Plasma Long Noncoding RNA LeXis is a Potential Diagnostic Marker for Non-Alcoholic Steatohepatitis
Abstract
:1. Introduction
2. Methods
2.1. Patient Samples
2.2. NAFLD Diagnois and Pathological Evaluation
2.3. RNA Extraction
2.4. Quantitative Real-Time PCR
2.5. Statistical Analysis
3. Results
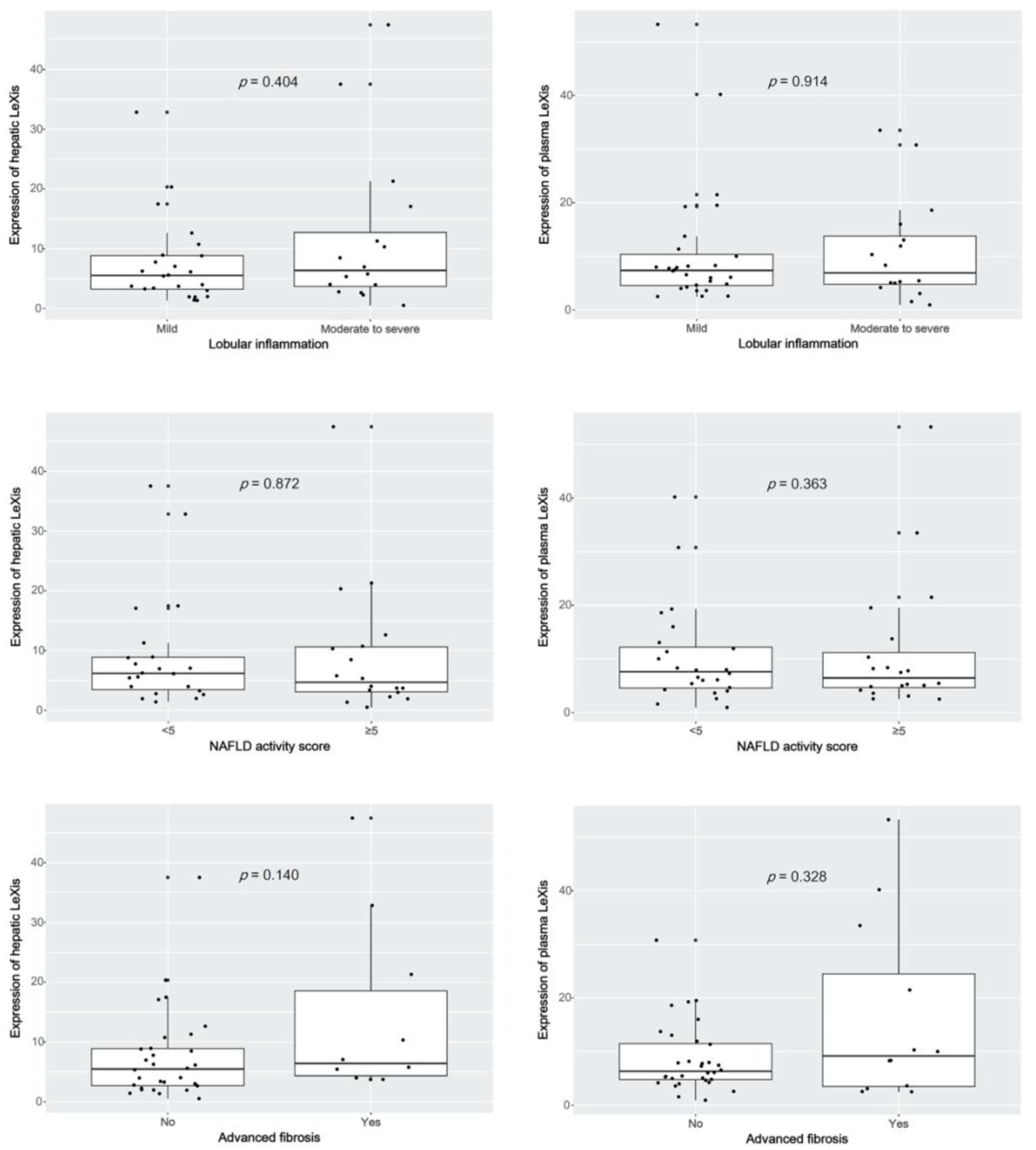
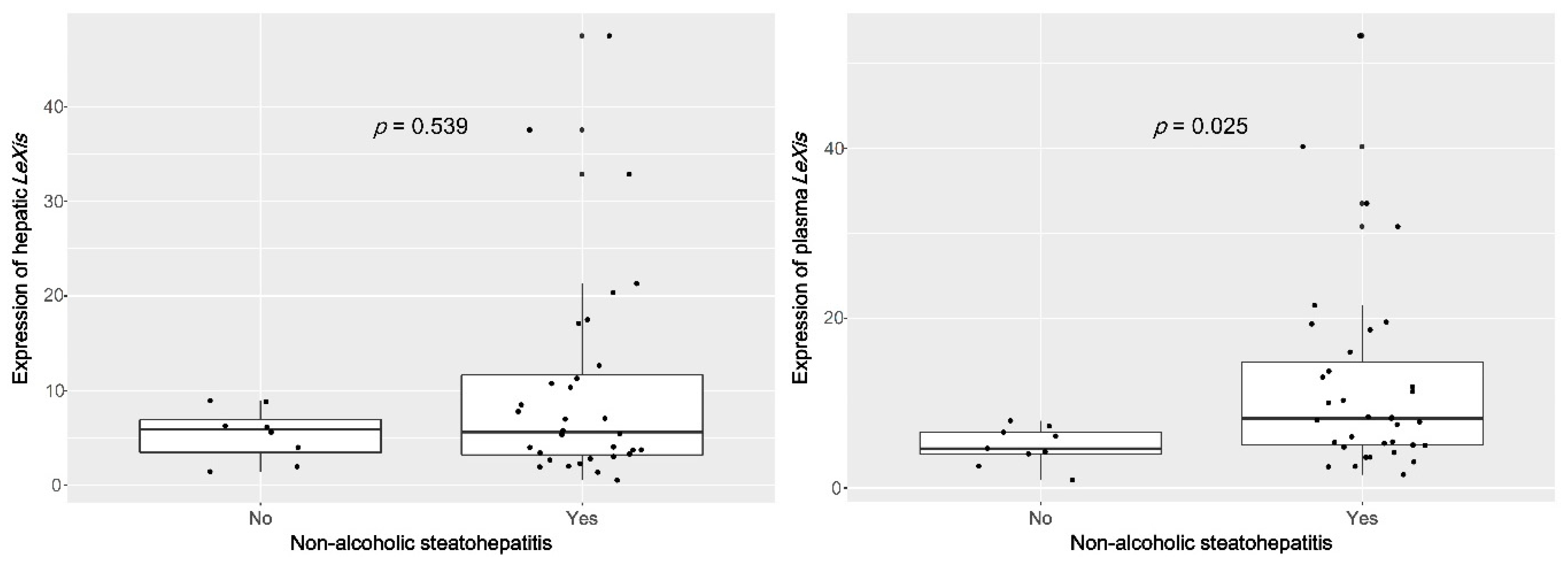
3.1. Hepatic lncRNA LeXis is Downregulated in Patients with Severe Hepatic Steatosis
3.2. Plasma lncRNA LeXis Levels are Increased in Patients with NASH
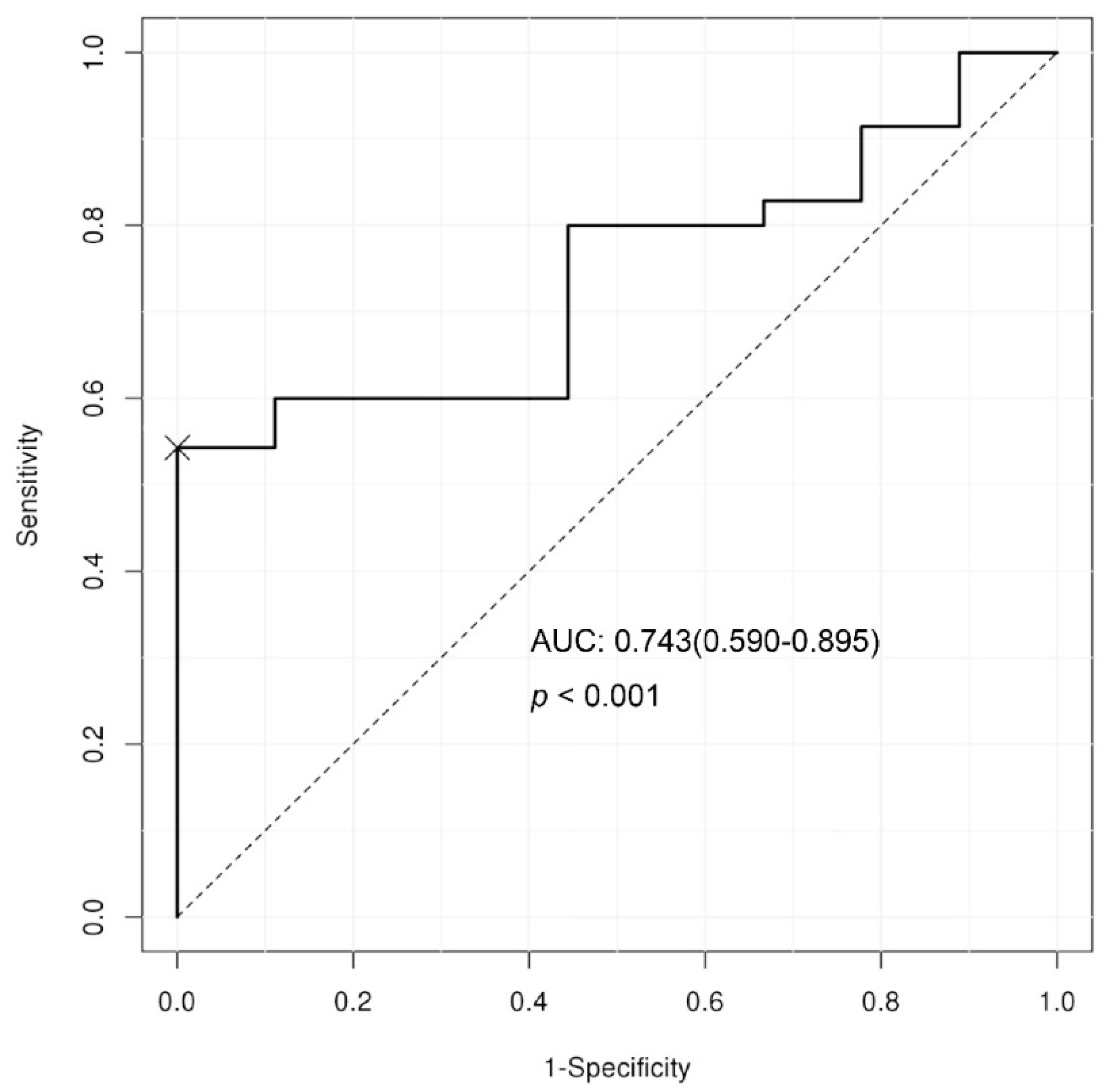
3.3. Predictive Cut-off Value and Diagnostic Performance of Plasma LeXis Levels for Diagnosing NASH.
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
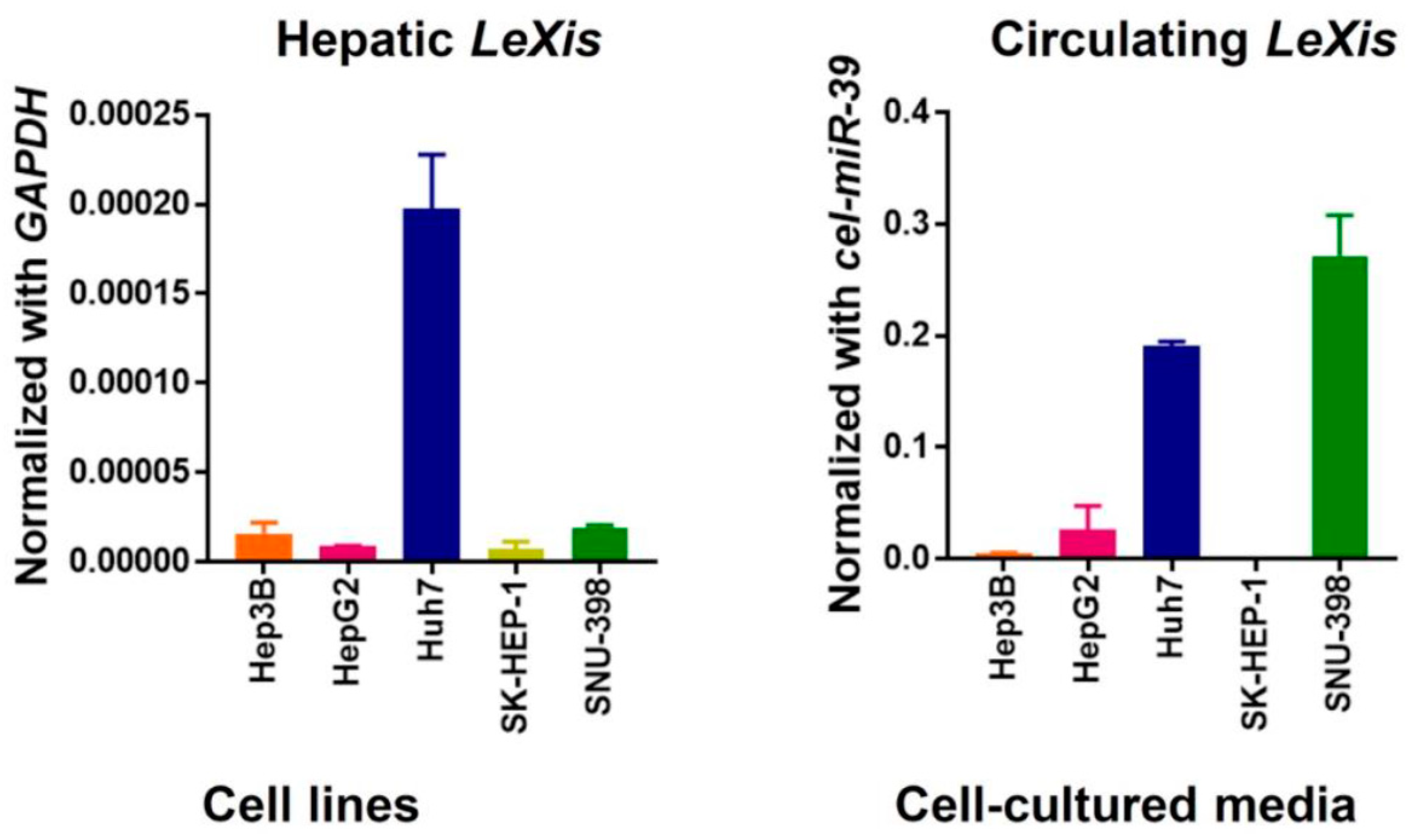
References
- Williams, R.; Aspinall, R.; Bellis, M.; Camps-Walsh, G.; Cramp, M.; Dhawan, A.; Ferguson, J.; Forton, D.; Foster, G.; Gilmore, I.; et al. Addressing liver disease in the UK: A blueprint for attaining excellence in health care and reducing premature mortality from lifestyle issues of excess consumption of alcohol, obesity, and viral hepatitis. Lancet 2014, 384, 1953–1997. [Google Scholar] [CrossRef]
- NatCen Social Research, University College London Department of Epidemiology and Public Health. Health Survey for England, 2012; UK Data Archive: Colchester, UK, 2014. [Google Scholar]
- Brunt, E.M.; Wong, V.W.; Nobili, V.; Day, C.P.; Sookoian, S.; Maher, J.J.; Bugianesi, E.; Sirlin, C.B.; Neuschwander-Tetri, B.A.; Rinella, M.E. Nonalcoholic fatty liver disease. Nat. Rev. Dis. Primers 2015, 1, 15080. [Google Scholar] [CrossRef] [PubMed]
- Ascha, M.S.; Hanouneh, I.A.; Lopez, R.; Tamimi, T.A.; Feldstein, A.F.; Zein, N.N. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology 2010, 51, 1972–1978. [Google Scholar] [CrossRef] [PubMed]
- Angulo, P.; Kleiner, D.E.; Dam-Larsen, S.; Adams, L.A.; Bjornsson, E.S.; Charatcharoenwitthaya, P.; Mills, P.R.; Keach, J.C.; Lafferty, H.D.; Stahler, A.; et al. Liver Fibrosis, but No Other Histologic Features, Is Associated With Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 2015, 149, 389–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekstedt, M.; Hagstrom, H.; Nasr, P.; Fredrikson, M.; Stal, P.; Kechagias, S.; Hultcrantz, R. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology 2015, 61, 1547–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, J.J.; Kim, W.; Kim, M.Y.; Jun, D.W.; Kim, S.G.; Yeon, J.E.; Lee, J.W.; Cho, Y.K.; Park, S.H.; Sohn, J.H.; et al. Recent research trends and updates on nonalcoholic fatty liver disease. Clin. Mol. Hepatol. 2019, 25, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, H.; Jun, D.W.; Saeed, W.K.; Nguyen, M.H. Non-alcoholic fatty liver diseases: Update on the challenge of diagnosis and treatment. Clin. Mol. Hepatol. 2016, 22, 327–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Steitz, J.A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5’-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998, 18, 6897–6909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sallam, T.; Jones, M.C.; Gilliland, T.; Zhang, L.; Wu, X.; Eskin, A.; Sandhu, J.; Casero, D.; Vallim, T.Q.; Hong, C.; et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 2016, 534, 124–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Diehl, A.M.; Brunt, E.M.; Cusi, K.; Charlton, M.; Sanyal, A.J. The diagnosis and management of non-alcoholic fatty liver disease: Practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 2012, 55, 2005–2023. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Hur, K.; Toiyama, Y.; Okugawa, Y.; Ide, S.; Imaoka, H.; Boland, C.R.; Goel, A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2017, 66, 654–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lackner, C.; Gogg-Kamerer, M.; Zatloukal, K.; Stumptner, C.; Brunt, E.M.; Denk, H. Ballooned hepatocytes in steatohepatitis: The value of keratin immunohistochemistry for diagnosis. J. Hepatol. 2008, 48, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Jung, E.S.; Hur, W.; Bae, S.H.; Choi, J.Y.; Song, M.J.; Kim, C.W.; Jo, S.H.; Lee, C.D.; Lee, Y.S.; et al. Noninvasive predictors of nonalcoholic steatohepatitis in Korean patients with histologically proven nonalcoholic fatty liver disease. Clin. Mol. Hepatol. 2013, 19, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Cusi, K.; Chang, Z.; Harrison, S.; Lomonaco, R.; Bril, F.; Orsak, B.; Ortiz-Lopez, C.; Hecht, J.; Feldstein, A.E.; Webb, A.; et al. Limited value of plasma cytokeratin-18 as a biomarker for NASH and fibrosis in patients with non-alcoholic fatty liver disease. J. Hepatol. 2014, 60, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Cui, T.; Sun, L.; Peng, N.; Yang, C. Long noncoding RNA LeXis promotes osteosarcoma growth through upregulation of CTNNB1 expression. Am. J. Cancer Res. 2017, 7, 1577–1587. [Google Scholar] [PubMed]



| Characteristics | Non-Alcoholic Fatty Liver Disease (NAFL) (n = 9) | Non-Alcoholic Steatohepatitis (NASH) (n = 35) | p Value |
|---|---|---|---|
| Male | 2 (22.2) | 20 (57.1) | 0.135 |
| Age, year | 38.3 ± 12.0 | 51.9 ± 15.5 | 0.020 * |
| Steatosis | 0.144 | ||
| 5-33 | 4 (44.4) | 16 (45.7) | |
| >33–66 | 1 (11.1) | 13 (37.1) | |
| >66% | 4 (44.4) | 6 (17.1) | |
| Lobular inflammation | 0.197 | ||
| <2 foci per 200× field | 8 (88.9) | 20 (57.1) | |
| 2–4 foci per 200× field | 1 (11.1) | 11 (31.4) | |
| >4 foci per 200× field | 0 (0.0) | 4 (11.4) | |
| Ballooning | 0.000 * | ||
| None | 9 (100.0) | 0 (0.0) | |
| Few ballooned cells | 0 (0.0) | 20 (57.1) | |
| Many cells/prominent ballooning | 0 (0.0) | 15 (42.9) | |
| Advanced fibrosis | 0 (0.0) | 12 (34.3) | 0.101 |
| NAS ≥ 5 | 0 (0.0) | 20 (57.1) | 0.007 * |
| Weight, kg | 83.3 ± 20.3 | 71.7 ± 11.6 | 0.182 |
| BMI, kg/m2 | 31.0 (26.8–33.1) | 27.1 (25.9–30.4) | 0.348 |
| Hypertension | 4 (44.4) | 17 (48.6) | 1.000 |
| Diabetes | 1 (11.1) | 15 (42.9) | 0.168 |
| Platelets, × 103/mm | 240.4 ± 51.3 | 219.8 ± 71.5 | 0.426 |
| AST, IU/L | 57.0 (43.0–95.0) | 78.0 (46.5–106.5) | 0.211 |
| ALT, IU/L | 90.0 (68.0–147.0) | 98.0 (66.5–120.5) | 0.816 |
| Bilirubin, mg/dL | 0.7 (0.5–0.8) | 0.6 (0.4–0.6) | 0.119 |
| Albumin, g/dL | 4.7 ± 0.3 | 4.5 ± 0.3 | 0.043 * |
| γ-GTP, mg/dL | 55.0 (43.0–95.0) | 77.0 (55.0–124.0) | 0.366 |
| Creatinine, mg/dL | 0.9 (0.8–1.0) | 0.8(0.6–0.9) | 0.134 |
| Fasting blood glucose, mg/dL | 107.0 (102.0–113.5) | 117.5 (104.0–136.0) | 0.133 |
| Total cholesterol, mg/dL | 179.0 (162.5–188.5) | 177.0 (159.0–210.0) | 0.835 |
| HDL, mg/dL | 47.5 (35.5–57.5) | 41.5 (33.0–50.0) | 0.441 |
| LDL, mg/dL | 117.0 ± 25.7 | 117.7 ± 38.3 | 0.963 |
| Triglyceride, mg/dL | 161.5 (115.5–194.5) | 142.5 (112.0–213.0) | 0.946 |
| Variable | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| Odds Ratio | p Value | Odds Ratio | p Value | |
| Sex | 4.67 (0.85–25.75) | 0.0771 | ||
| Age | 1.06 (1.01–1.12) | 0.0298 | ||
| Hypertension | 1.18 (0.27–5.15) | 0.8251 | ||
| Diabetes | 6.00 (0.68–53.29) | 0.1078 | ||
| Platelets | 1.00 (1.00–1.00) | 0.4175 | ||
| AST | 1.02 (0.99–1.05) | 0.1310 | 1.04 (0.99–1.08) | 0.136 |
| ALT | 1.00 (0.99–1.01) | 0.8114 | ||
| Bilirubin | 0.17 (0.02–1.78) | 0.1385 | 0.02 (0.00–0.68) | 0.029 * |
| Albumin | 0.07 (0.00–1.09) | 0.0574 | ||
| Creatinine | 0.10 (0.00–4.10) | 0.2263 | ||
| High plasma LeXis | 9.50 (1.07–84.25) | 0.0432 | 22.19 (1.25–395.22) | 0.035 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.G.; Kim, G.; Jang, S.Y.; Lee, Y.R.; Lee, E.; Lee, H.W.; Han, M.-H.; Chun, J.M.; Han, Y.S.; Yoon, J.S.; et al. Plasma Long Noncoding RNA LeXis is a Potential Diagnostic Marker for Non-Alcoholic Steatohepatitis. Life 2020, 10, 230. https://doi.org/10.3390/life10100230
Park JG, Kim G, Jang SY, Lee YR, Lee E, Lee HW, Han M-H, Chun JM, Han YS, Yoon JS, et al. Plasma Long Noncoding RNA LeXis is a Potential Diagnostic Marker for Non-Alcoholic Steatohepatitis. Life. 2020; 10(10):230. https://doi.org/10.3390/life10100230
Chicago/Turabian StylePark, Jung Gil, Gyeonghwa Kim, Se Young Jang, Yu Rim Lee, Eunhye Lee, Hye Won Lee, Man-Hoon Han, Jae Min Chun, Young Seok Han, Jun Sik Yoon, and et al. 2020. "Plasma Long Noncoding RNA LeXis is a Potential Diagnostic Marker for Non-Alcoholic Steatohepatitis" Life 10, no. 10: 230. https://doi.org/10.3390/life10100230
APA StylePark, J. G., Kim, G., Jang, S. Y., Lee, Y. R., Lee, E., Lee, H. W., Han, M.-H., Chun, J. M., Han, Y. S., Yoon, J. S., Kang, M. K., Kweon, Y. O., Tak, W. Y., Park, S. Y., & Hur, K. (2020). Plasma Long Noncoding RNA LeXis is a Potential Diagnostic Marker for Non-Alcoholic Steatohepatitis. Life, 10(10), 230. https://doi.org/10.3390/life10100230






