The Application of Next-Generation Sequencing to Define Factors Related to Oral Cancer and Discover Novel Biomarkers
Abstract
:1. Introduction
1.1. Next-Generation Sequencing
1.2. Oral Squamous Cell Carcinoma
2. Gene Alterations
3. Targeted Gene Therapy
4. Immune Checkpoint Inhibitors
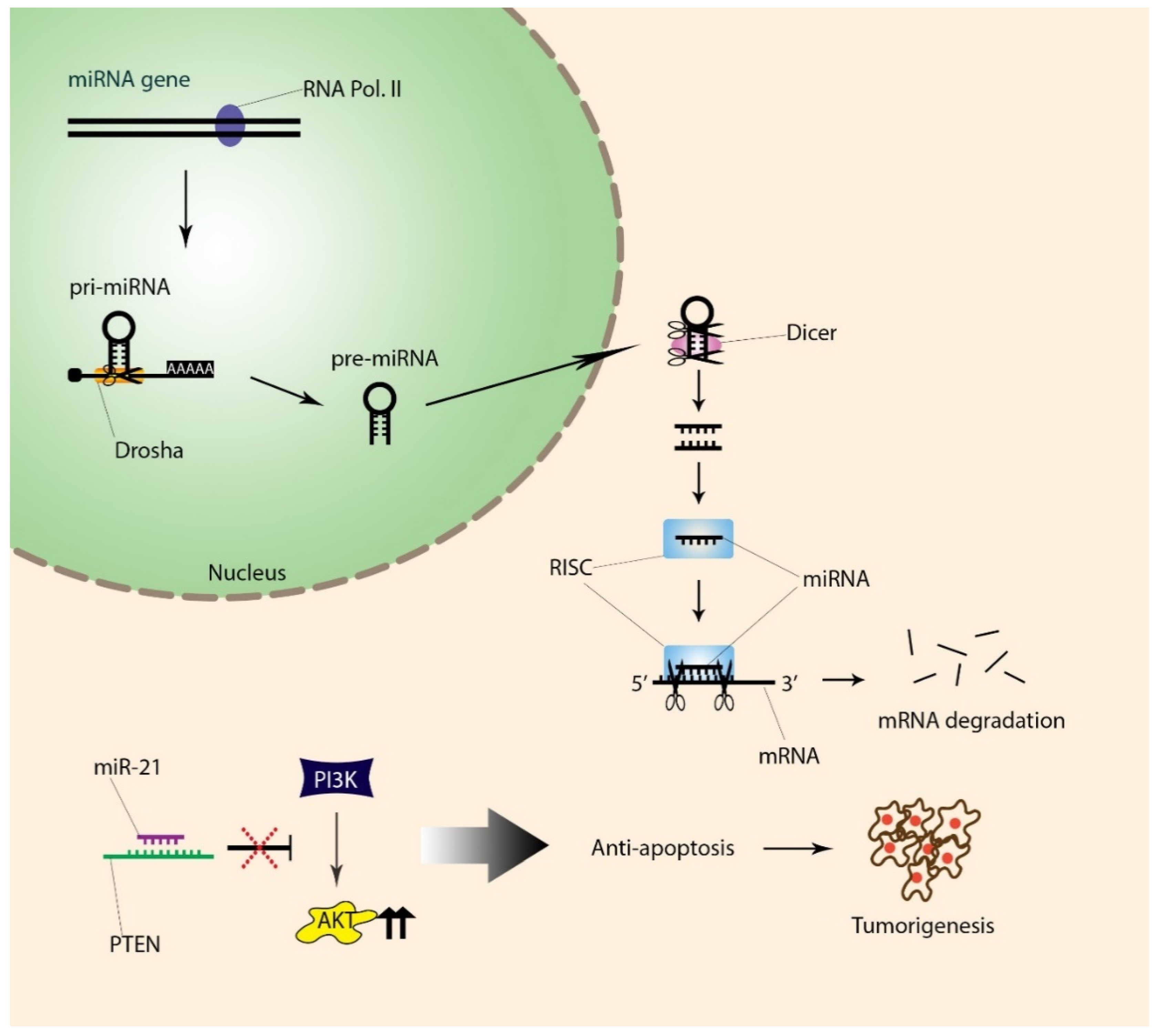
5. Differential Expressions of miRNAs
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Behjati, S.; Tarpey, P.S. What is next generation sequencing? Archives of disease in childhood. Education and practice edition. BMJ J. 2013, 98, 236–238. [Google Scholar]
- Sanger, F.; Air, G.M.; Barrell, B.G.; Brown, N.L.; Coulson, A.R.; Fiddes, J.C.; Hutchison, C.A.; Slocombe, P.M.; Smith, M.; Sanger, G.M.A.F. Nucleotide sequence of bacteriophage φX174 DNA. Nature 1977, 265, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxam, A.M.; Gilbert, W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 1977, 74, 560–564. [Google Scholar] [CrossRef] [Green Version]
- Hodzic, J.; Gurbeta, L.; Omanovic-Miklicanin, E.; Badnjevic, A. Overview of Next-generation Sequencing Platforms Used in Published Draft Plant Genomes in Light of Genotypization of Immortelle Plant (Helichrysium arenarium). Med. Arch. 2017, 71, 288–292. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of Next-Generation Sequencing Systems. J. Biomed. Biotechnol. 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Oxford Nanopore bests PacBio. Nat. Biotechnol. 2019, 37, 336. [CrossRef]
- Cui, J.; Shen, N.; Lu, Z.; Xu, G.; Wang, Y.; Jin, B. Analysis and comprehensive comparison of PacBio and nanopore-based RNA sequencing of the Arabidopsis transcriptome. Plant Methods 2020, 16, 85. [Google Scholar] [CrossRef]
- Li, Q.; Xia, Z.; Zhang, W.; Wang, L.; Wang, J.; Xu, D.; Mei, Z.; Liu, Q.; Du, S.; Li, Z.; et al. Reliable multiplex sequencing with rare index mis-assignment on DNB-based NGS platform. BMC Genom. 2019, 20, 215. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.-Y.; Chen, M.-X.; Ye, N.-H.; Qiao, W.-M.; Gao, B.; Law, W.-K.; Tian, Y.; Zhang, N.; Zhang, D.; Liu, T.-Y.; et al. Comparative performance of the BGISEQ-500 and Illumina HiSeq4000 sequencing platforms for transcriptome analysis in plants. Plant Methods 2018, 14, 69. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Liang, X.; Xuan, Y.; Geng, C.; Li, Y.; Lu, H.; Qu, S.; Mei, X.; Chen, H.; Yu, T.; et al. A reference human genome dataset of the BGISEQ-500 sequencer. GigaScience 2017, 6, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, S.S.T.; Gopalakrishnan, S.; Carøe, C.; Geng, C.; Liu, S.; Sinding, M.H.S.; Kuderna, L.; Zhang, W.; Fu, S.; Vieira, F.G.; et al. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. GigaScience 2017, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaura, E. Next-generation Sequencing Approaches to Understanding the Oral Microbiome. Adv. Dent. Res. 2012, 24, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Kilian, M.; Chapple, I.L.C.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.L.; Tonetti, N.S.; Wade, W.G.; Zaura, E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Deurenberg, R.H.; Bathoorn, E.; Chlebowicz, M.A.; Couto, N.; Ferdous, M.; García-Cobos, S.; Kooistra-Smid, A.M.; Raangs, E.C.; Rosema, S.; Veloo, A.C.; et al. Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol. 2017, 243, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. A comparative study of ddPCR and sanger sequencing for quantitative detection of low-frequency mutation rate. IOP Conf. Ser. Earth Environ. Sci. 2019, 332, 032023. [Google Scholar] [CrossRef]
- Lin, M.-T.; Mosier, S.L.; Thiess, M.; Beierl, K.F.; Debeljak, M.; Tseng, L.-H.; Chen, G.; Yegnasubramanian, S.; Ho, H.; Cope, L.; et al. Clinical Validation of KRAS, BRAF, and EGFR Mutation Detection Using Next-Generation Sequencing. Am. J. Clin. Pathol. 2014, 141, 856–866. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.J.; Osborn, A.M. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol. 2009, 67, 6–20. [Google Scholar] [CrossRef] [Green Version]
- Azim, F.S.; Houri, H.; Ghalavand, Z.; Nikmanesh, B. Next Generation Sequencing in Clinical Oncology: Applications, Challenges and Promises: A Review Article. Iran. J. Public Health 2018, 47, 1453–1457. [Google Scholar]
- Gabusi, A.; Gissi, D.B.; Tarsitano, A.; Asioli, S.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P.; Morandi, L. Intratumoral Heterogeneity in Recurrent Metastatic Squamous Cell Carcinoma of the Oral Cavity: New Perspectives Afforded by Multiregion DNA Sequencing and mtDNA Analysis. J. Oral Maxillofac. Surg. 2020, 77, 440–455. [Google Scholar] [CrossRef]
- Gabusi, A.; Gissi, D.B.; Montebugnoli, L.; Asioli, S.; Tarsitano, A.; Marchetti, C.; Balbi, T.; Helliwell, T.R.; Foschini, M.P.; Morandi, L. Prognostic impact of intra-field heterogeneity in oral squamous cell carcinoma. Virchows Arch. 2019, 476, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Johann, J.D.J.; Rodriguez-Canales, J.; Mukherjee, S.; Prieto, D.A.; Hanson, J.C.; Emmert-Buck, M.; Blonder, J. Approaching Solid Tumor Heterogeneity on a Cellular Basis by Tissue Proteomics Using Laser Capture Microdissection and Biological Mass Spectrometry. J. Proteome Res. 2009, 8, 2310–2318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oral Health. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/oral-health (accessed on 2 September 2020).
- Shah, J.P.; Gil, Z. Current concepts in management of oral cancer–surgery. Oral Oncol. 2009, 45, 394–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkin, D.M.; Läärä, E.; Muir, C.S. Estimates of the worldwide frequency of sixteen major cancers in 1980. Int. J. Cancer 1988, 41, 184–197. [Google Scholar] [CrossRef]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global Cancer Statistics, 2002. CA A Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Gupta, N.; Gupta, R.; Acharya, A.K.; Patthi, B.; Goud, V.; Reddy, S.; Garg, A.; Singla, A. Changing Trends in oral cancer—A global scenario. Nepal J. Epidemiol. 2016, 6, 613–619. [Google Scholar] [CrossRef]
- Curado, M.P.; Hashibe, M. Recent changes in the epidemiology of head and neck cancer. Curr. Opin. Oncol. 2009, 21, 194–200. [Google Scholar] [CrossRef]
- Sritippho, T.; Chotjumlong, P.; Iamaroon, A. Roles of Human Papillomaviruses and p16 in Oral Cancer. Asian Pac. J. Cancer Prev. 2015, 16, 6193–6200. [Google Scholar] [CrossRef] [Green Version]
- Alves, A.M.; Correa, M.B.; Da Silva, K.D.; De Araújo, L.M.A.; Vasconcelos, A.C.U.; Gomes, A.P.N.; Etges, A.; Tarquinio, S.B.C. Demographic and Clinical Profile of Oral Squamous Cell Carcinoma from a Service-Based Population. Braz. Dent. J. 2017, 28, 301–306. [Google Scholar] [CrossRef]
- Farah, C.S.; Woo, S.-B.; Zain, R.B.; Sklavounou, A.; McCullough, M.J.; Lingen, M. Oral Cancer and Oral Potentially Malignant Disorders. Int. J. Dent. 2014, 2014, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Yardimci, G.; Kutlubay, Z.; Engin, B.; Tuzun, Y. Precancerous lesions of oral mucosa. World J. Clin. Cases 2014, 2, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Irani, S. Pre-Cancerous Lesions in the Oral and Maxillofacial Region: A Literature Review with Special Focus on Etiopathogenesis. Iran. J. Pathol. 2016, 11, 303–322. [Google Scholar] [PubMed]
- Phookan, J.; Saikia, K.P. A clinicopathological study of the pre-malignant conditions of oral cavity. Indian J. Otolaryngol. Head Neck Surg. 1998, 50, 246–249. [Google Scholar] [PubMed]
- Silverman, S., Jr.; Gorsky, M.; Lozada, D.F. Oral leukoplakia and malignant transformation. A follow-up study of 257 patients. Cancer 1984, 53, 563–568. [Google Scholar] [CrossRef]
- Silverman, S. Demographics and occurrence of oral and pharyngeal cancers. J. Am. Dent. Assoc. 2001, 132, 7S–11S. [Google Scholar] [CrossRef]
- Okada, Y.; Mataga, I.; Katagiri, M.; Ishii, K. An analysis of cervical lymph nodes metastasis in oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2003, 32, 284–288. [Google Scholar] [CrossRef]
- Noguti, J.; De Moura, C.F.G.; De Jesus, G.P.P.; Da Silva, V.H.P.; Hossaka, T.A.; Oshima, C.T.F.; Ribeiro, D.A. Metastasis from oral cancer: An overview. Cancer Genom. Proteom. 2012, 9, 329–335. [Google Scholar]
- Greenberg, J.S.; Fowler, R.; Gomez, J.; Mo, V.; Roberts, D.; El-Naggar, A.K.; Myers, J.N. Extent of extracapsular spread. Cancer 2003, 97, 1464–1470. [Google Scholar] [CrossRef]
- Snow, G.B.; Brekel, M.W.M.V.D.; Leemans, C.R.; Patel, P. Surgical Management of Cervical Lymph Nodes in Patients with Oral and Oropharyngeal Cancer. Adv. Struct. Saf. Stud. 1994, 134, 43–55. [Google Scholar] [CrossRef]
- Ghantous, Y.; Abu Elnaaj, I. Global Incidence and Risk Factors of Oral Cancer. Harefuah 2017, 156, 645–649. [Google Scholar]
- Faden, D.L.; Arron, S.T.; Heaton, C.M.; DeRisi, J.L.; South, A.P.; Wang, S.J. Targeted next-generation sequencing of TP53 in oral tongue carcinoma from non-smokers. J. Otolaryngol. Head Neck Surg. 2016, 45, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S.C.; Carpenter, W.R.; Tyree, S.; Couch, M.E.; Weissler, M.; Hackman, T.; Hayes, D.N.; Shores, C.; Chera, B.S. Increasing Incidence of Oral Tongue Squamous Cell Carcinoma in Young White Women, Age 18 to 44 Years. J. Clin. Oncol. 2011, 29, 1488–1494. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Pan, Y.; Li, R.; Chi, A.C. Changing Trends in Oral Squamous Cell Carcinoma with Particular Reference to Young Patients: 1971–2006. The Emory University Experience. Head Neck Pathol. 2008, 2, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Schantz, S.P.; Yu, G.-P. Head and neck cancer incidence trends in young Americans, 1973–1997, with a special analysis for tongue cancer. Arch. Otolaryngol. Head Neck Surg. 2002, 128, 268–274. [Google Scholar] [CrossRef] [Green Version]
- Shiboski, C.H.; Schmidt, B.L.; Jordan, R.C.K. Tongue and tonsil carcinoma. Cancer 2005, 103, 1843–1849. [Google Scholar] [CrossRef]
- Goldstein, D.P.; Irish, J.C. Head and neck squamous cell carcinoma in the young patient. Curr. Opin. Otolaryngol. Head Neck Surg. 2005, 13, 207–211. [Google Scholar] [CrossRef]
- Song, X.; Xia, R.; Li, J.; Long, Z.; Ren, H.; Chen, W.; Mao, L. Common and complex Notch1 mutations in Chinese oral squamous cell carcinoma. Clin. Cancer Res. 2014, 20, 701–710. [Google Scholar] [CrossRef] [Green Version]
- Izumchenko, E.; Sun, K.; Jones, S.; Brait, M.; Agrawal, N.; Koch, W.; Mccord, C.L.; Riley, D.R.; Angiuoli, S.; Velculescu, V.E.; et al. Notch1 mutations are drivers of oral tumorigenesis. Cancer Prev. Res. 2014, 8, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.-X.; Zhang, J.; Wang, J.; et al. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef] [Green Version]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef] [Green Version]
- Tannock, I.F.; Hill, R.P.; Carey, R.W. The Basic Science of Oncology. Plast. Reconstr. Surg. 1989, 83, 920. [Google Scholar] [CrossRef]
- Nakagaki, T.; Tamura, M.; Kobashi, K.; Omori, A.; Koyama, R.; Idogawa, M.; Ogi, K.; Hiratsuka, H.; Tokino, T.; Sasaki, Y. Targeted next-generation sequencing of 50 cancer-related genes in Japanese patients with oral squamous cell carcinoma. Tumor Boil. 2018, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagaki, T.; Tamura, M.; Kobashi, K.; Koyama, R.; Fukushima, H.; Ohashi, T.; Idogawa, M.; Ogi, K.; Hiratsuka, H.; Tokino, T.; et al. Profiling cancer-related gene mutations in oral squamous cell carcinoma from Japanese patients by targeted amplicon sequencing. Oncotarget 2017, 8, 59113–59122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaprakash, C.; Varghese, V.K.; Jayaram, P.; Chakrabarty, S.; Kudva, A.; Ray, S.; Satyamoorthy, K. Relevance and actionable mutational spectrum in oral squamous cell carcinoma. J. Oral Pathol. Med. 2020, 49, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Fu, Y.; Tu, Y.-Y.; Liu, Y.; Tan, Y.-R.; Ju, W.-T.; Pickering, C.R.; Myers, J.N.; Zhang, Z.-Y.; Zhong, L.-P. Mutation allele frequency threshold does not affect prognostic analysis using next-generation sequencing in oral squamous cell carcinoma. BMC Cancer 2018, 18, 758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Er, T.K.; Wang, Y.-Y.; Chen, C.-C.; Herreros-Villanueva, M.; Liu, T.-C.; Yuan, S.-S.F. Molecular characterization of oral squamous cell carcinoma using targeted next-generation sequencing. Oral Dis. 2015, 21, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Batta, N.; Pandey, M. Mutational spectrum of tobacco associated oral squamous carcinoma and its therapeutic significance. World J. Surg. Oncol. 2019, 17, 1–12. [Google Scholar] [CrossRef]
- Chen, T.-W.; Lee, C.-C.; Liu, H.; Wu, C.-S.; Pickering, C.R.; Huang, P.-J.; Wang, J.; Chang, I.Y.-F.; Yeh, Y.-M.; Chen, C.-D.; et al. APOBEC3A is an oral cancer prognostic biomarker in Taiwanese carriers of an APOBEC deletion polymorphism. Nat. Commun. 2017, 8, 465. [Google Scholar] [CrossRef]
- Oikawa, Y.; Morita, K.-I.; Kayamori, K.; Tanimoto, K.; Sakamoto, K.; Katoh, H.; Ishikawa, S.; Inazawa, J.; Harada, H. Receptor tyrosine kinase amplification is predictive of distant metastasis in patients with oral squamous cell carcinoma. Cancer Sci. 2017, 108, 256–266. [Google Scholar] [CrossRef]
- Van Ginkel, J.H.; De Leng, W.W.; De Bree, R.; Van Es, R.J.; Willems, S.M. Targeted sequencing reveals TP53 as a potential diagnostic biomarker in the post-treatment surveillance of head and neck cancer. Oncotarget 2016, 7, 61575–61586. [Google Scholar] [CrossRef] [Green Version]
- Tabatabaeifar, S.; Larsen, M.J.; Larsen, S.R.; Kruse, T.A.; Thomassen, M.; Sørensen, J.A. Investigating a case of possible field cancerization in oral squamous cell carcinoma by the use of next-generation sequencing. Oral Oncol. 2017, 68, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-J.; Liu, H.; Liao, C.-T.; Huang, P.-J.; Huang, Y.; Hsu, A.; Tang, P.; Chang, Y.-S.; Chen, H.-C.; Yen, T.-C. Ultra-deep targeted sequencing of advanced oral squamous cell carcinoma identifies a mutation-based prognostic gene signature. Oncotarget 2015, 6, 18066–18080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, A.; Zhang, X.; Jones, D.; Veillard, A.-S.; Zhang, M.; Martin, A.; Lyons, J.G.; Lee, C.-S.; Rose, B. Relationships between p53 mutation, HPV status and outcome in oropharyngeal squamous cell carcinoma. Radiother. Oncol. 2016, 118, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Feldman, R.; Gatalica, Z.; Knezetic, J.; Reddy, S.; Nathan, C.-A.; Javadi, N.; Teknos, T. Molecular profiling of head and neck squamous cell carcinoma. Head Neck 2015, 38 (Suppl. S1), E1625–E1638. [Google Scholar] [CrossRef] [PubMed]
- Kozaki, K.-I.; Imoto, I.; Pimkhaokham, A.; Hasegawa, S.; Tsuda, H.; Omura, K.; Inazawa, J. PIK3CA mutation is an oncogenic aberration at advanced stages of oral squamous cell carcinoma. Cancer Sci. 2006, 97, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Guthrie, V.B.; Masica, D.L.; Tokheim, C.; Kang, H.; Richmon, J.; Agrawal, N.; Fakhry, C.; Quon, H.; Subramaniam, R.M.; et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann. Oncol. 2015, 26, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.R.; Zhang, J.; Yoo, S.Y.; Bengtsson, L.; Moorthy, S.; Neskey, D.M.; Zhao, M.; Alves, M.V.O.; Chang, K.; Drummond, J.; et al. Integrative Genomic Characterization of Oral Squamous Cell Carcinoma Identifies Frequent Somatic Drivers. Cancer Discov. 2013, 3, 770–781. [Google Scholar] [CrossRef] [Green Version]
- Tabatabaeifar, S.; Thomassen, M.; Larsen, M.J.; Larsen, S.R.; Kruse, T.A.; Sørensen, J.A. The subclonal structure and genomic evolution of oral squamous cell carcinoma revealed by ultra-deep sequencing. Oncotarget 2017, 8, 16571–16580. [Google Scholar] [CrossRef] [Green Version]
- Mohan, M.; Jagannathan, N. Oral field cancerization: An update on current concepts. Oncol. Rev. 2014, 8. [Google Scholar] [CrossRef] [Green Version]
- Slaughter, D.P.; Southwick, H.W.; Smejkal, W. “Field cancerization” in oral stratified squamous epithelium. Clin. Implic. Multicent. Orig. 1953, 6, 963–968. [Google Scholar]
- Zhang, X.C.; Xu, C.; Mitchell, R.M.; Zhang, B.; Zhao, D.; Li, Y.; Huang, X.; Fan, W.; Wang, H.; Lerma, L.A.; et al. Tumor Evolution and Intratumor Heterogeneity of an Oropharyngeal Squamous Cell Carcinoma Revealed by Whole-Genome Sequencing. Neoplasia 2013, 15, 1371-IN7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blandino, G.; Di Agostino, S. New therapeutic strategies to treat human cancers expressing mutant p53 proteins. J. Exp. Clin. Cancer Res. 2018, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant p53 as a target for cancer treatment. Eur. J. Cancer 2017, 83, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Perdrix, A.; Najem, A.; Saussez, S.; Awada, A.; Journe, F.; Ghanem, G.; Krayem, M. PRIMA-1 and PRIMA-1Met (APR-246): From Mutant/Wild Type p53 Reactivation to Unexpected Mechanisms Underlying Their Potent Anti-Tumor Effect in Combinatorial Therapies. Cancers 2017, 9, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roh, J.-L.; Ko, J.H.; Moon, S.J.; Ryu, C.H.; Choi, J.Y.; Koch, W.M. The p53-reactivating small-molecule RITA enhances cisplatin-induced cytotoxicity and apoptosis in head and neck cancer. Cancer Lett. 2012, 325, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Hang, W.; Yin, Z.-X.; Liu, G.; Zeng, Q.; Shen, X.-F.; Sun, Q.-H.; Li, D.-D.; Jian, Y.-P.; Zhang, Y.-H.; Wang, Y.-S.; et al. Piperlongumine and p53-reactivator APR-246 selectively induce cell death in HNSCC by targeting GSTP1. Oncogene 2018, 37, 3384–3398. [Google Scholar] [CrossRef]
- Castellanos, M.R.; Pan, Q. Novel p53 therapies for head and neck cancer. World J. Otorhinolaryngol. Head Neck Surg. 2016, 2, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhang, J.; Tong, J.H.M.; Chan, A.W.H.; Yu, J.; Kang, W.; To, K.F. Targeting the Oncogenic p53 Mutants in Colorectal Cancer and Other Solid Tumors. Int. J. Mol. Sci. 2019, 20, 5999. [Google Scholar] [CrossRef] [Green Version]
- Bykov, V.J.; Issaeva, N.; Shilov, A.; Hultcrantz, M.; Pugacheva, E.N.; Chumakov, P.M.; Bergman, J.; Wiman, K.; Selivanova, G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 2002, 8, 282–288. [Google Scholar] [CrossRef]
- Bykov, V.J.; Issaeva, N.; Zache, N.; Shilov, A.; Hultcrantz, M.; Bergman, J.; Selivanova, G.; Wiman, K. Reactivation of Mutant p53 and Induction of Apoptosis in Human Tumor Cells by Maleimide Analogs. J. Boil. Chem. 2005, 280, 30384–30391. [Google Scholar] [CrossRef] [Green Version]
- Lindemann, A.; Patel, A.A.; Silver, N.L.; Tang, L.; Liu, Z.; Wang, L.; Tanaka, N.; Rao, X.; Takahashi, H.; Maduka, N.K.; et al. COTI-2, A Novel Thiosemicarbazone Derivative, Exhibits Antitumor Activity in HNSCC through p53-dependent and -independent Mechanisms. Clin. Cancer Res. 2019, 25, 5650–5662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vareki, S.M.; Salim, K.Y.; Danter, W.R.; Koropatnick, J. Novel anti-cancer drug COTI-2 synergizes with therapeutic agents and does not induce resistance or exhibit cross-resistance in human cancer cell lines. PLoS ONE 2018, 13, e0191766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salim, K.Y.; Vareki, S.M.; Danter, W.R.; Koropatnick, J. COTI-2, a novel small molecule that is active against multiple human cancer cell lines in vitro and in vivo. Oncotarget 2016, 7, 41363–41379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Chen, J.; Cai, X.; Yao, Z.; Huang, J. Progress in targeted therapeutic drugs for oral squamous cell carcinoma. Surg. Oncol. 2019, 31, 90–97. [Google Scholar] [CrossRef]
- Oliva, M.; Spreafico, A.; Taberna, M.; Alemany, L.; Coburn, B.; Mesia, R.; Siu, L. Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann. Oncol. 2019, 30, 57–67. [Google Scholar] [CrossRef]
- GanjiBakhsh, M.; Monshizadeh, R.; Nasimian, A.; Aminishakib, P.; Farzaneh, P.; Shiraji, S.T.; Gharajei, A.; Rahrotaban, S.; Baghaei, F.; Gohari, N.S. Anti-angiogenic efficacy of aflibercept and bevacizumab in primary oral squamous cell carcinoma cells. J. Oral Pathol. Med. 2018, 47, 575–582. [Google Scholar] [CrossRef]
- Ramakrishnan, M.S.; Eswaraiah, A.; Crombet, T.; Piedra, P.; Saurez, G.; Iyer, H.; Arvind, A. Nimotuzumab, a promising therapeutic monoclonal for treatment of tumors of epithelial origin. MAbs 2010, 1, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Li, Y.; Zhou, Q.; Xu, Z.; Sun, C.; Tan, X.; Lu, L. Cetuximab inhibits oral squamous cell carcinoma invasion and metastasis via degradation of epidermal growth factor receptor. J. Oral Pathol. Med. 2013, 43, 250–257. [Google Scholar] [CrossRef]
- Khalil, A.; Jameson, M.J. The EGFR Inhibitor Gefitinib Enhanced the Response of Human Oral Squamous Cell Carcinoma to Cisplatin In Vitro. Drugs R&D 2017, 17, 545–555. [Google Scholar] [CrossRef] [Green Version]
- Gupta, D.; Rao, R. OP0012 Chemotherapy with gefitinib in recurrent and metastatic head and neck cancer. Eur. J. Cancer 2014, 50, e4. [Google Scholar] [CrossRef]
- Macha, M.A.; Rachagani, S.; Qazi, A.K.; Jahan, R.; Gupta, S.; Patel, A.; Seshacharyulu, P.; Lin, C.; Li, S.; Wang, S.; et al. Afatinib radiosensitizes head and neck squamous cell carcinoma cells by targeting cancer stem cells. Oncotarget 2017, 8, 20961–20973. [Google Scholar] [CrossRef] [PubMed]
- William, W.N.; Papadimitrakopoulou, V.; Lee, J.J.; Mao, L.; Cohen, E.E.W.; Lin, H.Y.; Gillenwater, A.M.; Martin, J.W.; Lingen, M.W.; Boyle, J.O.; et al. Erlotinib and the Risk of Oral Cancer. JAMA Oncol. 2016, 2, 209–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mărgăritescu, C.; Pirici, D.; Stîngă, A.; Simionescu, C.; Raica, M.; Mogoantă, L.; Stepan, A.; Ribatti, D. VEGF expression and angiogenesis in oral squamous cell carcinoma: An immunohistochemical and morphometric study. Clin. Exp. Med. 2010, 10, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Yoshimura, H.; Matsuda, S.; Ryoke, T.; Kiyoshima, T.; Kobayashi, M.; Sano, K. Effects of peritumoral bevacizumab injection against oral squamous cell carcinoma in a nude mouse xenograft model: A preliminary study. Oncol. Lett. 2018, 15, 8627–8634. [Google Scholar] [CrossRef]
- Hsu, F.-T.; Chang, B.; Chen, J.C.-H.; Chiang, I.-T.; Liu, Y.-C.; Kwang, W.-K.; Hwang, J. Synergistic Effect of Sorafenib and Radiation on Human Oral Carcinoma in vivo. Sci. Rep. 2015, 5, 15391. [Google Scholar] [CrossRef] [Green Version]
- Wedge, S.R.; Ogilvie, D.J.; Dukes, M.; Kendrew, J.; Chester, R.; Jackson, J.A.; Boffey, S.J.; Valentine, P.J.; Curwen, J.; Musgrove, H.L.; et al. ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Cancer Res. 2002, 62, 4645–4655. [Google Scholar]
- Ciardiello, F.; Caputo, R.; Damiano, V.; Caputo, R.; Troiani, T.; Vitagliano, D.; Carlomagno, F.; Veneziani, B.M.; Fontanini, G.; Bianco, A.R.; et al. Antitumor effects of ZD6474, a small molecule vascular endothelial growth factor receptor tyrosine kinase inhibitor, with additional activity against epidermal growth factor receptor tyrosine kinase. Clin. Cancer Res. 2003, 9, 1546–1556. [Google Scholar]
- Chu, P.L.; Shihabuddeen, W.A.; Low, K.P.; Poon, D.J.; Ramaswamy, B.; Liang, Z.-G.; Nei, W.L.; Chua, K.L.; Thong, P.S.; Soo, K.C.; et al. Vandetanib sensitizes head and neck squamous cell carcinoma to photodynamic therapy through modulation of EGFR-dependent DNA repair and the tumour microenvironment. Photodiagnosis Photodyn. Ther. 2019, 27, 367–374. [Google Scholar] [CrossRef]
- Zhou, G.; Hasina, R.; Wroblewski, K.; Mankame, T.P.; Doçi, C.L.; Lingen, M.W. Dual inhibition of vascular endothelial growth factor receptor and epidermal growth factor receptor is an effective chemopreventive strategy in the mouse 4-NQO model of oral carcinogenesis. Cancer Prev. Res. 2010, 3, 1493–1502. [Google Scholar] [CrossRef] [Green Version]
- Tan, F.H.; Bai, Y.; Saintigny, P.; Darido, C. mTOR Signalling in Head and Neck Cancer: Heads Up. Cells 2019, 8, 333. [Google Scholar] [CrossRef] [Green Version]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballou, L.M.; Lin, R.Z. Rapamycin and mTOR kinase inhibitors. J. Chem. Boil. 2008, 1, 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rini, B.I. Temsirolimus, an Inhibitor of Mammalian Target of Rapamycin. Clin. Cancer Res. 2008, 14, 1286–1290. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, K.H.; Apelo, S.I.A.; Yu, D.; Brinkman, J.A.; Velarde, M.C.; Syed, F.A.; Liao, C.-Y.; Baar, E.L.; Carbajal, K.A.; Sherman, D.S.; et al. A novel rapamycin analog is highly selective for mTORC1 in vivo. Nat. Commun. 2019, 10, 3194. [Google Scholar] [CrossRef] [PubMed]
- Houghton, P.J. Everolimus, Clinical cancer research. Off. J. Am. Assoc. Cancer Res. 2010, 16, 1368–1372. [Google Scholar] [CrossRef] [Green Version]
- Okui, T.; Shimo, T.; Fukazawa, T.; Kurio, N.; Hassan, N.M.M.; Honami, T.; Takaoka, M.; Naomoto, Y.; Sasaki, A. Antitumor Effect of Temsirolimus against Oral Squamous Cell Carcinoma Associated with Bone Destruction. Mol. Cancer Ther. 2010, 9, 2960–2969. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z. Simultaneously Targeting Epidermal Growth Factor Receptor Tyrosine Kinase and Cyclooxygenase-2, an Efficient Approach to Inhibition of Squamous Cell Carcinoma of the Head and Neck. Clin. Cancer Res. 2004, 10, 5930–5939. [Google Scholar] [CrossRef] [Green Version]
- Huo, J.Y.; Chang, Y.-L.; Li, M.; Ma, Y.-Z. Study status and prospective of multiview video coding. Tongxin Xuebao/J. Commun. 2010, 31, 113–121. [Google Scholar]
- Chiang, S.-L.; Velmurugan, B.K.; Chung, C.-M.; Lin, S.-H.; Wang, Z.-H.; Hua, C.-H.; Tsai, M.-H.; Kuo, T.-M.; Yeh, K.-T.; Chang, P.-Y.; et al. Preventive effect of celecoxib use against cancer progression and occurrence of oral squamous cell carcinoma. Sci. Rep. 2017, 7, 6235. [Google Scholar] [CrossRef]
- Kwak, Y.E.; Jeon, N.K.; Kim, J.; Lee, E.J. The Cyclooxygenase-2 Selective Inhibitor Celecoxib Suppresses Proliferation and Invasiveness in the Human Oral Squamous Carcinoma. Ann. NY Acad. Sci. 2007, 1095, 99–112. [Google Scholar] [CrossRef]
- Cohen, E.E.W.; Bell, R.B.; Bifulco, C.B.; Burtness, B.; Gillison, M.L.; Harrington, K.J.; Le, Q.-T.; Lee, N.Y.; Leidner, R.; Lewis, R.L.; et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC). J. Immunother. Cancer 2019, 7, 184. [Google Scholar] [CrossRef] [Green Version]
- Fessas, P.; Lee, H.; Ikemizu, S.; Janowitz, T. A molecular and preclinical comparison of the PD-1–targeted T-cell checkpoint inhibitors nivolumab and pembrolizumab. Semin. Oncol. 2017, 44, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Syed, Y.Y. Durvalumab: First Global Approval. Drugs 2017, 77, 1369–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Larkins, E.; Blumenthal, G.M.; Yuan, W.; He, K.; Sridhara, R.; Subramaniam, S.; Zhao, H.; Liu, C.; Yu, J.; Goldberg, K.B.; et al. FDA Approval Summary: Pembrolizumab for the Treatment of Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma with Disease Progression on or After Platinum-Containing Chemotherapy. Oncologist 2017, 22, 873–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Jiao, D.; Xu, H.; Liu, Q.; Zhao, W.; Han, X.; Wu, K. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer 2018, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. New Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef]
- Lui, V.W.Y.; Hedberg, M.L.; Li, H.; Vangara, B.S.; Pendleton, K.; Zeng, Y.; Lu, Y.; Zhang, Q.; Du, Y.; Gilbert, B.R.; et al. Frequent Mutation of the PI3K Pathway in Head and Neck Cancer Defines Predictive Biomarkers. Cancer Discov. 2013, 3, 761–769. [Google Scholar] [CrossRef] [Green Version]
- Tang, F.; Zheng, P. Tumor cells versus host immune cells: Whose PD-L1 contributes to PD-1/PD-L1 blockade mediated cancer immunotherapy? Cell Biosci. 2018, 8, 34. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.M.; Robert, L.; Chmielowski, B.; Spasić, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-Y.; Wu, C.-T.; Wang, C.-W.; Lan, K.-H.; Liang, H.-K.; Huang, B.-S.; Chang, Y.-L.; Kuo, S.-H.; Cheng, A.-L. Prognostic significance of tumor-infiltrating lymphocytes in patients with operable tongue cancer. Radiat. Oncol. 2018, 13, 157. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Salm, M.; Dangl, M. Adoptive cell therapy with tumour-infiltrating lymphocytes: The emerging importance of clonal neoantigen targets for next-generation products in non-small cell lung cancer. Immuno-Oncol. Technol. 2019, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Federico, L.; Haymaker, C.L.; Forget, M.-A.; Ravelli, A.; Bhatta, A.; Karpinets, T.; Zhang, R.; Weissferdt, A.; Fang, B.; Zhang, J.; et al. A preclinical study of tumor-infiltrating lymphocytes in NSCLC. J. Clin. Oncol. 2018, 36 (Suppl. S5), 161. [Google Scholar] [CrossRef]
- Rohaan, M.W.; Berg, J.H.V.D.; Kvistborg, P.; Haanen, J.B.A.G. Adoptive transfer of tumor-infiltrating lymphocytes in melanoma: A viable treatment option. J. Immunother. Cancer 2018, 6, 102. [Google Scholar] [CrossRef]
- Fancello, L.; Gandini, S.; Pelicci, P.G.; Mazzarella, L. Tumor mutational burden quantification from targeted gene panels: Major advancements and challenges. J. Immunother. Cancer 2019, 7, 183. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Li, B.; Peng, Y.; Wu, F.; Li, Q.; Lin, Z.; Xie, S.; Xiao, L.; Lin, X.; Ou, Z.; et al. The prognostic value of TMB and the relationship between TMB and immune infiltration in head and neck squamous cell carcinoma: A gene expression-based study. Oral Oncol. 2020, 110, 104943. [Google Scholar] [CrossRef]
- Berland, L.; Heeke, S.; Humbert, O.; Macocco, A.; Long-Mira, E.; Lassalle, S.; Lespinet-Fabre, V.; Lalvée, S.; Bordone, O.; Cohen, C.; et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J. Thorac. Dis. 2019, 11 (Suppl. S1), S71–S80. [Google Scholar] [CrossRef]
- Adachi, K.; Tamada, K. Microbial biomarkers for immune checkpoint blockade therapy against cancer. J. Gastroenterol. 2018, 53, 999–1005. [Google Scholar] [CrossRef] [Green Version]
- Abdi, J.; Yang, Y.; Meyer-Erlach, P.; Chang, H. Bone Marrow Stromal Cells Induce Bortezomib Resistance in Multiple Myeloma Cells through Downregulation of miRNA-101-3p Targeting Survivin. Blood 2015, 126, 1772. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L.X. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Jennings, S.F.; Tong, W.; Hong, H. Next generation sequencing for profiling expression of miRNAs: Technical progress and applications in drug development. J. Biomed. Sci. Eng. 2011, 4, 666–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.-F.; Wu, Z.-P.; Chen, Y.; Zhu, Q.-S.; Hamidi, S.; Navab, R. MicroRNA-21 (miR-21) Regulates Cellular Proliferation, Invasion, Migration, and Apoptosis by Targeting PTEN, RECK and Bcl-2 in Lung Squamous Carcinoma, Gejiu City, China. PLoS ONE 2014, 9, e103698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.-H.; Tsao, C.-J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.-X.; Huang, X.-F.; Shao, Q.; Huang, M.-Y.; Deng, L.; Wu, Q.-L.; Zeng, Y.-X.; Shao, J.-Y. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008, 14, 2348–2360. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Li, G.; Wang, Z.; Yao, Y.; Chen, R.; Pu, X.; Wang, J. Circulating MicroRNA-21 Is a Potential Diagnostic Biomarker in Gastric Cancer. Dis. Markers 2015, 2015, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Tseng, H.-H.; Tseng, Y.-K.; You, J.-J.; Kang, B.-H.; Wang, T.-H.; Yang, C.-M.; Chen, H.-C.; Liou, H.-H.; Liu, P.-F.; Ger, L.-P.; et al. Next-generation Sequencing for microRNA Profiling: MicroRNA-21-3p Promotes Oral Cancer Metastasis. Anticancer. Res. 2017, 37, 1059–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Wang, X.; Venø, M.T.; Bakholdt, V.; Sørensen, J.A.; Krogdahl, A.; Sun, Z.; Gao, S.; Kjems, J. Circulating miRNAs as biomarkers for oral squamous cell carcinoma recurrence in operated patients. Oncotarget 2017, 8, 8206–8214. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, N.J.; Jensen, D.H.; Lelkaitis, G.; Kiss, K.; Charabi, B.W.; Ullum, H.; Specht, L.; Schmidt, A.Y.; Nielsen, F.C.; Von Buchwald, C. MicroRNA-based classifiers for diagnosis of oral cavity squamous cell carcinoma in tissue and plasma. Oral Oncol. 2018, 83, 46–52. [Google Scholar] [CrossRef]
- Xu, G.-Q.; Li, L.-H.; Wei, J.-N.; Xiao, L.-F.; Wang, X.-T.; Pang, W.-B.; Yan, X.-Y.; Chen, Z.-Y.; Song, G. Identification and profiling of microRNAs expressed in oral buccal mucosa squamous cell carcinoma of Chinese hamster. Sci. Rep. 2019, 9, 15616. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.-A.; Weng, S.-L.; Yang, S.-F.; Chou, C.; Huang, W.-C.; Tu, S.-J.; Chang, T.-H.; Huang, C.; Jong, Y.-J.; Huang, H.-D. A Three–MicroRNA Signature as a Potential Biomarker for the Early Detection of Oral Cancer. Int. J. Mol. Sci. 2018, 19, 758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, A.J.; Wang, S.; Shen, J.; Robine, N.; Philipone, E.; Oster, M.W.; Nam, A.; Santella, R.M. Prognostic value of miR-375 and miR-214-3p in early stage oral squamous cell carcinoma. Am. J. Transl. Res. 2014, 6, 580–592. [Google Scholar] [PubMed]
- Wiklund, E.D.; Gao, S.; Hulf, T.; Sibbritt, T.; Nair, S.; Costea, D.E.; Villadsen, S.B.; Bakholdt, V.; Bramsen, J.B.; Sørensen, J.A.; et al. MicroRNA Alterations and Associated Aberrant DNA Methylation Patterns across Multiple Sample Types in Oral Squamous Cell Carcinoma. PLoS ONE 2011, 6, e27840. [Google Scholar] [CrossRef] [PubMed]
- Goberdhan, D.C.; Wilson, C. PTEN: Tumour suppressor, multifunctional growth regulator and more. Hum. Mol. Genet. 2003, 12 (Suppl. S2), R239–R248. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Henson, R.; Wehbe–Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Xu, S.; Jin, W.; Yi, Q.; Wei, W.; Wei, W. Effect of the PTEN gene on adhesion, invasion and metastasis of osteosarcoma cells. Oncol. Rep. 2014, 32, 1741–1747. [Google Scholar] [CrossRef] [Green Version]
- Gissi, D.B.; Morandi, L.; Gabusi, A.; Tarsitano, A.; Marchetti, C.; Curà, F.; Palmieri, A.; Montebugnoli, L.; Asioli, S.; Foschini, M.P.; et al. A Noninvasive Test for MicroRNA Expression in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2018, 19, 1789. [Google Scholar] [CrossRef] [Green Version]
- Li, J.-S.; Huang, H.; Sun, L.; Yang, M.; Pan, C.; Chen, W.; Wu, N.; Lin, Z.; Zeng, C.; Yao, Y.; et al. MiR-21 Indicates Poor Prognosis in Tongue Squamous Cell Carcinomas as an Apoptosis Inhibitor. Clin. Cancer Res. 2009, 15, 3998–4008. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.-X.; Wu, Q.-N.; Zhang, Y.; Li, Y.Y.; Liao, D.Z.; Hou, J.H.; Fu, J.; Zeng, M.; Yun, J.; Wu, Q.-L.; et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011, 13, R2. [Google Scholar] [CrossRef] [Green Version]
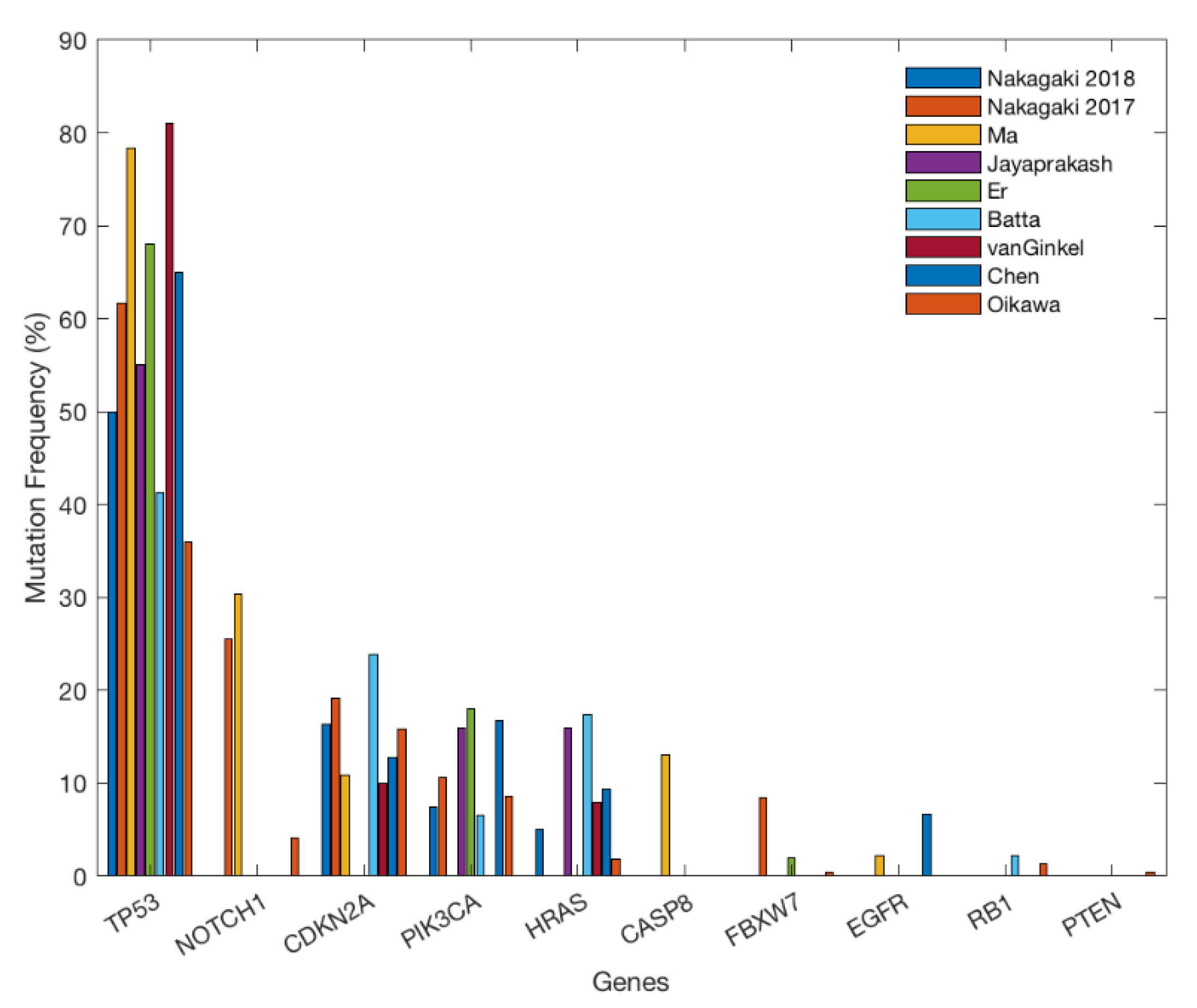
| Genes/Driver Mutations | Research Method | Type of Variant | Location of Mutation | Regulatory Function | References |
|---|---|---|---|---|---|
| TP53 NM_000546.5 (p.Arg248Trp) (p.Arg213Gln) (p.Pro72Arg) (p.Pro152Leu) (p.Val274Gly) (p.Val274Phe) (p.Glu204Ter*) (p.Gln165Ter*) | Ion Torrent [48,53,54,56,58,61,62,63] Ion AmpliSeq [55] Ion Proton [60] Illumina [57,59,62] | Missense [48,53,54,55,56,57,58,59,60,61,63] Nonsense [48,53,54,56,57,58,59,60,63] Deletion [54,55,56,57,58,60] Insertion [55,56,57,60] Splice-site mutation [53,54,56,59,60,63] | DNA-binding domain [53,54,56] | p53 induces apoptosis (tumor suppressor) | [48,53,54,55,56,57,58,59,60,61,62,63] |
| CDKN2A NM_000077 (p.Pro81His) (p.Arg80Ter*) (p.Trp110Ter*) | Ion Torrent [53,54,56,58,61,63] Ion Proton [60] Illumina [59] | Missense [54,56,59,60] Nonsense [54,56,58,60,63] Deletion [54,58,60,63] Insertion [54,56,60,63] Splice-site mutation [54,56,60,63] | Ankyrin repeats [56] | p16 and p14 function as tumor suppressors | [53,54,56,58,59,60,61,63] |
| NOTCH1 NM_017617.5 (p.Gly310Arg) (p.Asp352Gly) (p.Arg365Cys) (p.Thr1014Met) (p.Cys1383Tyr) (p.Gln1957Pro) | Ion Torrent[48,54,56] Ion Proton[60] Illumina[49,59] | Missense [48,54,56,59,60] Nonsense [48,60] Insertions [54,56] Deletions [56] Splice-site [54,56] | EGF-like repeats [48,49,54,56] | Cell growth and division | [48,49,54,56,58,59,60] |
| HRAS NM_005343 (p.Gly12Ser) (p.Gly13Ser) (p.Gly13Arg) (p.Gly13Val) | Ion Torrent [53,58,61,63] Ion AmpliSeq [55] Ion Proton [60] Illumina [59] | Missense [55,58,60,63] | Cell growth and division | [53,55,58,59,60,61,63] | |
| PIK3CA NM_006213.2 NM_006218.1 (p.Glu545Lys) (p.His1048Ser) (p.His1047Arg) (p.Glu542Ala) (p.Glu542Lys) (p.His1047Arg) | Ion Torrent [53,54,58,63] Ion AmpliSeq [55] Ion Proton [60] Illumina [57,59] | Missense [55,56,57,58,59,60,63] Nonsense [63] Splice-site [60] | Cell growth and division | [53,54,55,57,58,59,60,63] | |
| CASP8 NM_001080125 (p.Ile354Asn) (p.Arg472Ter) (p.Cys404Tyr) | Ion Torrent [56,62] | Nonsense [56] Deletion [56] | Caspase homologues domain [56] | Regulates cell apoptosis | [56,62] |
| FBXW7 NM_033632.3 (p.Ser462Phe) | Illumina [57] Ion Torrent [54,61] Ion Proton [60] | Missense [54,57,60] | Tumor suppressor | [57,60,61] | |
| RB1 NM_000321.3 (p.Ile680Thr) | Ion Torrent [58,61] Ion Proton [60] | Missense [60] Nonsense [60] | Tumor suppressor | [58,60,61] | |
| PTEN NM_000314.4 (p.Arg161Lys) (p.His185Tyr) (p.Val249Met) | Ion AmpliSeq [55] Ion Proton [60] | Missense [55] Nonsense [60] Deletion [55] | Cell growth and division | [55,60,63] | |
| EGFR NM_005228 (p.Ile107Val) | Ion Torrent [54,56,58,59,63] Ion AmpliSeq [55] Illumina [57] | Missense [56] Insertion [59] Deletion [58] | Furin-like repeats [56] | Regulates cell proliferation | [53,54,55,56,57,58,59,63] |
| p53 Targeted | EGFR Targeted | VEGF Targeted | mTOR Inhibitors | PD-1 Targeted | Others |
|---|---|---|---|---|---|
| PRIMA-1 | Cetuximab | Bevacizumab | Rapamycin | Pembrolizumab | COX-2 inhibitor |
| PRIMA-1MET (APR-246) | Nimotuzumab | Aflibercept | Temsirolimus | Nivolumab | - |
| MIRA-1 | Gefitinib | Sorafenib | Everolimus | Durvalumab | - |
| STIMA-1 | Erlotinib | Vandetanib | Sirolimus | Atezolizumab | - |
| COTI-2 | - | - | - | - | - |
| Oncogenic (Up-Regulated) miRNA | Suppressive (Down-Regulated) miRNA |
|---|---|
| miR-21: [138,139,140,141] | miR-92b: [139] |
| miR-22: [140] | miR-199: [143] |
| miR-26a: [139] | miR-214: [143] |
| miR-34c: [141] | miR-375: [139] |
| miR-34c: [141] | miR-486: [139] |
| miR-34b: [141] | miR-504: [141] |
| miR-117: [141] | miR-499: [141] |
| miR-118: [141] | miR-486: [141] |
| miR-130b: [141,142] | |
| miR-135: [141] | |
| miR-142: [141] | |
| miR-143: [140] | |
| miR-148a: [139] | |
| miR-150: [142] | |
| miR-221: [142] | |
| miR-222: [142] | |
| miR-423: [142] | |
| miR-542: [141] | |
| miR-1269a: [143] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Lee, J.W.; Park, Y.-S. The Application of Next-Generation Sequencing to Define Factors Related to Oral Cancer and Discover Novel Biomarkers. Life 2020, 10, 228. https://doi.org/10.3390/life10100228
Kim S, Lee JW, Park Y-S. The Application of Next-Generation Sequencing to Define Factors Related to Oral Cancer and Discover Novel Biomarkers. Life. 2020; 10(10):228. https://doi.org/10.3390/life10100228
Chicago/Turabian StyleKim, Soyeon, Joo Won Lee, and Young-Seok Park. 2020. "The Application of Next-Generation Sequencing to Define Factors Related to Oral Cancer and Discover Novel Biomarkers" Life 10, no. 10: 228. https://doi.org/10.3390/life10100228
APA StyleKim, S., Lee, J. W., & Park, Y.-S. (2020). The Application of Next-Generation Sequencing to Define Factors Related to Oral Cancer and Discover Novel Biomarkers. Life, 10(10), 228. https://doi.org/10.3390/life10100228




