Maternal Obesity Modulates Expression of Satb2 in Hypothalamic VMN of Female Offspring
Abstract
1. Introduction
2. Results
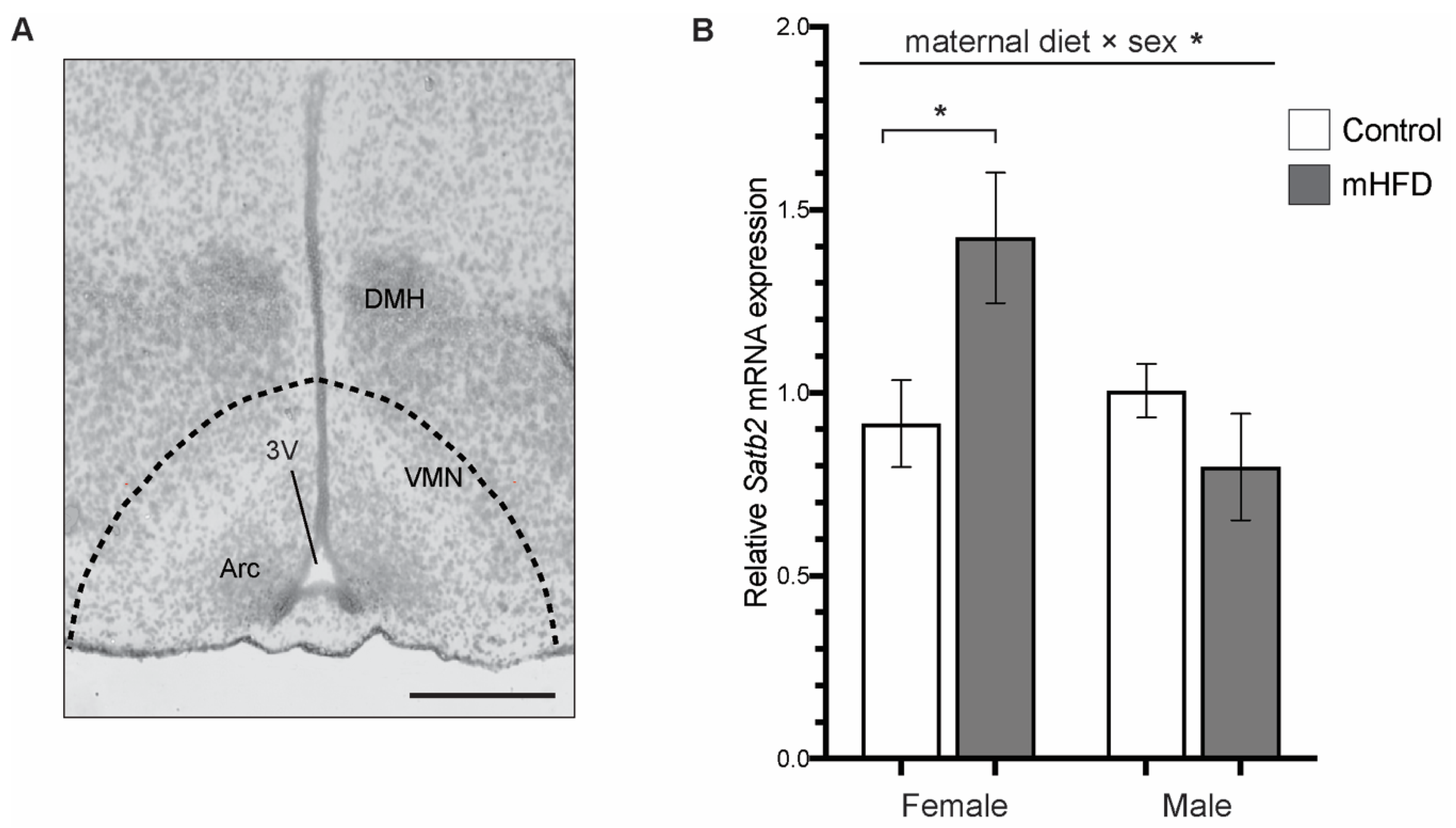
2.1. Satb2 mRNA is Upregulated in Mediobasal Hypothalamus of GD17.5 Female Offspring from mFD-Induced Obese Dams
2.2. mHFD Alters the Number of SATB2 Expressing Cells in the Rostral VMN of Female Offspring
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. RNA Extraction and qPCR Analysis
4.3. Immunohistochemistry
4.4. Imaging and Analysis
4.5. Statistics
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 3V | Third ventricle |
| Arc | Arcuate Nucleus |
| DMH | Dorsomedial hypothalamic nucleus |
| GD | Gestational Day |
| mHFD | Maternal high fat diet |
| qPCR | Quantitative PCR |
| VMN | Ventromedial nucleus of the hypothalamus |
References
- Schack-Nielsen, L.; Michaelsen, K.F.; Gamborg, M.; Mortensen, E.L.; Sorensen, T.I.A. Gestational weight gain in relation to offspring body mass index and obesity from infancy through adulthood. Int. J. Obes. 2010, 34, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Krakowiak, P.; Walker, C.K.; Bremer, A.A.; Baker, A.S.; Ozonoff, S.; Hansen, R.L.; Hertz-Picciotto, I. Maternal metabolic conditions and risk for autism and other neurodevelopmental disorders. Pediatrics 2012, 129, e1121–e1128. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A. Maternal pre-pregnancy obesity and risk for inattention and negative emotionality in children. J. Child Psychol. Psychiatr. Allied Discip. 2010, 51, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Talbot, C.P.J.; Dolinsky, V.W. Sex differences in the developmental origins of cardiometabolic disease following exposure to maternal obesity and gestational diabetes (1). Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2019, 44, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Dearden, L.; Bouret, S.G.; Ozanne, S.E. Sex and gender differences in developmental programming of metabolism. Mol. Metab. 2018, 15, 8–19. [Google Scholar] [CrossRef]
- Dearden, L.; Balthasar, N. Sexual dimorphism in offspring glucose-sensitive hypothalamic gene expression and physiological responses to maternal high-fat diet feeding. Endocrinology 2014, 155, 2144–2154. [Google Scholar] [CrossRef]
- Bilbo, S.D.; Tsang, V. Enduring consequences of maternal obesity for brain inflammation and behavior of offspring. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 2104–2115. [Google Scholar] [CrossRef]
- Glendining, K.A.; Fisher, L.C.; Jasoni, C.L. Maternal high fat diet alters offspring epigenetic regulators, amygdala glutamatergic profile and anxiety. Psychoneuroendocrinology 2018, 96, 132–141. [Google Scholar] [CrossRef]
- Kang, S.S.; Kurti, A.; Fair, D.A.; Fryer, J.D. Dietary intervention rescues maternal obesity induced behavior deficits and neuroinflammation in offspring. J. Neuroinflammation 2014, 11, 156. [Google Scholar] [CrossRef]
- Robb, J.L.; Messa, I.; Lui, E.; Yeung, D.; Thacker, J.; Satvat, E.; Mielke, J.G. A maternal diet high in saturated fat impairs offspring hippocampal function in a sex-specific manner. Behav. Brain Res. 2017, 326, 187–199. [Google Scholar] [CrossRef]
- Sullivan, E.L.; Grayson, B.; Takahashi, D.; Robertson, N.; Maier, A.; Bethea, C.L.; Smith, M.S.; Coleman, K.; Grove, K.L. Chronic consumption of a high-fat diet during pregnancy causes perturbations in the serotonergic system and increased anxiety-like behavior in nonhuman primate offspring. J. Neurosci. Off. J. Soc. Neurosci. 2010, 30, 3826–3830. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.J.; Chen, H. Established maternal obesity in the rat reprograms hypothalamic appetite regulators and leptin signaling at birth. Int. J. Obes. 2009, 33, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Glendining, K.A.; Grattan, D.R.; Jasoni, C.L. Maternal obesity leads to increased proliferation and numbers of astrocytes in the developing fetal and neonatal mouse hypothalamus. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 2016, 53, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Samuelsson, A.M.; Matthews, P.A.; Argenton, M.; Christie, M.R.; McConnell, J.M.; Jansen, E.H.; Piersma, A.H.; Ozanne, S.E.; Twinn, D.F.; Remacle, C.; et al. Diet-induced obesity in female mice leads to offspring hyperphagia, adiposity, hypertension, and insulin resistance: A novel murine model of developmental programming. Hypertension 2008, 51, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Sanders, T.R.; Kim, D.W.; Glendining, K.A.; Jasoni, C.L. Maternal obesity and IL-6 lead to aberrant developmental gene expression and deregulated neurite growth in the fetal arcuate nucleus. Endocrinology 2014, 155, 2566–2577. [Google Scholar] [CrossRef]
- Vucetic, Z.; Kimmel, J.; Reyes, T.M. Chronic high-fat diet drives postnatal epigenetic regulation of mu-opioid receptor in the brain. Neuropsychopharmacology 2011, 36, 1199–1206. [Google Scholar] [CrossRef]
- Bayless, D.W.; Shah, N.M. Genetic dissection of neural circuits underlying sexually dimorphic social behaviours. Philos. Trans. R Soc. Lond. B Biol. Sci. 2016, 371, 20150109. [Google Scholar] [CrossRef]
- Yang, C.F.; Chiang, M.C.; Gray, D.C.; Prabhakaran, M.; Alvarado, M.; Juntti, S.A.; Unger, E.K.; Wells, J.A.; Shah, N.M. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell 2013, 153, 896–909. [Google Scholar] [CrossRef]
- Bingham, N.C.; Anderson, K.K.; Reuter, A.L.; Stallings, N.R.; Parker, K.L. Selective loss of leptin receptors in the ventromedial hypothalamic nucleus results in increased adiposity and a metabolic syndrome. Endocrinology 2008, 149, 2138–2148. [Google Scholar] [CrossRef]
- Dhillon, H.; Zigman, J.M.; Ye, C.; Lee, C.E.; McGovern, R.A.; Tang, V.; Kenny, C.D.; Christiansen, L.M.; White, R.D.; Edelstein, E.A.; et al. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron 2006, 49, 191–203. [Google Scholar] [CrossRef]
- Cheung, C.C.; Krause, W.C.; Edwards, R.H.; Yang, C.F.; Shah, N.M.; Hnasko, T.S.; Ingraham, H.A. Sex-dependent changes in metabolism and behavior, as well as reduced anxiety after eliminating ventromedial hypothalamus excitatory output. Mol. Metab. 2015, 4, 857–866. [Google Scholar] [CrossRef]
- Talalaenko, A.N.; Pankrat’ev, D.V.; Goncharenko, N.V. Neurochemical characteristics of the ventromedial hypothalamus in mediating the antiaversive effects of anxiolytics in different models of anxiety. Neurosci. Behave. Physiol. 2003, 33, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.; Ye, C.; McCrimmon, R.J.; Dhillon, H.; Choi, B.; Kramer, M.D.; Yu, J.; Yang, Z.; Christiansen, L.M.; Lee, C.E.; et al. Synaptic glutamate release by ventromedial hypothalamic neurons is part of the neurocircuitry that prevents hypoglycemia. Cell Metab. 2007, 5, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, D.R.; Cullinan, W.E.; Herman, J.P. Distribution of vesicular glutamate transporter mRNA in rat hypothalamus. J. Comp. Neurol. 2002, 448, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Song, N.N.; Lan, W.; Hu, L.; Su, C.J.; Ding, Y.Q.; Zhang, L. Expression of transcription factor Satb2 in adult mouse brain. Anat. Rec. 2013, 296, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Kurrasch, D.M.; Cheung, C.C.; Lee, F.Y.; Tran, P.V.; Hata, K.; Ingraham, H.A. The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 13624–13634. [Google Scholar] [CrossRef]
- Chowdhury, G.M.I.; Wang, P.; Ciardi, A.; Mamillapalli, R.; Johnson, J.; Zhu, W.; Eid, T.; Behar, K.; Chan, O. Impaired glutamatergic neurotransmission in the ventromedial hypothalamus may contribute to defective counterregulation in recurrently hypoglycemic rats. Diabetes 2017, 66, 1979–1989. [Google Scholar] [CrossRef]
- Allen Developing Mouse Brain Atlas. Seattle (WA): Allen Institute for Brain Science. ©2009. Available online: http://developingmouse.brain-map.org. (accessed on 24 April 2020).
- Page, K.C.; Jones, E.K.; Anday, E.K. Maternal and postweaning high-fat diets disturb hippocampal gene expression, learning, and memory function. Am. J. Physiol. Reg. I 2014, 306, R527–R537. [Google Scholar] [CrossRef]
- Li, J.; Liu, C. Coding or noncoding, the converging concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef]
- Murphy, S.J.; Lusardi, T.A.; Phillips, J.I.; Saugstad, J.A. Sex differences in microRNA expression during development in rat cortex. Neurochem. Int. 2014, 77, 24–32. [Google Scholar] [CrossRef]
- Schumann, C.M.; Sharp, F.R.; Ander, B.P.; Stamova, B. Possible sexually dimorphic role of miRNA and other sncRNA in ASD brain. Mol. Autism 2017, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Ziats, M.N.; Rennert, O.M. Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatr. 2014, 19, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Azuma, R.; Akumuo, R.; Goetzl, L.; Pinney, S.E. Gestational diabetes and maternal obesity are associated with sex-specific changes in miRNA and target gene expression in the fetus. Int. J. Obes. 2019. [Google Scholar] [CrossRef] [PubMed]
- Hu, N.; Feng, C.; Jiang, Y.; Miao, Q.; Liu, H. Regulative effect of Mir-205 on osteogenic differentiation of bone mesenchymal stem cells (BMSCs): Possible role of SATB2/Runx2 and ERK/MAPK pathway. Int. J. Mol. Sci. 2015, 16, 10491–10506. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Shi, Y.; Zheng, L.; Zhou, B.; Inose, H.; Wang, J.; Guo, X.E.; Grosschedl, R.; Karsenty, G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell Boil. 2012, 197, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Koga, S.; Kojima, A.; Ishikawa, C.; Kuwabara, S.; Arai, K.; Yoshiyama, Y. Effects of diet-induced obesity and voluntary exercise in a tauopathy mouse model: Implications of persistent hyperleptinemia and enhanced astrocytic leptin receptor expression. Neurobiol. Dis. 2014, 71, 180–192. [Google Scholar] [CrossRef]
- Liu, Z.J.; Bian, J.; Liu, J.; Endoh, A. Obesity reduced the gene expressions of leptin receptors in hypothalamus and liver. Horm. Metab. Res. 2007, 39, 489–494. [Google Scholar] [CrossRef]
- Mellati, A.A.; Mazloomzadeh, S.; Anjomshoaa, A.; Alipour, M.; Karimi, F.; Mazloomi, S.; Naghi Kazemi, S.A. Multiple correlations between cord blood leptin concentration and indices of neonatal growth. Arch. Med. Res. 2010, 41, 26–32. [Google Scholar] [CrossRef]
- Tessier, D.R.; Ferraro, Z.M.; Gruslin, A. Role of leptin in pregnancy: Consequences of maternal obesity. Placenta 2013, 34, 205–211. [Google Scholar] [CrossRef]
- Valleau, J.C.; Sullivan, E.L. The impact of leptin on perinatal development and psychopathology. J. Chem. Neuroanat. 2014, 61–62, 221–232. [Google Scholar] [CrossRef]
- Glendining, K.A.; Higgins, M.B.A.; Fisher, L.C.; Jasoni, C.L. Maternal obesity modulates sexually dimorphic epigenetic regulation and expression of leptin receptor in offspring hippocampus. Brain Behav. Immun. 2020. [Google Scholar] [CrossRef] [PubMed]
- Glendining, K.A.; Markie, D.; Gardner, R.J.; Franz, E.A.; Robertson, S.P.; Jasoni, C.L. A novel role for the DNA repair gene Rad51 in Netrin-1 signalling. Sci. Rep. 2017, 7, 39823. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]

| ANOVA Table | SS | DF | MS | F (DFn, DFd) | p Value | Significance |
|---|---|---|---|---|---|---|
| Region | 353,820 | 2 | 176,910 | F (2, 24) = 39.36 | p < 0.0001 | **** |
| Sex | 396,375 | 1 | 396,375 | F (1, 24) = 88.20 | p < 0.0001 | **** |
| Diet | 17,270 | 1 | 17,270 | F (1, 24) = 3.843 | p = 0.0617 | ns |
| Region × Sex | 277,058 | 2 | 138,529 | F (2, 24) = 30.82 | p < 0.0001 | **** |
| Region × Diet | 84,658 | 2 | 42,329 | F (2, 24) = 9.418 | p = 0.0010 | *** |
| Sex × Diet | 885.1 | 1 | 885.1 | F (1, 24) = 0.1969 | p = 0.6612 | ns |
| Region × Sex × Diet | 52,894 | 2 | 26,447 | F (2, 24) = 5.885 | p = 0.0083 | ** |
| Residual | 107862 | 24 | 4494 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glendining, K.A.; Fisher, L.C.; Jasoni, C.L. Maternal Obesity Modulates Expression of Satb2 in Hypothalamic VMN of Female Offspring. Life 2020, 10, 48. https://doi.org/10.3390/life10040048
Glendining KA, Fisher LC, Jasoni CL. Maternal Obesity Modulates Expression of Satb2 in Hypothalamic VMN of Female Offspring. Life. 2020; 10(4):48. https://doi.org/10.3390/life10040048
Chicago/Turabian StyleGlendining, Kelly A., Lorryn C. Fisher, and Christine L. Jasoni. 2020. "Maternal Obesity Modulates Expression of Satb2 in Hypothalamic VMN of Female Offspring" Life 10, no. 4: 48. https://doi.org/10.3390/life10040048
APA StyleGlendining, K. A., Fisher, L. C., & Jasoni, C. L. (2020). Maternal Obesity Modulates Expression of Satb2 in Hypothalamic VMN of Female Offspring. Life, 10(4), 48. https://doi.org/10.3390/life10040048





