Synthetic Biology Approaches to Engineer Saccharomyces cerevisiae towards the Industrial Production of Valuable Polyphenolic Compounds
Abstract
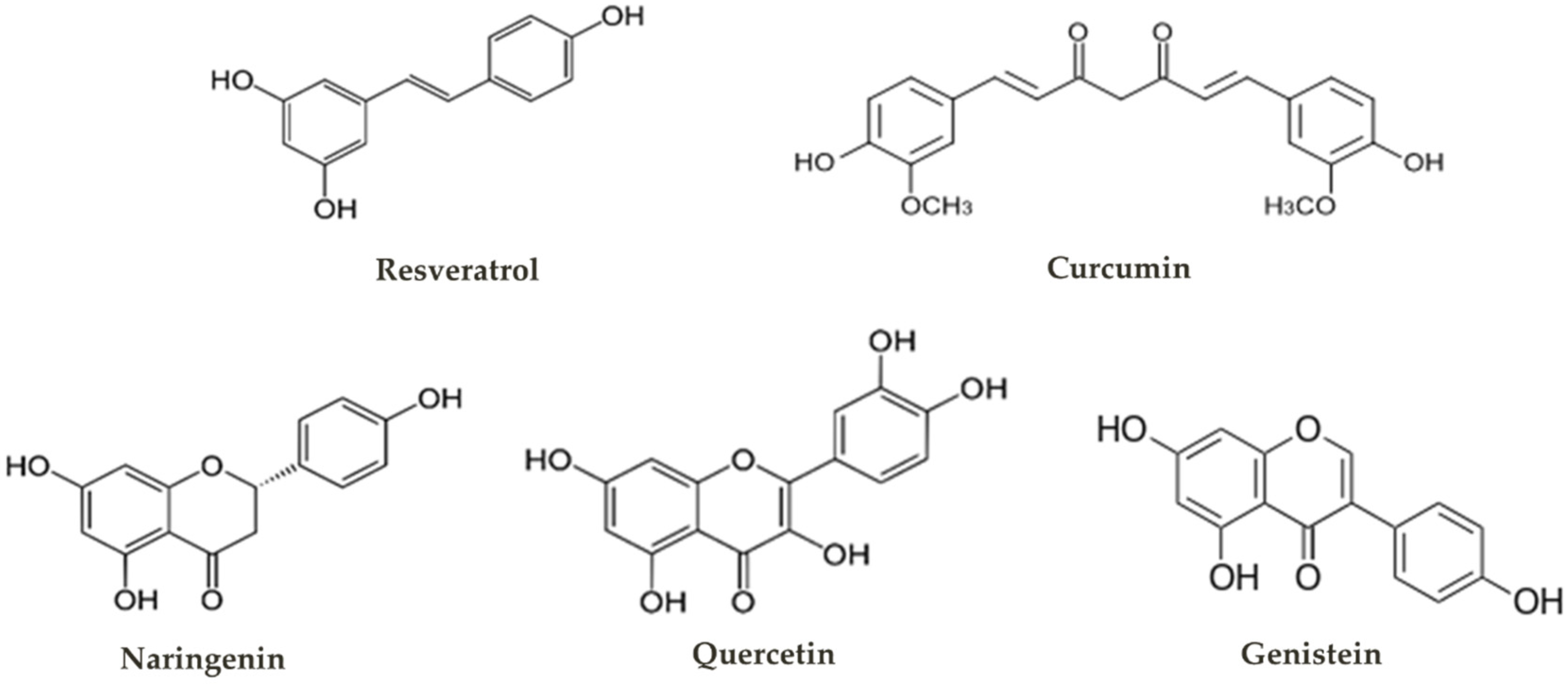
1. Plant Polyphenols
1.1. Biological Activities
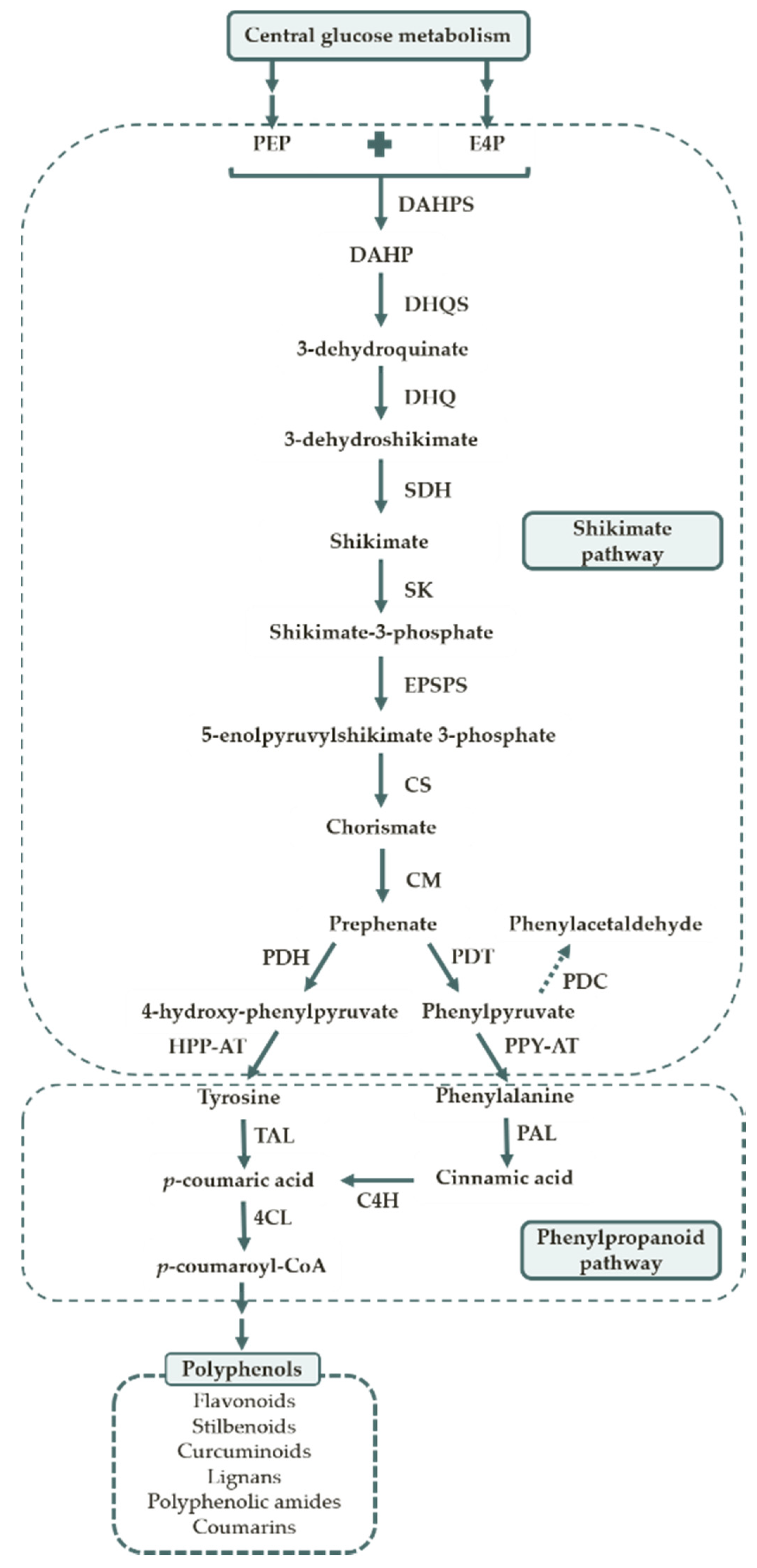
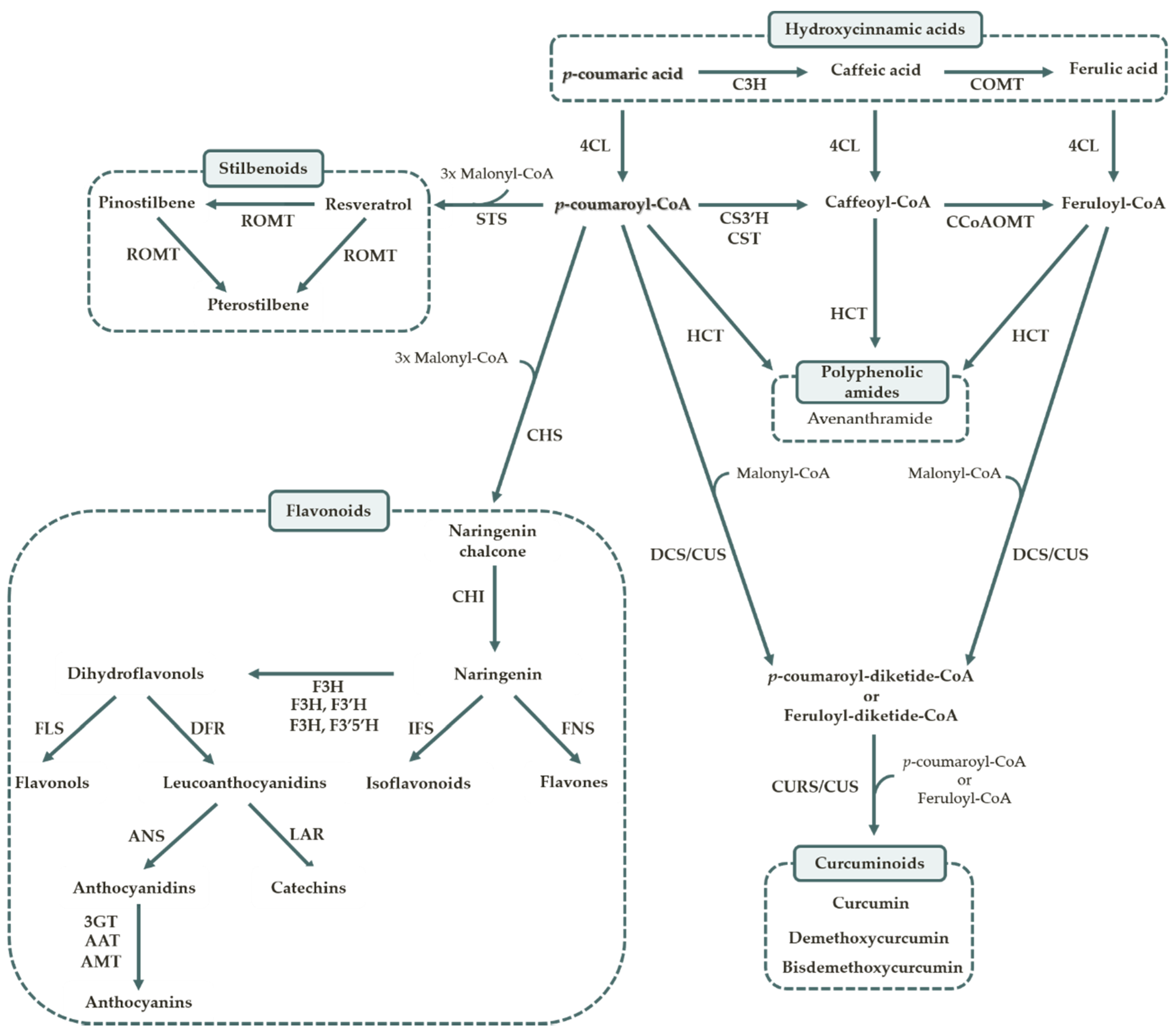
1.2. Polyphenol Biosynthesis
1.3. Polyphenol Heterologous Production
2. Genetic Engineering of Saccharomyces Cerevisiae
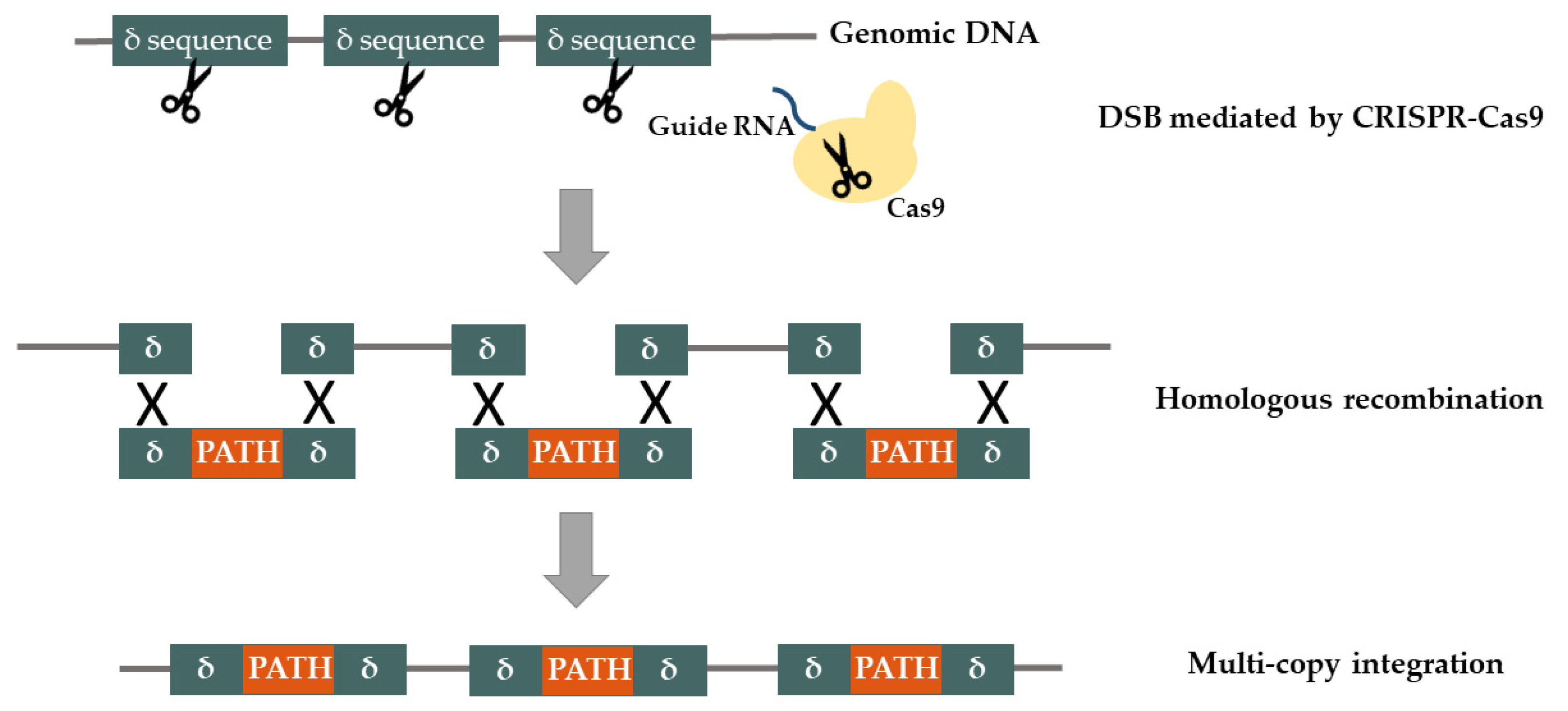
2.1. Synthetic Biology Tools for Genome Engineering
| Phenolic Compound | Genes 1 | Genetic System | Chassis/Chassis Modification 2 | Substrate 3 | Conditions | Titer (mg/L) | Reference |
|---|---|---|---|---|---|---|---|
| Coumaric acid | FjTAL | One copy genome integration | S. cerevisiae CEN.PK102-5B/Aromatic amino acids overproducing strain | Synthetic fed-batch medium | Microtiter plates | 1900 | [47] |
| Coumaric acid | FjTAL | One copy genome integration | S. cerevisiae CEN.PK102-5B/Knockout of the downregulated transporter TAT1 | Glucose (10 g/L) | Fed-batch | 2400 | [48] |
| Coumaric acid | FjTAL, xylose utilization genes | One copy genome integration | S. cerevisiae CMB.GS010/Aromatic amino acids overproducing strain | Xylose (15 g/L) | Batch bioreactor | 242 | [49] |
| Coumaric acid | AtPAL, AtC4H | One copy genome integration | S. cerevisiae IMX/Aromatic amino acids overproducing strain; Overexpression of CYB5 and expression of AtCPR to enhance cytochrome P450 activity | Glucose (20 g/L) | Fed-batch | 12500 | [50] |
| Caffeic acid | RtTAL, PaHpaB, SeHpaC | Episomal plasmid | S. cerevisiae BY4741/- | Tyrosine (0.5 g/L) | Shake flask | 289.4 | [51] |
| Caffeic acid | RcTAL, AtC3H, AtCPR | One copy genome integration | S. cerevisiae BY4742/Aromatic amino acids overproducing strain; Expression of AtCPR to enhance cytochrome P450 activity | Glucose (40 g/L) | Shake flask | 11.4 | [52] |
| Resveratrol | Ph4CL216, VvVST1 | Episomal plasmid | S. cerevisiae FY23/- | Coumaric acid (10 mg/L) | Shake flask | 0.0014 | [53] |
| Resveratrol | Nt4CL2, VvSTS | One copy genome integration | S. cerevisiae CEN-PK113-3B/- | Coumaric acid (820 mg/L) | Shake flask | 5.8 | [54] |
| Resveratrol | At4CL1, VvSTS | Episomal plasmid | S. cerevisiae strain isolated from a Brazilian sugar cane plantation. | Coumaric acid (2.46 g/L) | Shake flask | 391 | [55] |
| Resveratrol | At4CL1, AhSTS | Episomal plasmid | S. cerevisiae W303-1A/PAD knock-out | Coumaric acid (16 mg/L) | Shake flask | 3.1 | [56] |
| Resveratrol | At4CL1 and VvSTS fusion protein | One copy genome integration | S. cerevisiae WAT11/Expression of araE transporter | Coumaric acid (10 mg/L) every 24 h | Shake flask | 2.3 | [57] |
| Resveratrol | RtPAL, AtC4H, At4CL1, AhSTS | Episomal plasmid | S. cerevisiae W303-1A/Overexpression of ACC1 | Tyrosine (2.17 g/L) | Batch bioreactor | 5.8 | [58] |
| Resveratrol | At4CL1 and VvSTS synthetic scaffold | Episomal plasmid | S. cerevisiae WAT11/- | Coumaric acid (20 mg/L) | Shake flask | 14.4 | [59] |
| Resveratrol | HaTAL, At4CL1, VvVST | Multiple copy genome integration | S. cerevisiae CEN.PK102-5B/Aromatic amino acids overproducing strain | Glucose (40 g/L) | Fed-batch bioreactor | 531.4 | [60] |
| Resveratrol | AtPAL, AtC4H, At4CL2, VvVST1 | Multiple copy genome integration | S. cerevisiae CEN.PK102-5B/Aromatic amino acids overproducing strain; Expression of AtCPR and CYB5 to enhance cytochrome P450 activity | Glucose (88 g/L) | Fed-batch bioreactor | 800 | [61] |
| Pinostilbene | AtPAL, AtC4H, At4CL2, VvVST1, VvROMT | 1.96 | |||||
| Pterostilbene | AtPAL, AtC4H, At4CL2, VvVST1, VvROMT | 34.9 | |||||
| Pterostilbene | At4CL, VvSTS, VvROMT | Episomal plasmids | S. cerevisiae WAT11 | Coumaric acid (10 mg/L) | Shake flask | 2.2 | [62] |
| Naringenin chalcone | RtPAL, At4CL1, HaCHS | Episomal plasmids | S. cerevisiae AH22/PAD knock-out | Galactose (20 g/L) | Shake flask | 7 | [63] |
| Naringenin | AtPAL, AtCHI, AtCHS, At4CL3, AtTAL, AtC4H, AtCHS3 | One copy genome integration | S. cerevisiae CEN.PK717.5A/Aromatic amino acids overproducing strain; Expression of AtCPR to enhance cytochrome P450 activity | Glucose (20 g/L) | Batch bioreactor | 108.9 | [64] |
| Naringenin | PhPAL, Gm4CL2, GmCHS, GmCHI | One copy genome integration | S. cerevisiae YPH499/Expression of PhCPR to enhance cytochrome P450 activity | Phenylalanine (1.7 g/L) | Shake flask | 8.9 | [65] |
| Genistein | PhPAL, Gm4CL2, GmCHS, GmCHI, GmIFS | 0.1 | |||||
| Kaempferol | PhPAL, Gm4CL2, GmCHS, GmCHI, GmF3′H, StFLS | 1.3 | |||||
| Quercetin | PhPAL, GmC4H, Gm4CL2, GmCHS, GmCHI, GmF3′H, StFLS | 0.26 | |||||
| Naringenin | FjTAL, Pc4CL, PhCHS, MsCHI | One copy genome integration | S. cerevisiae CEN.PK102-5B/Aromatic amino acids overproducing strain; Expression of CrCPR to enhance cytochrome P450 activity | Synthetic fed-batch medium | Microtiter plates | 1.55 | [66] |
| Liquiritigenin | FjTAL, Pc4CL, PhCHS, MsCHI, AmCHR | 5.31 | |||||
| Kaempferol | FjTAL, Pc4CL, PhCHS, MsCHI, AmF3H, AtFLS | 26.57 | |||||
| Resokaempferol | FjTAL, Pc4CL, PhCHS, AmCHR, MsCHI, AmF3H, AtFLS | 0.51 | |||||
| Quercetin | FjTAL, Pc4CL, PhCHS, MsCHI, AmF3H, AtFLS, PhFMO | 20.38 | |||||
| Fisetin | FjTAL, Pc4CL, PhCHS, AmCHR, MsCHI, AmF3H, AtFLS, PhFMO | 1.65 | |||||
| Naringenin | FjTAL, At4CL2, HaCHS, MsCHI | One copy genome integration | S. cerevisiae BY4741/Aromatic amino acids overproducing strain; Overexpression of ACC1 and decrease of the flux towards fatty acid biosynthesis | Sucrose (10 g/L) | Shake flask | 90 | [67] |
| Kaempferol | EgPAL, EgC4H, Eg4CL, EgCHS, EgCHI, AtF3H, PdFLS | One copy genome integration | S. cerevisiae W303-1A/Increased flux towards malonyl-CoA synthesis | Glucose (20 g/L) | Fed-batch bioreactor | 66.3 | [68] |
| Naringenin | FjTAL, At4CL1, HaCHS, PhCHI | Episomal plasmids | S. cerevisiae BY4741/Aromatic amino acids overproducing strain | Glucose (20 g/L) | Shake flask | 220 | [69] |
| Kaempferol | FjTAL; At4CL1; HaCHS; CuF3H, AtFLS | 86 | |||||
| Naringenin | AtPAL, AtTAL, AtC4H, At4CL3, AtCHI, AtCHS3, SfFPT | One copy genome integration and episomal plasmids | S. cerevisiae IMK393/Aromatic amino acids overproducing strain; Expression of AtCPR to enhance cytochrome P450 activity; replacement of TSC13 by an orthologue gene | Glucose (20 g/L) | Shake flask | 100 | [70] |
| 8-Prenylnaringenin | 0.12 | ||||||
| Liquiritin | GuPAL, GuC4H, Gu4CL1, GuCHS, GuCHR, GuCHI, GuUGT | Episomal plasmids | S. cerevisiae WM4-3 | Glucose (20 g/L) | Fed-batch bioreactor | 0.42 | [71] |
| Phloterin | AtPAL, AmmC4H, At4CL2, HaCHS, ScTSC13 | Episomal plasmids | S. cerevisiae BG/Overexpression of ScCPR1 to enhance cytochrome P450 activity | Glucose (20 g/L) | Microtiter plates | 42.7 | [72] |
| Pinocembrin | AtPAL, At4CL2, HaCHS, ScTSC13 | 2.6 | |||||
| Phlorizin | AtPAL, AmmC4H, At4CL2, HaCHS, ScTSC13, PycUGT | Episomal plasmids | S. cerevisiae BG/Overexpression of ScCPR1 to enhance cytochrome P450 activity | Glucose (20 g/L) | Microtiter plates | 65 | [72] |
| Nothofagin | AtPAL, AmmC4H, At4CL2, HaCHS, ScTSC13, OsUGT | 59 | |||||
| Trilobatin | AtPAL, AmmC4H, At4CL2, HaCHS, ScTSC13, AtUGT | 32.8 | |||||
| Naringin dihydrochalcone | AtPAL, AmmC4H, At4CL2, HaCHS, ScTSC13, AtUGT, CmRHAT, AtRHM | 11.6 | |||||
| 3-Hydroxyphloretin | AtPAL, AmmC4H, At4CL2, HaCHS, ScTSC13, CsCH3H, AtATR | 28.8 | |||||
| Afzelechin | AtPAL, AnmC4H, MsCHI, At4CL2, MdF3H, AaDFR, VvLAR | One copy genome integration | S. cerevisiae BG/Expression of AtCPR1 to enhance cytochrome P450 activity | Glucose (20 g/L) | Microtiter plates | ~80 4 | [73] |
| Catechin | AtPAL, AnmC4H, MsCHI, At4CL2, MdF3H, MdF3′H, AaDFR, VvLAR | ~90 4 | |||||
| Gallocatechin | AtPAL, AnmC4H, MsCHI, At4CL2, MdF3H, MdF3′5′H, AaDFR, VvLAR | ~25 4 | |||||
| Pelargonidin-3-O-glucosidase | AtPAL, AnmC4H, MsCHI, At4CL2, MdF3H, AaDFR, PhANS, DcA3GT | 0.85 | |||||
| Cyanidin-3-O-glucosidase | AtPAL, AnmC4H, MsCHI, At4CL2, MdF3H, MdF3′H, AaDFR, PhANS, FaA3GT | 1.55 | |||||
| Delphinidin-3-O-glucosidase | AtPAL, AnmC4H, MsCHI, At4CL2, MdF3H, MdF3′5´H, AaDFR, PhANS, FaA3GT | 1.86 | |||||
| Apigenin | AtC4H, Pc4CL2, PehCHI, PehCHS, PcFSI | Episomal plasmids | S. cerevisiae INVSc1/Overexpression of ScCPR and AnmCPR to enhance cytochrome P450 activity | Cinnamic acid (74 mg/L) | Shake flask | 0.4 | [74] |
| Naringenin | AtC4H, Pc4CL2, PehCHI, PehCHS, PcFSI | 0.07 | |||||
| Luteolin | AtC4H, Pc4CL2, PehCHI, PehCHS, PcFSI | 1.1 | |||||
| Eriodictoyl | AtC4H, Pc4CL2, PehCHI, PehCHS, PcFSI | 2.5 | |||||
| Pinocembrin | AtC4H, Pc4CL2, PehCHI, PehCHS, PcFSI | 1.4 | |||||
| Naringenin | FjTAL, At4CL1, AtCHS, AtCHI | One copy genome integration | S. cerevisiae BY4741/Increased flux towards aromatic amino acids and malonyl-CoA biosynthesis | Glucose (20 g/L) | Shake flask | 144.1 | [75] |
| Kaempferol | FjTAL, At4CL1, AtCHS, AtCHI, NtF3H, AtFLS | 168.1 | |||||
| Quercetin | FjTAL, At4CL1, AtCHS, AtCHI, NtF3H, PhF3′H, AtFLS | 154.2 | |||||
| Myricetin | FjTAL, At4CL1, AtCHS, AtCHI, NtF3H, PhF3′H, SlF3′5′H, AtFLS | 145 | |||||
| Pelargodinin | FjTAL, At4CL1, AtCHS, AtCHI, NtF3H, AaDFR, GeANS | 33.3 | |||||
| Cyanidin | FjTAL, At4CL1, AtCHS, AtCHI, NtF3H, PhF3′H, GeDFR, GeANS | 31.7 | |||||
| p-Coumaroyl-3-hydroxyanthranilic acid | Nt4CL2, CcHCT | Episomal plasmids | S. cerevisiae CENPK113-5d/- | Coumaric acid (462 mg/L) and HAA (77 mg/L) | Batch bioreactor | 120 | [38] |
| Caffeoyl-3-hydroxyanthranilic acid | Caffeic acid (540 mg/L) and HAA (77 mg/L) | 22 |
Genome Integration
2.2. Pathway Enzyme Engineering
Spatial Location of Heterologous Enzymes
2.3. Strain Optimization
2.3.1. Amino Acids Overproducing Strains
2.3.2. Extender Substrates Availability
2.3.3. Transporter Expression
3. Key Messages
Author Contributions
Funding
Conflicts of Interest
References
- Pagare, S.; Bhatia, M.; Tripathi, N.; Pagare, S.; Bansal, Y.K. Secondary metabolites of plants and their role: Overview. Curr. Trends Biotechnol. Pharm. 2016, 9, 293–304. [Google Scholar]
- Yang, L.; Wen, K.S.; Ruan, X.; Zhao, Y.X.; Wei, F.; Wang, Q. Response of plant secondary metabolites to environmental factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef]
- Galanakis, C.M. Polyphenols: Properties, Recovery, and Applications; Woodhead Publishing: Sawston, UK; Cambrigde, UK, 2018. [Google Scholar]
- Global Polyphenols Market Will Register a CAGR of Around 9.0% by 2027. Available online: https://proficientmarket.com/press-release/1225/global-polyphenols-market (accessed on 21 April 2020).
- Global Flavonoids Market Will Reach USD 1047.63 Million in 2021. Available online: https://www.zionmarketresearch.com/news/global-flavonoids-market (accessed on 21 April 2020).
- Resveratrol Market—Global Industry Trends and Forecast to 2027|Data Bridge Market Research. Available online: https://www.databridgemarketresearch.com/reports/global-resveratrol-market (accessed on 21 April 2020).
- Curcumin Market Size Worth $151.9 Million By 2027. Available online: https://www.grandviewresearch.com/press-release/curcumin-market (accessed on 21 April 2020).
- Ezzati, M.; Yousefi, B.; Velaei, K.; Safa, A. A review on anti-cancer properties of quercetin in breast cancer. Life Sci. 2020, 248, 117463. [Google Scholar] [CrossRef] [PubMed]
- Tomeh, M.A.; Hadianamrei, R.; Zhao, X. A review of curcumin and its derivatives as anticancer agents. Int. J. Mol. Sci. 2019, 20, 1033. [Google Scholar] [CrossRef]
- Khan, H.; Reale, M.; Ullah, H.; Sureda, A.; Tejada, S.; Wang, Y.; Zhang, Z.J.; Xiao, J. Anti-cancer effects of polyphenols via targeting p53 signaling pathway: Updates and future directions. Biotechnol. Adv. 2020, 38, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Teplova, V.V.; Isakova, E.P.; Klein, O.I.; Dergachova, D.I.; Gessler, N.N.; Deryabina, Y.I. Natural polyphenols: Biological activity, pharmacological potential, means of metabolic engineering (Review). Appl. Biochem. Microbiol. 2018, 54, 221–237. [Google Scholar] [CrossRef]
- Rodríguez-García, C.; Sánchez-Quesada, C.; Gaforio, J.J. Dietary flavonoids as cancer chemopreventive agents: An updated review of human studies. Antioxidants 2019, 8, 137. [Google Scholar] [CrossRef]
- Abbaszadeh, H.; Keikhaei, B.; Mottaghi, S. A review of molecular mechanisms involved in anticancer and antiangiogenic effects of natural polyphenolic compounds. Phytother. Res. 2019, 33, 2002–2014. [Google Scholar] [CrossRef]
- Kumar Singh, S.; Patra, A. Evaluation of phenolic composition, antioxidant, anti-inflammatory and anticancer activities of Polygonatum verticillatum (L.). J. Integr. Med. 2018, 16, 273–282. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Wang, Z. Antioxidant activities of leaf extract of Salvia miltiorrhiza Bunge and related phenolic constituents. Food Chem. Toxicol. 2010, 48, 2656–2662. [Google Scholar] [CrossRef]
- Gallardo, M.J.; Suwalsky, M.; Ramírez, D.; Tapia, J.; Sepulveda, B. Antioxidant effect of resveratrol in single red blood cells measured by thermal fluctuation spectroscopy. Arch. Biochem. Biophys. 2016, 665, 30–35. [Google Scholar] [CrossRef] [PubMed]
- García-Alonso, J.; Ros, G.; Vidal-Guevara, M.L.; Periago, M.J. Acute intake of phenolic-rich juice improves antioxidant status in healthy subjects. Nutr. Res. 2006, 26, 330–339. [Google Scholar] [CrossRef]
- Cruz-Correa, M.; Shoskes, D.A.; Sanchez, P.; Zhao, R.; Hylind, L.M.; Wexner, S.D.; Giardiello, F.M. Combination treatment with curcumin and quercetin of adenomas in familial Adenomatous polyposis. Clin. Gastroenterol. Hepatol. 2006, 4, 1035–1038. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Y.; Shi, C.B.; Wen, H.; Li, F.L.; Wang, B.L.; Wang, J. Upregulation of p53 expression in patients with colorectal cancer by administration of curcumin. Cancer Investig. 2011, 29, 208–213. [Google Scholar] [CrossRef]
- Kundur, S.; Prayag, A.; Selvakumar, P.; Nguyen, H.; McKee, L.; Cruz, C.; Srinivasan, A.; Shoyele, S.; Lakshmikuttyamma, A. Synergistic anticancer action of quercetin and curcumin against triple-negative breast cancer cell lines. J. Cell. Physiol. 2019, 234, 11103–11118. [Google Scholar] [CrossRef]
- Xiong, H.; Cheng, J.; Jiang, S.; Wen, J.; Jian, Y.; Wei, L.; Zhe, Z.; Jiang, F.-Q.; Peng, X. The antitumor effect of resveratrol on nasopharyngeal carcinoma cells. Front. Biosci. 2019, 24, 961–970. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Chang, Y.M.; Wang, K.Y.; Chen, P.N.; Hseu, Y.C.; Chen, K.M.; Yeh, K.T.; Chen, S.J.; Hsu, L.S. Naringenin inhibited migration and invasion of glioblastoma cells through multiple mechanisms. Environ. Toxicol. 2019, 34, 233–239. [Google Scholar] [CrossRef]
- Panahi, Y.; Hosseini, M.S.; Khalili, N.; Naimi, E.; Majeed, M.; Sahebkar, A. Antioxidant and anti-inflammatory effects of curcuminoid-piperine combination in subjects with metabolic syndrome: A randomized controlled trial and an updated meta-analysis. Clin. Nutr. 2015, 34, 1101–1108. [Google Scholar] [CrossRef]
- Pinho-Ribeiro, F.A.; Zarpelon, A.C.; Fattori, V.; Manchope, M.F.; Mizokami, S.S.; Casagrande, R.; Verri, W.A. Naringenin reduces inflammatory pain in mice. Neuropharmacology 2016, 105, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Porro, C.; Cianciulli, A.; Trotta, T.; Lofrumento, D.D.; Panaro, M.A. Curcumin regulates anti-inflammatory responses by JAK/STAT/SOCS signaling pathway in BV-2 microglial cells. Biology 2019, 8, 51. [Google Scholar] [CrossRef]
- Lecher, J.C.; Diep, N.; Krug, P.W.; Hilliard, J.K. Genistein has antiviral activity against herpes B virus and acts synergistically with antiviral treatments to reduce effective dose. Viruses 2019, 11, 499. [Google Scholar] [CrossRef]
- Zhao, X.; Cui, Q.; Fu, Q.; Song, X.; Jia, R.; Yang, Y.; Zou, Y.; He, C.; Liang, X.; Yin, L.; et al. Antiviral properties of resveratrol against pseudorabies virus are associated with the inhibition of IκB kinase activation. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Lima, M.C.; Paiva de Sousa, C.; Fernandez-Prada, C.; Harel, J.; Dubreuil, J.D.; de Souza, E.L. A review of the current evidence of fruit phenolic compounds as potential antimicrobials against pathogenic bacteria. Microb. Pathog. 2019, 130, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-del-Río, I.; Fernández, J.; Lombó, F. Plant nutraceuticals as antimicrobial agents in food preservation: Terpenoids, polyphenols and thiols. Int. J. Antimicrob. Agents 2018, 52, 309–315. [Google Scholar] [CrossRef]
- Moussa, C.; Hebron, M.; Huang, X.; Ahn, J.; Rissman, R.A.; Aisen, P.S.; Turner, R.S. Resveratrol regulates neuro-inflammation and induces adaptive immunity in Alzheimer’s disease. J. Neuroinflamm. 2017, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Small, G.W.; Siddarth, P.; Li, Z.; Miller, J.K.; Ercoli, L.; Emerson, D.N.; Martinez, J.; Wong, K.-P.; Liu, J.; Merril, A.D.; et al. Memory and brain amyloid and Tau effects of a bioavailable form of curcumin in non-demented adults: A double-blind, placebo-controlled 18-month trial. Am. J. Geriatr. Psychiatry 2018, 26, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Enogieru, A.B.; Haylett, W.; Hiss, D.C.; Bardien, S.; Ekpo, O.E. Rutin as a potent antioxidant: Implications for neurodegenerative disorders. Oxidative Med. Cell. Longev. 2018, 2018, 6241017. [Google Scholar] [CrossRef] [PubMed]
- Erkkola, R.; Vervarcke, S.; Vansteelandt, S.; Rompotti, P.; De Keukeleire, D.; Heyerick, A. A randomized, double-blind, placebo-controlled, cross-over pilot study on the use of a standardized hop extract to alleviate menopausal discomforts. Phytomedicine 2010, 17, 389–396. [Google Scholar] [CrossRef]
- Luo, D.; Kang, L.; Ma, Y.; Chen, H.; Kuang, H.; Huang, Q.; He, M.; Peng, W. Effects and mechanisms of 8-prenylnaringenin on osteoblast MC3T3-E1 and osteoclast-like cells RAW264.7. Food Sci. Nutr. 2014, 2, 341–350. [Google Scholar] [CrossRef]
- Averesch, N.J.H.; Krömer, J.O. Metabolic engineering of the shikimate pathway for production of aromatics and derived compounds—Present and future strain construction strategies. Front. Bioeng. Biotechnol. 2018, 6, 1–19. [Google Scholar] [CrossRef]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III polyketide synthases in natural product biosynthesis. Iubmb Life 2012, 64, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Takahashi, S.; Waki, T. Formation of flavonoid metabolons: Functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front. Plant Sci. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Moglia, A.; Goitre, L.; Gianoglio, S.; Baldini, E.; Trapani, E.; Genre, A.; Scattina, A.; Dondo, G.; Trabalzini, L.; Beekwilder, J.; et al. Evaluation of the bioactive properties of avenanthramide analogs produced in recombinant yeast. BioFactors 2015, 41, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jian, X.; Wu, J.; Huang, W.; Huang, C.; Luo, J.; Kong, L. Elucidation of the biosynthesis pathway and heterologous construction of a sustainable route for producing umbelliferone. J. Biol. Eng. 2019, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Satake, H.; Shiraishi, A.; Koyama, T.; Matsumoto, E.; Morimoto, K.; Bahabadi, S.E.; Ono, E.; Murata, J. Lignan biosynthesis for food bioengineering. Food Biosynth. 2017, 351–379. [Google Scholar] [CrossRef]
- Wang, J.; Guleria, S.; Koffas, M.A.G.; Yan, Y. Microbial production of value-added nutraceuticals. Curr. Opin. Biotechnol. 2016, 37, 97–104. [Google Scholar] [CrossRef]
- Rodrigues, J.L.; Prather, K.L.J.; Kluskens, L.D.; Rodrigues, L.R. Heterologous production of curcuminoids. Microbiol. Mol. Biol. Rev. 2015, 79, 39–60. [Google Scholar] [CrossRef]
- Wilson, S.A.; Roberts, S.C. Recent advances towards development and commercialization of plant cell culture processes for the synthesis of biomolecules. Plant Biotechnol. J. 2012, 10, 249–268. [Google Scholar] [CrossRef]
- Braga, A.; Rocha, I.; Faria, N. Microbial Hosts as a Promising Platform for Polyphenol Production. In Natural Bio-active Compounds; Akhtar, M., Swamy, M., Sinniah, U., Eds.; Springer: Singapore, 2019; Volume 1, pp. 71–103. [Google Scholar]
- Hanson, P.K. Saccharomyces cerevisiae: A unicellular model genetic organism of enduring importance. Curr. Protoc. Essent. Lab. Tech. 2018, 16, 1–15. [Google Scholar] [CrossRef]
- Shao, Z.; Zhao, H. Manipulating natural product biosynthetic pathways via DNA assembler. Curr. Protoc. Chem. Biol. 2014, 6, 65–100. [Google Scholar] [CrossRef]
- Rodriguez, A.; Kildegaard, K.; Li, M.; Borodina, I.; Nielsen, J. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab. Eng. 2015, 31, 181–188. [Google Scholar] [CrossRef]
- Rodriguez, A.; Chen, Y.; Khoomrung, S.; Özdemir, E.; Borodina, I.; Nielsen, J. Comparison of the metabolic response to over-production of p-coumaric acid in two yeast strains. Metab. Eng. 2017, 44, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Borja, G.M.; Rodriguez, A.; Campbell, K.; Borodina, I.; Chen, Y.; Nielsen, J. Metabolic engineering and transcriptomic analysis of Saccharomyces cerevisiae producing p-coumaric acid from xylose. Microb. Cell Factories 2019, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yu, T.; Li, X.; Chen, Y.; Campbell, K.; Nielsen, J.; Chen, Y. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Liu, L.; Liu, H.; Zhang, W.; Yao, M.; Li, B.; Liu, D.; Yuan, Y. Engineering the biosynthesis of caffeic acid in Saccharomyces cerevisiae with heterologous enzyme combinations. Engineering 2019, 5, 287–295. [Google Scholar] [CrossRef]
- Li, Y.; Mao, J.; Liu, Q.; Song, X.; Wu, Y.; Cai, M.; Xu, H.; Qiao, M. De novo biosynthesis of caffeic acid from glucose by engineered Saccharomyces cerevisiae. ACS Synth. Biol. 2020, 9, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.V.W.; Armstrong, G.O.; van der Merwe, M.J.; Lambrechts, M.G.; Vivier, M.A.; Pretorius, I.S. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res. 2003, 4, 79–85. [Google Scholar] [CrossRef]
- Beekwilder, J.; Wolswinkel, R.; Jonker, H.; Hall, R.; De Rie Vos, C.H.; Bovy, A. Production of resveratrol in recombinant microorganisms. Appl. Environ. Microbiol. 2006, 72, 5670–5672. [Google Scholar] [CrossRef]
- Sydor, T.; Schaffer, S.; Boles, E. Considerable increase in resveratrol production by recombinant industrial yeast strains with use of rich medium. Appl. Environ. Microbiol. 2010, 76, 3361–3363. [Google Scholar] [CrossRef]
- Shin, S.Y.; Han, N.S.; Park, Y.C.; Kim, M.D.; Seo, J.H. Production of resveratrol from p-coumaric acid in recombinant Saccharomyces cerevisiae expressing 4-coumarate:coenzyme A ligase and stilbene synthase genes. Enzym. Microb. Technol. 2011, 48, 48–53. [Google Scholar] [CrossRef]
- Wang, Y.; Halls, C.; Zhang, J.; Matsuno, M.; Zhang, Y.; Yu, O. Stepwise increase of resveratrol biosynthesis in yeast Saccharomyces cerevisiae by metabolic engineering. Metab. Eng. 2011, 13, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.-Y.; Jung, S.-M.; Kim, M.-D.; Han, N.S.; Seo, J.-H. Production of resveratrol from tyrosine in metabolically engineered Saccharomyces Cerevisiae. Enzym. Microb. Technol. 2012, 51, 211–216. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, O. Synthetic scaffolds increased resveratrol biosynthesis in engineered yeast cells. J. Biotechnol. 2012, 157, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Kildegaard, K.R.; Chen, Y.; Rodriguez, A.; Borodina, I.; Nielsen, J. De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae. Metab. Eng. 2015, 32, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Schneider, K.; Kristensen, M.; Borodina, I.; Nielsen, J. Engineering yeast for high-level production of stilbenoid antioxidants. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bhuiya, M.W.; Zhou, R.; Yu, O. Pterostilbene production by microorganisms expressing resveratrol O-methyltransferase. Ann. Microbiol. 2015, 65, 817–826. [Google Scholar] [CrossRef]
- Jiang, H.; Wood, K.V.; Morgan, J.A. Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2962–2969. [Google Scholar] [CrossRef]
- Koopman, F.; Beekwilder, J.; Crimi, B.; van Houwelingen, A.; Hall, R.D.; Bosch, D.; van Maris, A.J.A.; Pronk, J.T.; Daran, J.M. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb. Cell Factories 2012, 11, 1–15. [Google Scholar] [CrossRef]
- Trantas, E.; Panopoulos, N.; Ververidis, F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces Cerevisiae. Metab. Eng. 2009, 11, 355–366. [Google Scholar] [CrossRef]
- Rodriguez, A.; Strucko, T.; Stahlhut, S.G.; Kristensen, M.; Svenssen, D.K.; Forster, J.; Nielsen, J.; Borodina, I. Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour. Technol. 2017, 245, 1645–1654. [Google Scholar] [CrossRef]
- Lyu, X.; Ng, K.R.; Lee, J.L.; Mark, R.; Chen, W.N. Enhancement of naringenin biosynthesis from tyrosine by metabolic engineering of Saccharomyces cerevisiae. J. Agric. Food Chem. 2017, 65, 6638–6646. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Ding, W.; Liu, X.; Cheng, X.; Cai, J.; Hua, E.; Jiang, H. Biosynthesis and engineering of kaempferol in Saccharomyces cerevisiae. Microb. Cell Factories 2017, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Zhao, G.; Ng, K.R.; Mark, R.; Chen, W.N. Metabolic engineering of Saccharomyces cerevisiae for de Novo production of kaempferol. J. Agric. Food Chem. 2019, 67, 5596–5606. [Google Scholar] [CrossRef] [PubMed]
- Levisson, M.; Araya-Cloutier, C.; De Bruijn, W.J.C.; Van Der Heide, M.; Salvador López, J.M.; Daran, J.M.; Vincken, J.P.; Beekwilder, J. Toward developing a yeast cell factory for the production of prenylated flavonoids. J. Agric. Food Chem. 2019, 67, 13478–13486. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Li, Y.; Jiang, D.; Zhang, X.; Gao, W.; Liu, C. De novo biosynthesis of liquiritin in Saccharomyces cerevisiae. Acta Pharm. Sin. B 2019, 10, 711–721. [Google Scholar] [CrossRef]
- Eichenberger, M.; Lehka, B.J.; Folly, C.; Fischer, D.; Martens, S.; Simón, E.; Naesby, M. Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab. Eng. 2017, 39, 80–89. [Google Scholar] [CrossRef]
- Eichenberger, M.; Hansson, A.; Fischer, D.; Dürr, L.; Naesby, M. De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, 1–13. [Google Scholar] [CrossRef]
- Leonard, E.; Yan, Y.; Lim, K.H.; Koffas, M.A.G. Investigation of two distinct flavone synthases for plant-specific flavone biosynthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 8241–8248. [Google Scholar] [CrossRef]
- Du, Y.; Yang, B.; Yi, Z.; Hu, L.; Li, M. Engineering Saccharomyces cerevisiae coculture platform for the production of flavonoids. J. Agric. Food Chem. 2020, 68, 2146–2154. [Google Scholar] [CrossRef]
- Gnügge, R.; Rudolf, F. Saccharomyces cerevisiae shuttle vectors. Yeast 2017, 34, 205–221. [Google Scholar] [CrossRef]
- Karim, A.; Curran, K.; Alper, H. Characterization of plasmid burden and copy number in Saccharomyces cerevisiae for optimization of metabolic engineering applications. FEMS Yeast Res. 2013, 13, 107–116. [Google Scholar] [CrossRef]
- Lee, F.W.F.; Da Silva, N.A. Sequential δ-integration for the regulated insertion of cloned genes in Saccharomyces cerevisiae. Biotechnol. Prog. 1997, 13, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Ching, C.B. Combinatorial assembly of large biochemical pathways into yeast chromosomes for improved production of value-added compounds. ACS Synth. Biol. 2015, 4, 23–31. [Google Scholar] [CrossRef] [PubMed]
- DiCarlo, J.; Norville, J.; Mali, P.; Rios, X.; Aach, J.; Church, G. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Liang, Y.; Zhang, M.; Ang, E.; Zhao, H. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab. Eng. 2016, 33, 19–27. [Google Scholar] [CrossRef]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef]
- Levisson, M.; Patinios, C.; Hein, S.; de Groot, P.A.; Daran, J.M.; Hall, R.D.; Martens, S.; Beekwilder, J. Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb. Cell Factories 2018, 17, 1–16. [Google Scholar] [CrossRef]
- Lv, X.; Wang, F.; Zhou, P.; Ye, L.; Xie, W.; Xu, H.; Yu, H. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae. Nat. Commun. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Farhi, M.; Marhevka, E.; Masci, T.; Marcos, E.; Eyal, Y.; Ovadis, M.; Abeliovich, H.; Vainstein, A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab. Eng. 2011, 13, 474–481. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.Z.; Li, J.; Pan, X.; Cahoon, R.E.; Jaworski, J.G.; Wang, X.; Jez, J.M.; Chen, F.; Yu, O. Using unnatural protein fusions to engineer resveratrol biosynthesis in yeast and mammalian cells. J. Am. Chem. Soc. 2006, 128, 13030–13031. [Google Scholar] [CrossRef]
- Gorman, S.D.; Boehr, D.D. Energy and enzyme activity landscapes of yeast chorismate mutase at cellular concentrations of allosteric effectors. Biochemistry 2019, 58, 4058–4069. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Keasling, J. Engineering cellular metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, X.; Shen, Y.; Hou, J.; Bao, X. Increasing malonyl-CoA derived product through controlling the transcription regulators of phospholipid synthesis in Saccharomyces Cerevisiae. ACS Synth. Biol. 2017, 6, 905–912. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.; Shen, Y.; Hou, J.; Bao, X. Screening phosphorylation site mutations in yeast acetyl-CoA carboxylase using malonyl-CoA sensor to improve malonyl-CoA-derived product. Front. Microbiol. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Shi, S.; Chen, Y.; Siewers, V.; Nielsen, J. Improving production of malonyl coenzyme A-derived metabolites by abolishing snf1-dependent regulation of Acc1. mBio 2014, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant. Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Rodrigues, J.L.; Gomes, D.; Rodrigues, L.R. A combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli. Front. Bioeng. Biotechnol. 2020, 8, 1–15. [Google Scholar] [CrossRef]
| Biological Activity | Mechanism of Action | References |
|---|---|---|
| Anticancer |
| [18,19,20,21,22] |
| Anti- inflammatory |
| [23,24,25] |
| Antiviral |
| [26,27] |
| Antimicrobial |
| [28,29] |
| Anti-ageing |
| [30,31,32] |
| Estrogenic |
| [33,34] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rainha, J.; Gomes, D.; Rodrigues, L.R.; Rodrigues, J.L. Synthetic Biology Approaches to Engineer Saccharomyces cerevisiae towards the Industrial Production of Valuable Polyphenolic Compounds. Life 2020, 10, 56. https://doi.org/10.3390/life10050056
Rainha J, Gomes D, Rodrigues LR, Rodrigues JL. Synthetic Biology Approaches to Engineer Saccharomyces cerevisiae towards the Industrial Production of Valuable Polyphenolic Compounds. Life. 2020; 10(5):56. https://doi.org/10.3390/life10050056
Chicago/Turabian StyleRainha, João, Daniela Gomes, Lígia R. Rodrigues, and Joana L. Rodrigues. 2020. "Synthetic Biology Approaches to Engineer Saccharomyces cerevisiae towards the Industrial Production of Valuable Polyphenolic Compounds" Life 10, no. 5: 56. https://doi.org/10.3390/life10050056
APA StyleRainha, J., Gomes, D., Rodrigues, L. R., & Rodrigues, J. L. (2020). Synthetic Biology Approaches to Engineer Saccharomyces cerevisiae towards the Industrial Production of Valuable Polyphenolic Compounds. Life, 10(5), 56. https://doi.org/10.3390/life10050056







