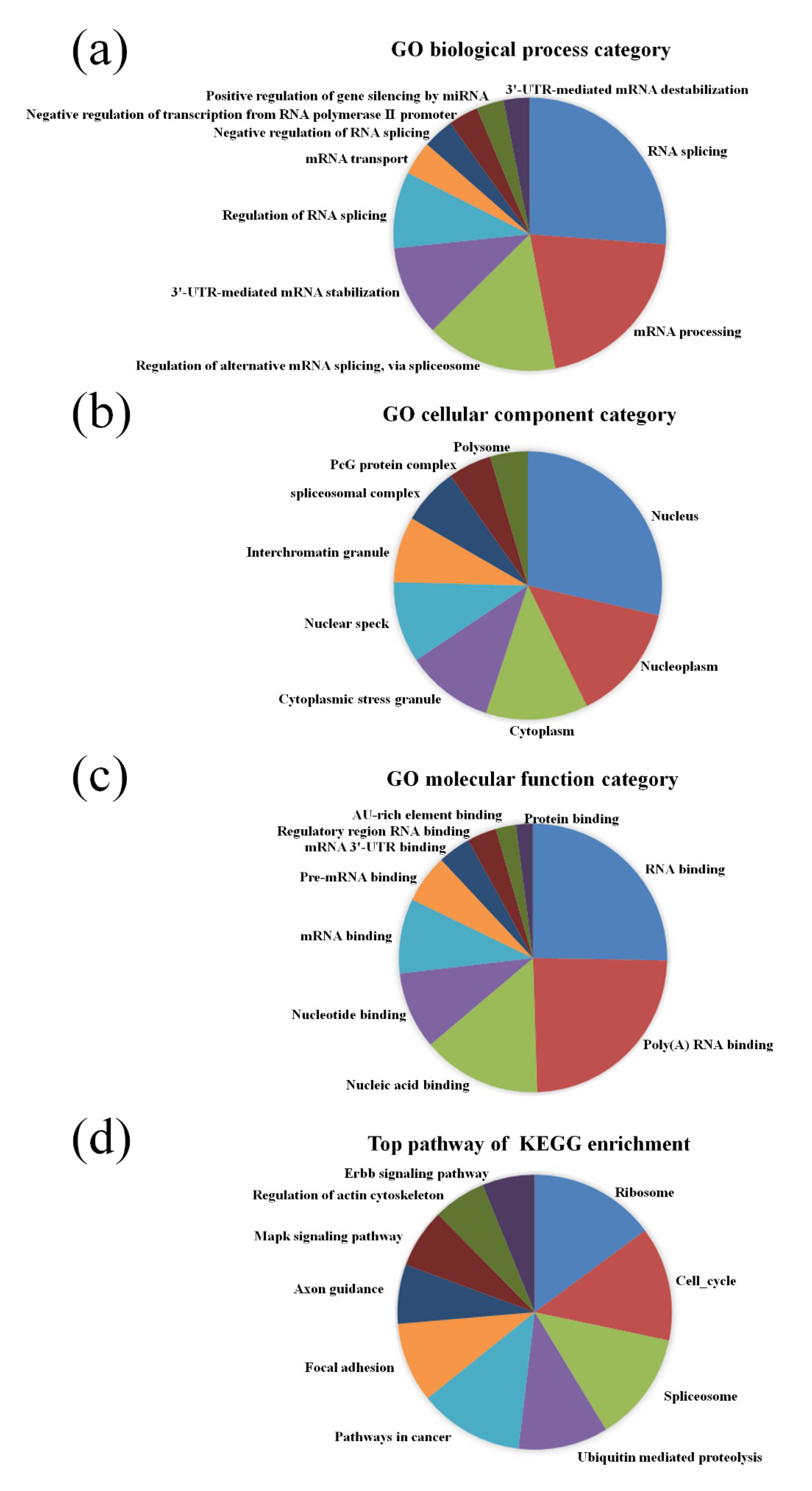
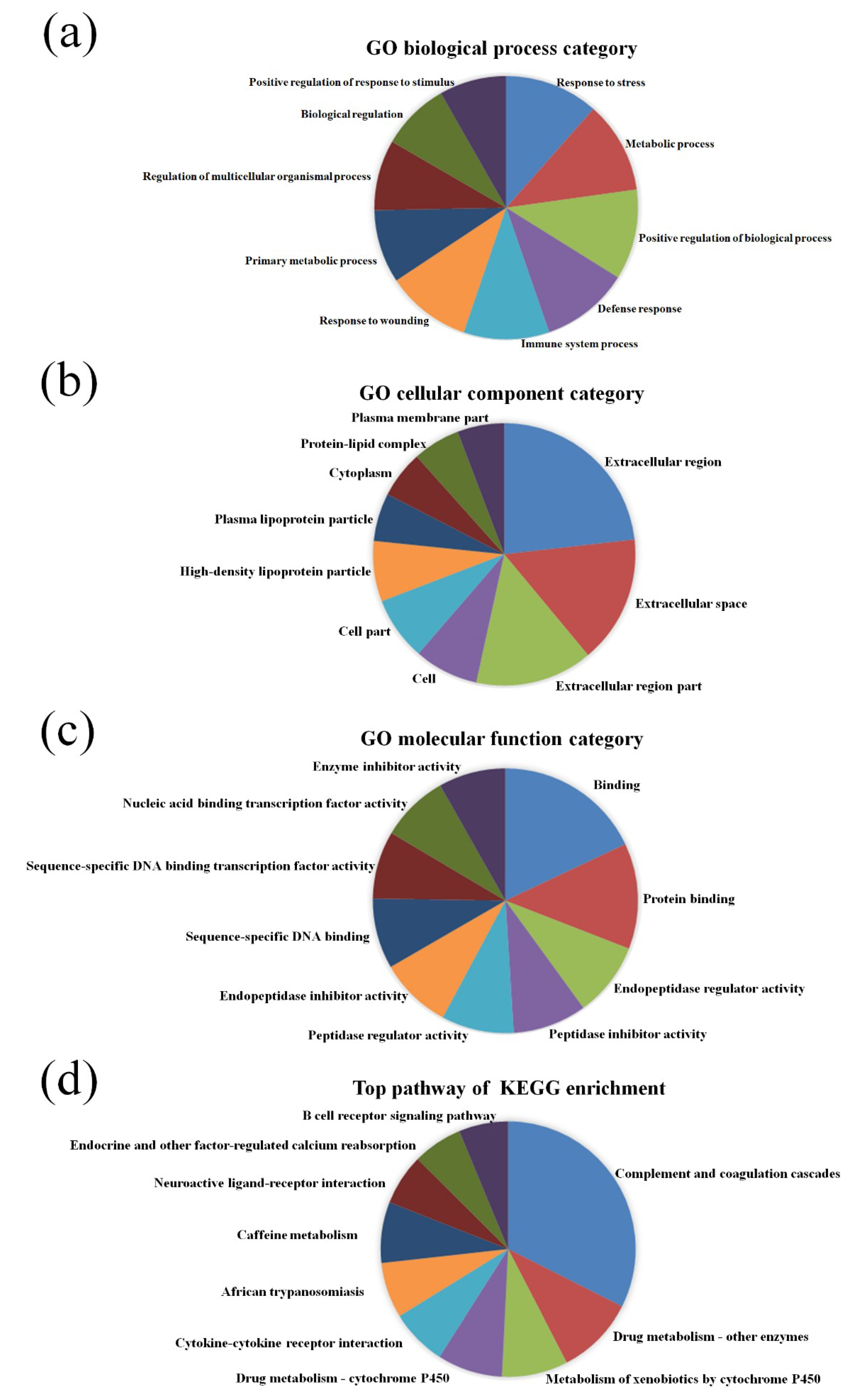
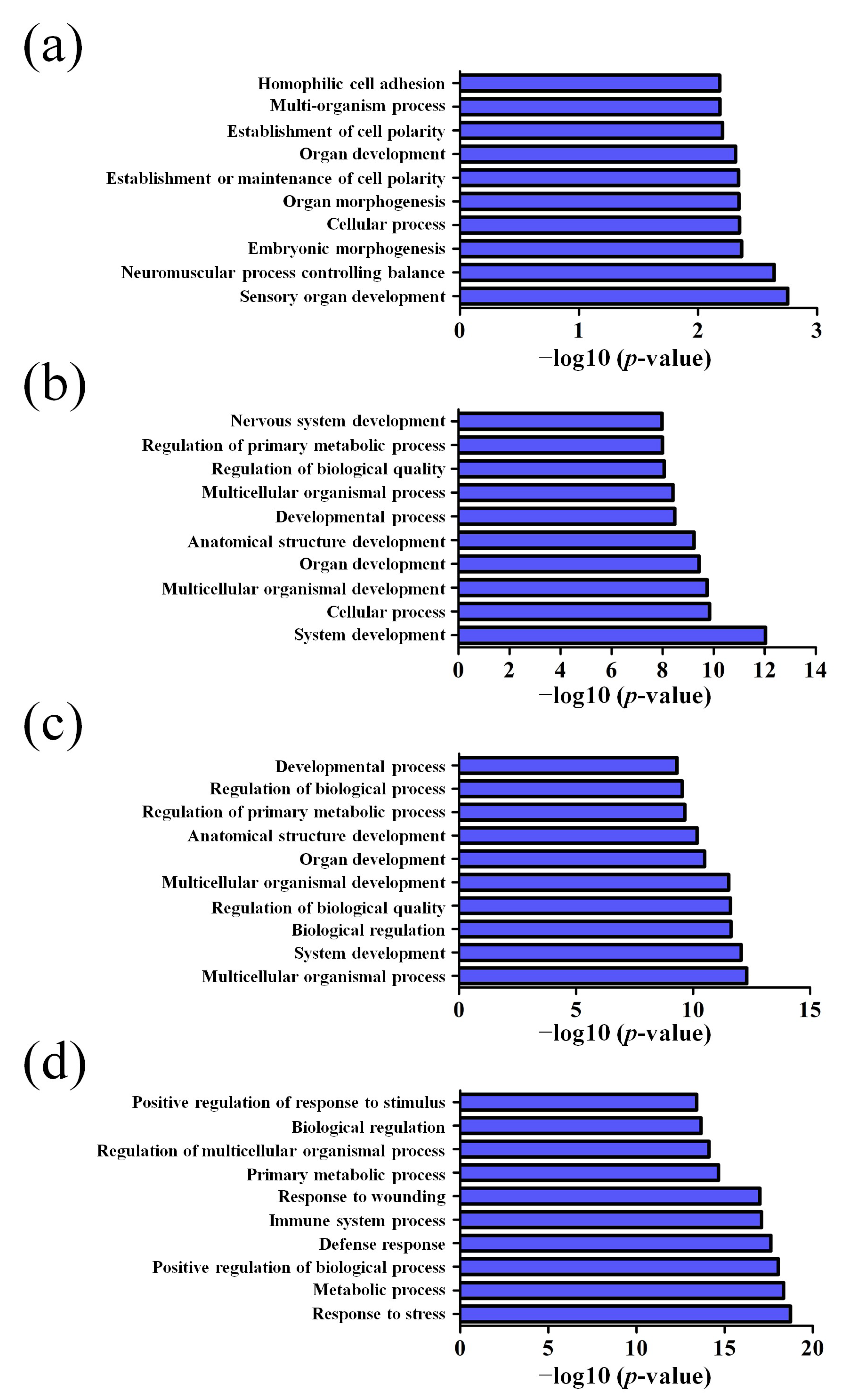
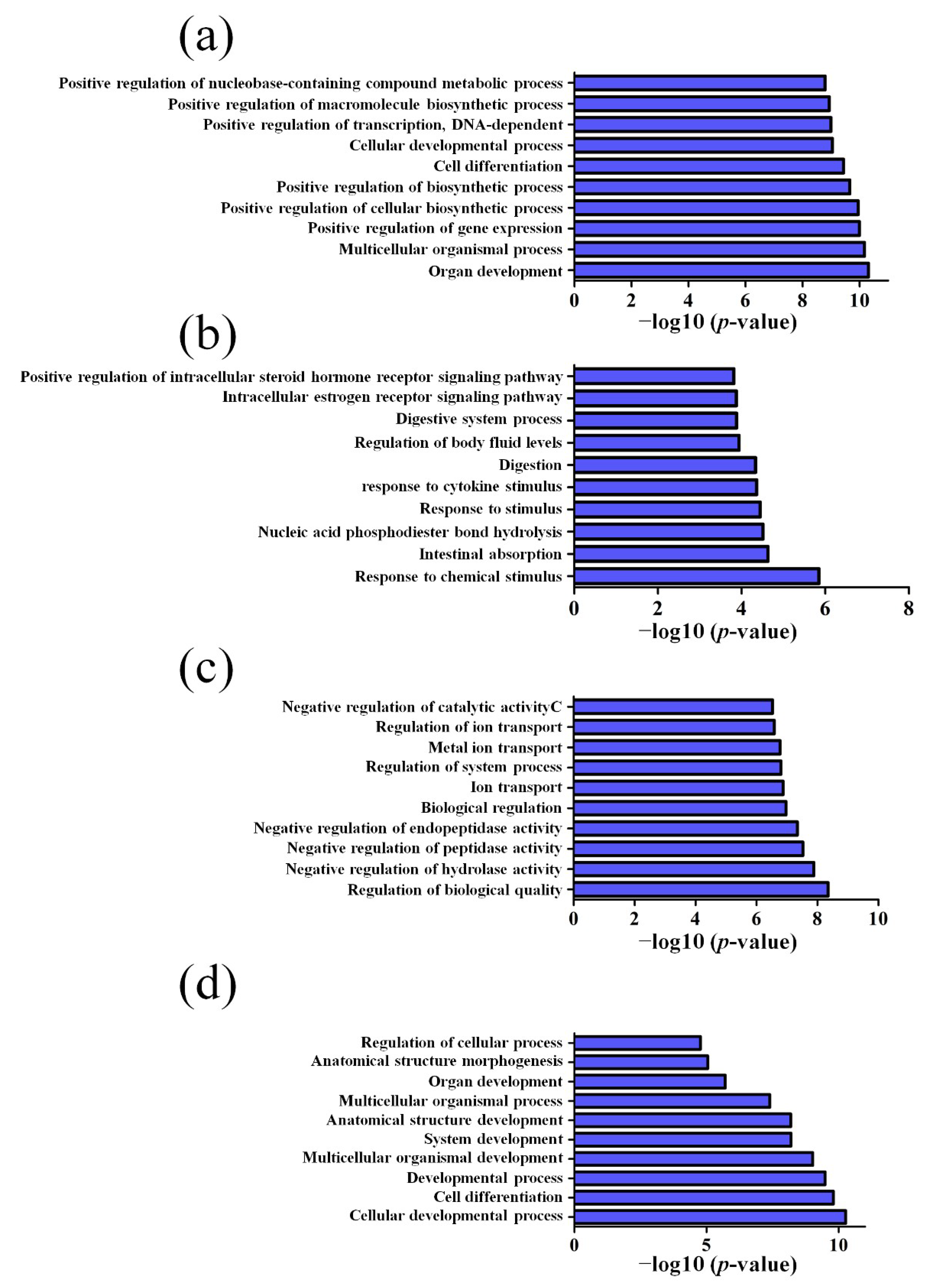
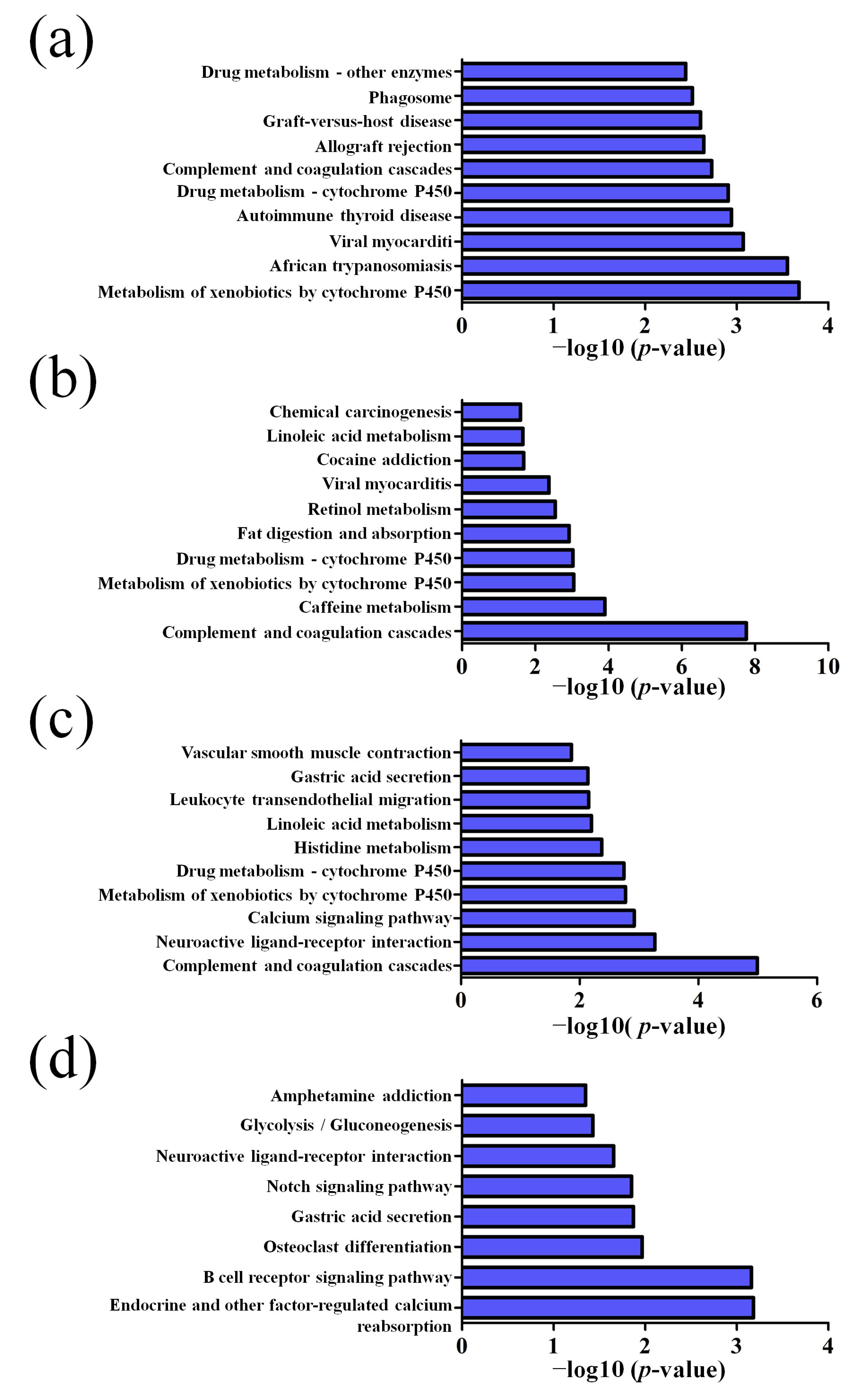
2.4. GO and Pathway Analysis
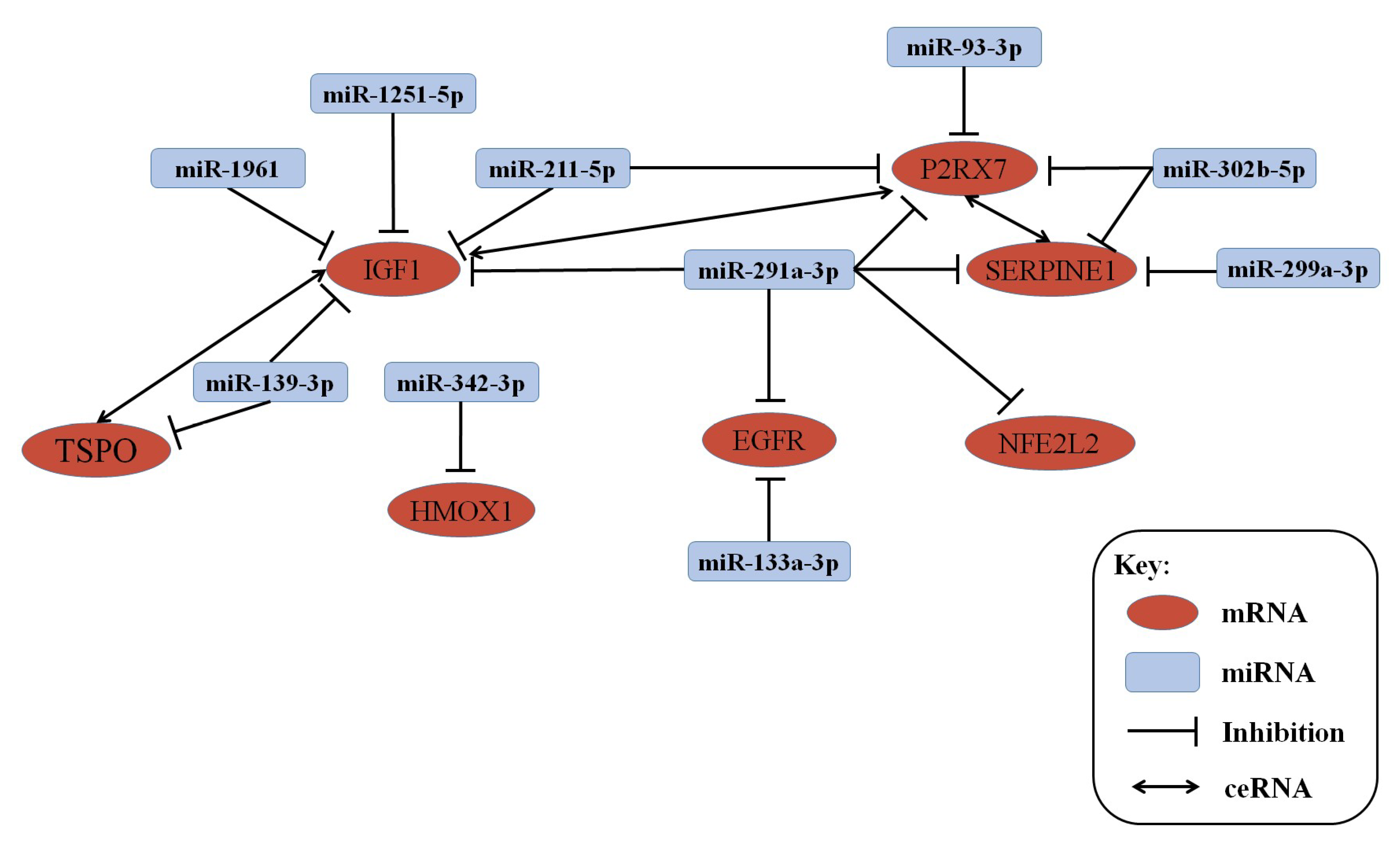
2.5. lncRNA–mRNA–miRNA Co-Expression Network Construction
2.6. Statistical Analysis
3. Results
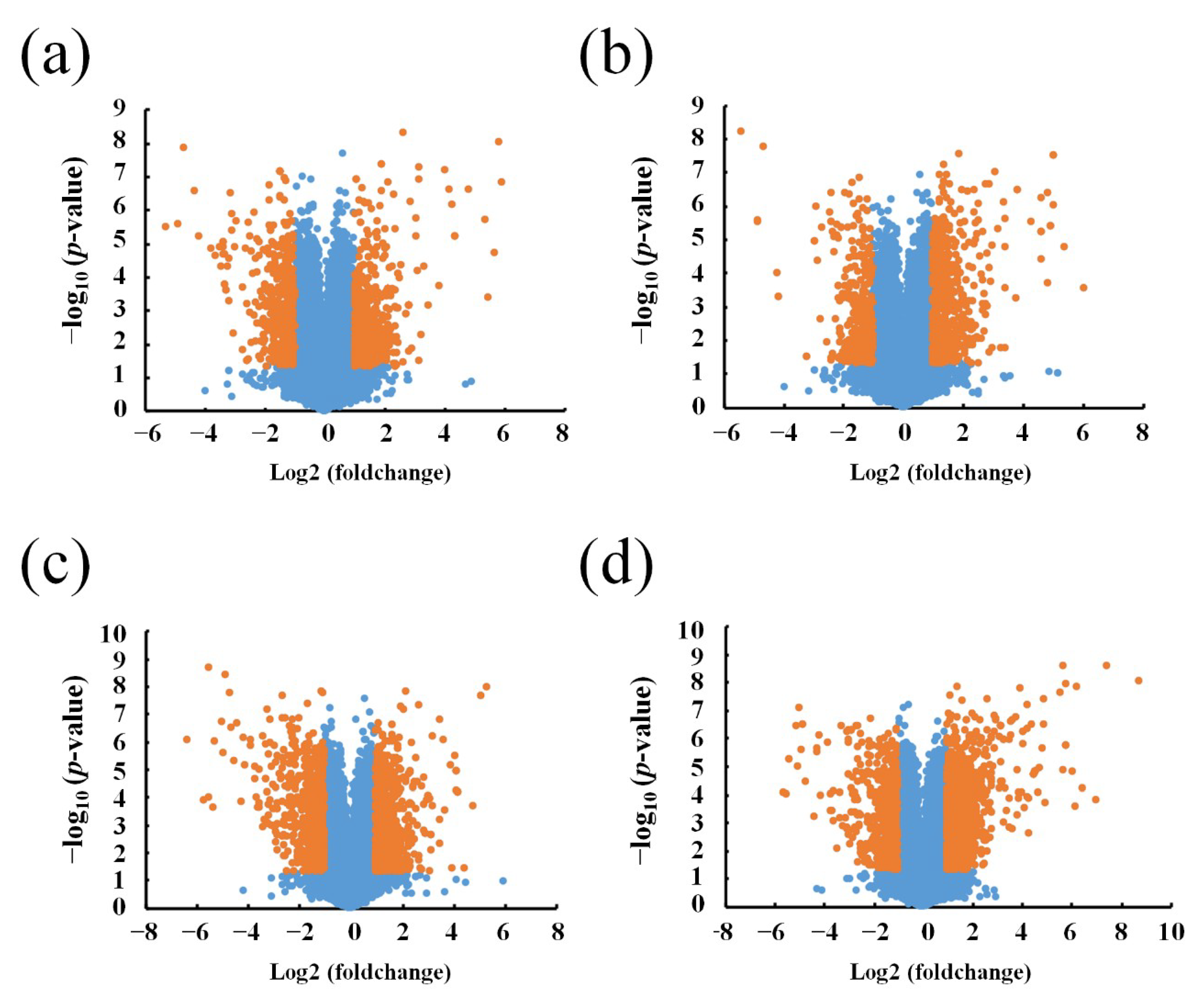
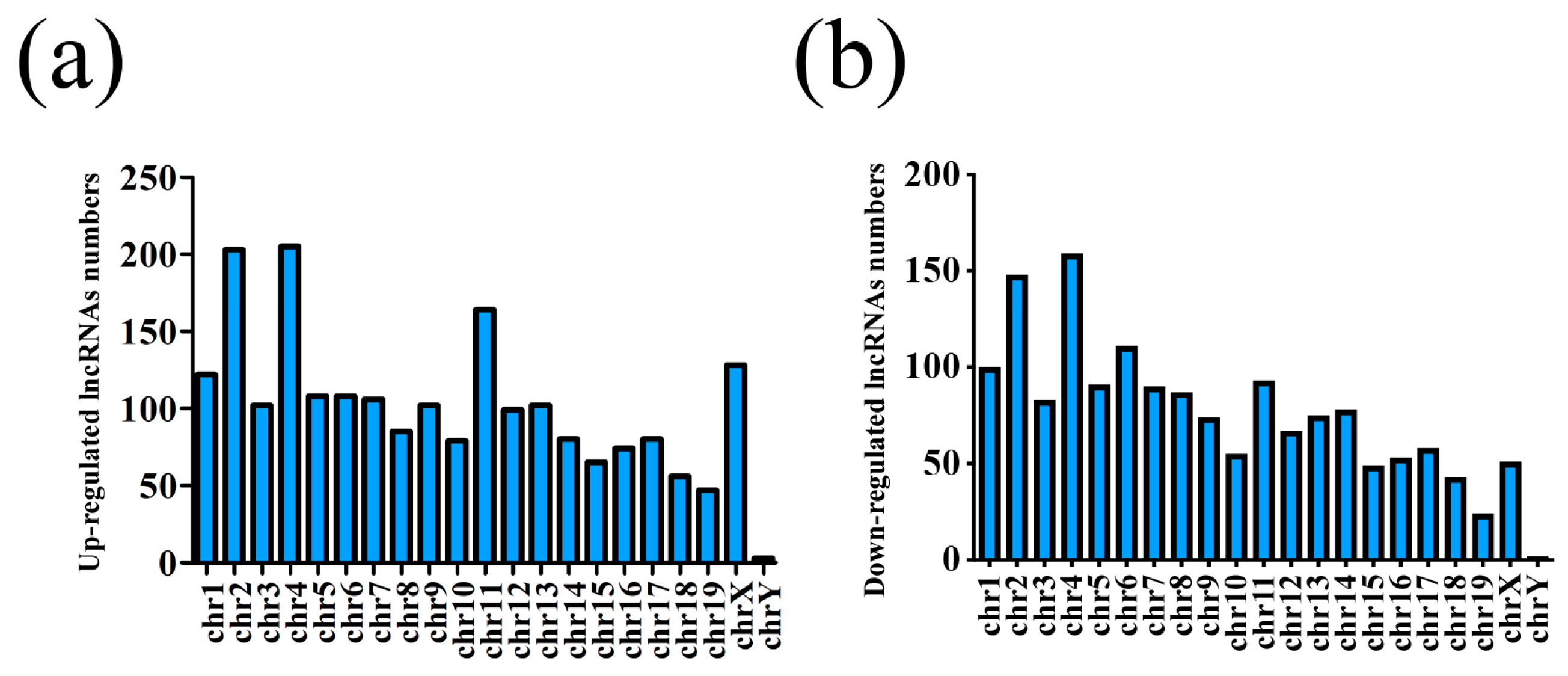
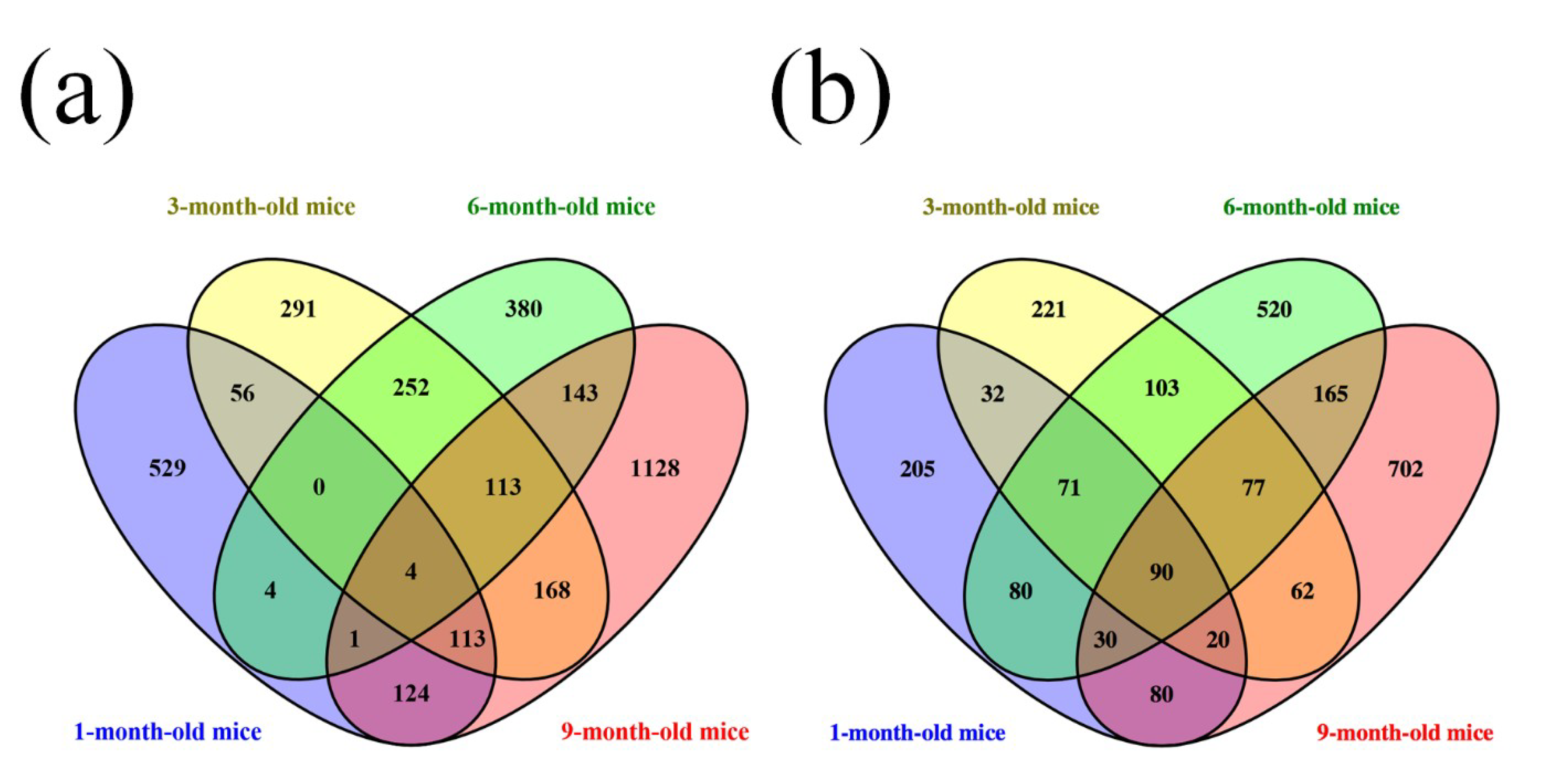
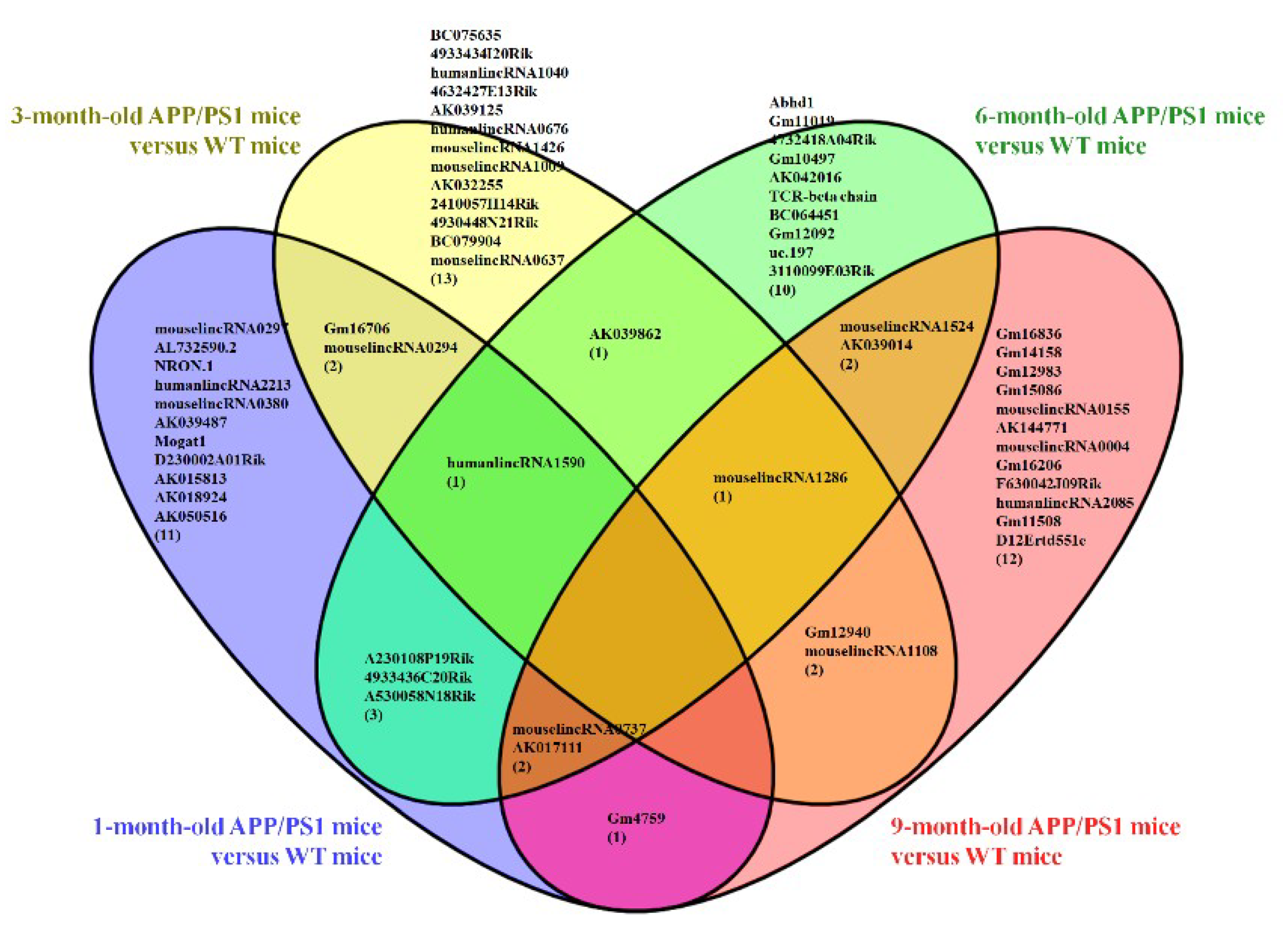
3.1. Different Expression Profile of lncRNA and mRNA
3.2. GO and KEGG Pathway Analysis
3.3. lncRNA–mRNA–miRNA Co-Expression Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ansari, S.A.; Satar, R.; Perveen, A.; Ashraf, G.M. Current opinion in Alzheimer’s disease therapy by nanotechnology-based approaches. Curr. Opin. Psychiatr. 2017, 30, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, A.; Kamal, M.A.; Ashraf, G.M. The Alzheimer’s disease challenge. Front. Neurosci. 2019, 13, 768. [Google Scholar] [CrossRef] [PubMed]
- Mamun, A.A.; Uddin, M.S.; Mathew, B.; Ashraf, G.M. Toxic tau: Structural origins of tau aggregation in Alzheimer’s disease. Neural Regen. Res. 2020, 15, 1417–1420. [Google Scholar] [PubMed]
- Uddin, M.S.; Mamun, A.A.; Labu, Z.K.; Hidalgo-Lanussa, O.; Barreto, G.E.; Ashraf, G.M. Autophagic dysfunction in Alzheimer’s disease: Cellular and molecular mechanistic approaches to halt Alzheimer’s pathogenesis. J. Cell Physiol. 2019, 234, 8094–8112. [Google Scholar] [CrossRef] [PubMed]
- 2018 Alzheimer’s disease facts and figures. Alzheimer Dement. 2018, 14, 367–429. [CrossRef]
- Gasiorowski, K.; Brokos, B.; Leszek, J.; Tarasov, V.V.; Ashraf, G.M.; Aliev, G. Insulin resistance in alzheimer disease: p53 and micrornas as important players. Curr. Top Med. Chem. 2017, 17, 1429–1437. [Google Scholar] [CrossRef]
- Ashraf, G.M.; Tabrez, S.; Jabir, N.R.; Firoz, C.K.; Ahmad, S.; Hassan, I.; Alexiou, A.; Kamal, M.A. An overview on global trends in nanotechnological approaches for alzheimer therapy. Curr. Drug Metab. 2015, 16, 719–727. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, L.; Qin, C. Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain Res. Bull. 2017, 132, 160–169. [Google Scholar] [CrossRef]
- Wang, K.L.; Chen, F.X.; He, D.D.; Li, Y.; Fu, J. Dissection of functional lncRNAs in Alzheimer’s disease by construction and analysis of lncRNA-mRNA networks based on competitive endogenous RNAs. Biochem. Biophys. Res. Commun. 2017, 485, 569–576. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Huang, Y.; Chen, J.; Chi, H.; Yu, Z.; Wang, J.; Chen, C. Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1AS expression. Mol. Med. Rep. 2014, 10, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zheng, L.; Jiang, A.; Mo, Y.; Gong, Q. Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. Neuroreport 2018, 29, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Ciarlo, E.; Massone, S.; Penna, I.; Nizzari, M.; Gigoni, A.; Dieci, G.; Russo, C.; Florio, T.; Cancedda, R.; Pagano, A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis. Model. Mech. 2013, 6, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Massone, S.; Vassallo, I.; Fiorino, G.; Castelnuovo, M.; Barbieri, F.; Borghi, R.; Tabaton, M.; Robello, M.; Gatta, E.; Russo, C.; et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 2011, 41, 308–317. [Google Scholar] [CrossRef]
- Massone, S.; Ciarlo, E.; Vella, S.; Nizzari, M.; Florio, T.; Russo, C.; Cancedda, R.; Pagano, A. NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid b secretion. Biochim. Biophys. Acta 2012, 1823, 1170–1177. [Google Scholar] [CrossRef]
- Parenti, R.; Paratore, S.; Torrisi, A.; Cavallaro, S. A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during b-amyloid-induced apoptosis. Eur. J. Neurosci. 2007, 26, 2444–2457. [Google Scholar] [CrossRef]
- Sosinska, P.; Mikula-Pietrasik, J.; Ksiazek, K. The double-edged sword of long non-coding RNA: The role of human brain-specific BC200 RNA in translational control, neurodegenerative diseases, and cancer. Mutation research. Rev. Mutat. Res. 2015, 766, 58–67. [Google Scholar] [CrossRef]
- Shen, Z.; Lei, J.; Li, X.; Wang, Z.; Bao, X.; Wang, R. Multifaceted assessment of the APP/PS1 mouse model for Alzheimer’s disease: Applying MRS, DTI, and ASL. Brain Res. 2018, 1698, 114–120. [Google Scholar] [CrossRef]
- Gene Ontology. Available online: http://www.geneontology.org (accessed on 6 January 2017).
- Kyoto Encyclopedia of Genes and Genomes. Available online: http://www.genome.jp/kegg/ (accessed on 15 January 2017).
- miRWalk. Available online: http://www.ma.uni-heidelberg.de/ (accessed on 25 January 2017).
- TargetScan. Available online: http://www.targetscan.org/ (accessed on 25 January 2017).
- miRBase. Available online: http://www.mirbase.org/ (accessed on 25 January 2017).
- miRanda. Available online: http://www.microrna.org/ (accessed on 25 January 2017).
- Wang, L.L.; Min, L.; Guo, Q.D.; Zhang, J.X.; Jiang, H.L.; Shao, S.; Xing, J.G.; Yin, L.L.; Liu, J.H.; Liu, R. Profiling microRNA from Brain by Microarray in a Transgenic Mouse Model of Alzheimer’s Disease. BioMed Res. Int. 2017, 8030369. [Google Scholar] [CrossRef]
- Uddin, M.S.; Kabir, M.T.; Al Mamun, A.; Abdel-Daim, M.M.; Barreto, G.E.; Ashraf, G.M. APOE and Alzheimer’s Disease: Evidence Mounts that Targeting APOE4 may Combat Alzheimer’s Pathogenesis. Mol. Neurobiol. 2019, 56, 2450–2465. [Google Scholar] [CrossRef] [PubMed]
- Al Mamun, A.; Uddin, M.S.; Kabir, M.T.; Khanum, S.; Sarwar, M.S.; Mathew, B.; Rauf, A.; Ahmed, M.; Ashraf, G.M. Exploring the promise of targeting ubiquitin-proteasome system to combat alzheimer’s disease. Neurotox. Res. 2020. [Google Scholar] [CrossRef]
- Zhou, D.; Liu, H.; Li, C.; Wang, F.; Shi, Y.; Liu, L.; Zhao, X.; Liu, A.; Zhang, J.; Wang, C.; et al. Atorvastatin ameliorates cognitive impairment, Aβ1-42 production and Tau hyperphosphorylation in APP/PS1 transgenic mice. Metab. Brain Dis. 2016, 31, 693–703. [Google Scholar] [CrossRef]
- Delatour, B.; Guegan, M.; Volk, A.; Dhenain, M. In vivo MRI and histological evaluation of brain atrophy in APP/PS1 transgenic mice. Neurobiol. Aging 2006, 27, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Poisnel, G.; Dhilly, M.; Moustie, O.; Delamare, J.; Abbas, A.; Guilloteau, D.; Louisa, B. PET imaging with [18F] AV-45 in an APP/PS1-21 murine model of amyloid plaque deposition. Neurobiol. Aging 2012, 33, 2561–2571. [Google Scholar] [CrossRef] [PubMed]
- Lagadec, S.; Rotureau, L.; Hemar, A.; Macrez, N.; Delcasso, S.; Jeantet, Y.; Yoon, H.C. Early temporal short-term memory deficits in double transgenic APP/PS1 mice. Neurobiol. Aging 2012, 33, e201–e211. [Google Scholar] [CrossRef]
- Ferguson, A.S.; Sarkar, S.; Schmued, C.L. Longitudinal behavioral changes in the APP/PS1 transgenic Alzheimer’s disease model. Behav. Brain Res. 2013, 242C, 125–134. [Google Scholar] [CrossRef]
- Bao, J.; Zhang, H.; Hu, K.; Liu, H.; Wang, H.; Li, A.; Lin, F.; Zhang, L.; Sun, X.; Du, Z.; et al. Silencing DNA methyltransferase 1 (DNMT1) inhibits proliferation metastasis and invasion in ESCC by suppressing methylation of RASSF1A and DAPK. Oncotarget 2016, 7, 44129–44141. [Google Scholar] [CrossRef]
- West, R.L.; Lee, J.M.; Maroun, L.E. Hypomethylation of the Amyloid Precursor Protein Gene in the Brain of an Alzheimer’s Disease Patient. J. Mol. Neurosci. Lett. 1995, 6, 141–146. [Google Scholar] [CrossRef]
- Fuso, A.; Seminara, L.; Cavallaro, R.A.; Anselmi, F.; Scarpa, S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol. Cell. Neurosci. 2005, 28, 195–204. [Google Scholar] [CrossRef]
- Yang, B.; Xia, Z.A.; Zhong, B.; Xiong, X.; Sheng, C.; Wang, Y.; Gong, W.; Cao, Y.; Wang, Z.; Peng, W. Distinct hippocampal expression profiles of long non-coding RNAs in an Alzheimer’s disease model. Mol. Neurobiol. 2017, 54, 4833–4846. [Google Scholar] [CrossRef] [PubMed]
- Frater, J.; Lie, D.; Bartlett, P.; McGrath, J.J. Insulin-like Growth Factor 1 (IGF-1) as a marker of cognitive decline in normal ageing: A review. Ageing Res. Rev. 2018, 42, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Sperlagh, B.; Illes, P. P2X7 receptor: An emerging target in central nervous system diseases. Trends Pharmacol. Sci. 2014, 35, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E. Numb and Numblike control cell number during vertebrate neurogenesis. Trends Neurosci. 2003, 26, 395–396. [Google Scholar] [CrossRef]
- Diaz-Hernandez, J.; Gomez-Villafuertes, R. In vivo P2X7 inhibition reduces amyloid plaques in Alzheimer’sdisease through GSK3 and secretases. Neurobiol. Aging 2012, 33, 1816–1828. [Google Scholar] [CrossRef]
- Cabezas, R.; Baez-Jurado, E.; Hidalgo-Lanussa, O.; Echeverria, V.; Ashraf, G.M.; Sahebkar, A.; Barreto, G.E. Growth Factors and Neuroglobin in Astrocyte Protection Against Neurodegeneration and Oxidative Stress. Mol. Neurobiol. 2019, 56, 2352. [Google Scholar] [CrossRef]
- Repalli, J. Translocator Protein (TSPO) Role in Aging and Alzheimer’s Disease. Curr. Aging Sci. 2012, 7, 168–175. [Google Scholar] [CrossRef]
- Barker, R.; Kehoe, P.G.; Love, S. Activators and inhibitors of the plasminogen system in Alzheimer’s disease. J. Cell. Mol. Med. 2012, 16, 865–876. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zeng, L.; Jiang, H.; Li, Z.; Liu, R. Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease. Life 2020, 10, 64. https://doi.org/10.3390/life10050064
Wang L, Zeng L, Jiang H, Li Z, Liu R. Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease. Life. 2020; 10(5):64. https://doi.org/10.3390/life10050064
Chicago/Turabian StyleWang, Linlin, Li Zeng, Hailun Jiang, Zhuorong Li, and Rui Liu. 2020. "Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease" Life 10, no. 5: 64. https://doi.org/10.3390/life10050064
APA StyleWang, L., Zeng, L., Jiang, H., Li, Z., & Liu, R. (2020). Microarray Profile of Long Noncoding RNA and Messenger RNA Expression in a Model of Alzheimer’s Disease. Life, 10(5), 64. https://doi.org/10.3390/life10050064




