First, we analyze the estimators’ accuracy for given parameters under various link functions. Subsequently, we compare the algorithms. Then, we investigate the likelihood ratio test, Wald test, and score test under different link functions in terms of the type I error rates (TIEs) and powers.
3.1. Accuracy of the Estimators
In this subsection, we apply bias, average standard errors (ASE), and coverage probability (CP) to assess the point and interval estimates of the parameters . The bias measures the difference between the average MLEs of the parameters and the corresponding actual values. The average standard errors mean the average of the standard errors of the MLEs. The coverage probability is a proportion of the asymptotic 95% confidence intervals for the parameters .
Take the sample size and correlation coefficient . Other parameter settings are given according to the cases below:
Case (1). , , , where .
Case (2). , , , where , , and .
The samples
are generated randomly from the discrete distribution
, where
depends on the specified link function. Once the samples are generated, the MLEs of the parameters
are computed following the estimation procedures detailed in
Section 2 for each corresponding link function. In addition, the standard errors associated with the MLEs are also calculated to assess the variability of the parameter estimates. This simulation process is repeated 10,000 times for each link function, and the resulting values for bias, average standard error (ASE), and coverage probability (CP) are presented in
Table 3 and
Table 4.
Table 3 shows that the values of the three evaluation metrics, bias, ASE, and CP, fall within acceptable and reasonable ranges. As the sample size increases, while keeping other parameters constant, both the bias and ASE of the parameters
decrease under all five link functions. A closer analysis of the three metrics across different link functions reveals that, for a fixed sample size and correlation coefficient, the logistic link function tends to exhibit the highest ASE for the parameter
. However, when using the Fisher bounded algorithm to estimate
, the ASE is lower than the other link functions, suggesting that the Fisher bounded algorithm performs better than the QLB algorithm in estimation accuracy.
Further examination of the probit link function shows that the estimated parameters demonstrate smaller bias and lower ASE, with the CP ranging from 94% to 96%, which is within an acceptable range. In contrast, under the DE link function, when = 0.7 and n = 400, the bias and ASE for the parameters are higher. Additionally, for smaller sample sizes, under the log–log and complementary log–log link functions, the CP for the parameters ranges from 93% to 97%, which is slightly wider than the CP observed for the probit link function.
As shown in
Table 4, the overall performance for Case (2) is quite similar to that observed in Case (1). The logistic link function shows the largest ASE for
. The three metrics demonstrate a more pronounced advantage for the probit link function than other link functions. While the CP for
improves under the complementary log–log link function, the bias and ASE are still higher than those for the probit link function. The performance of the double exponential and log–log link functions remains similar to that observed in Case (1), with comparable results for the three metrics.
In summary, all five link functions perform within an acceptable range, validating the algorithm’s convergence. The probit link function demonstrates the most robust performance and can be considered the preferred choice. The logistic link function exhibits a higher ASE related to the algorithm. The performance of the CLL and LL link functions fluctuates significantly, reflecting each link function’s characteristic suitability for extreme event probabilities. The DE link function shows poor stability under high correlation and large sample sizes.
3.2. Comparison of Algorithms
Under various link functions, we compare the effectiveness of algorithms by the average iteration count and the computing time. An algorithm is considered more efficient if it achieves a desired level of accuracy with fewer iterations or in less computation time.
Let , and , and the other parameter settings refer to Case (1). The samples are randomly generated from the discrete distribution , where depends on the specified link function. Regarding parameter estimations, we use the QLB-based NR algorithm with the logistic link function, while for the other four link functions, we employ the Fisher-bound-based NR algorithm. All experiments were repeated 500 times.
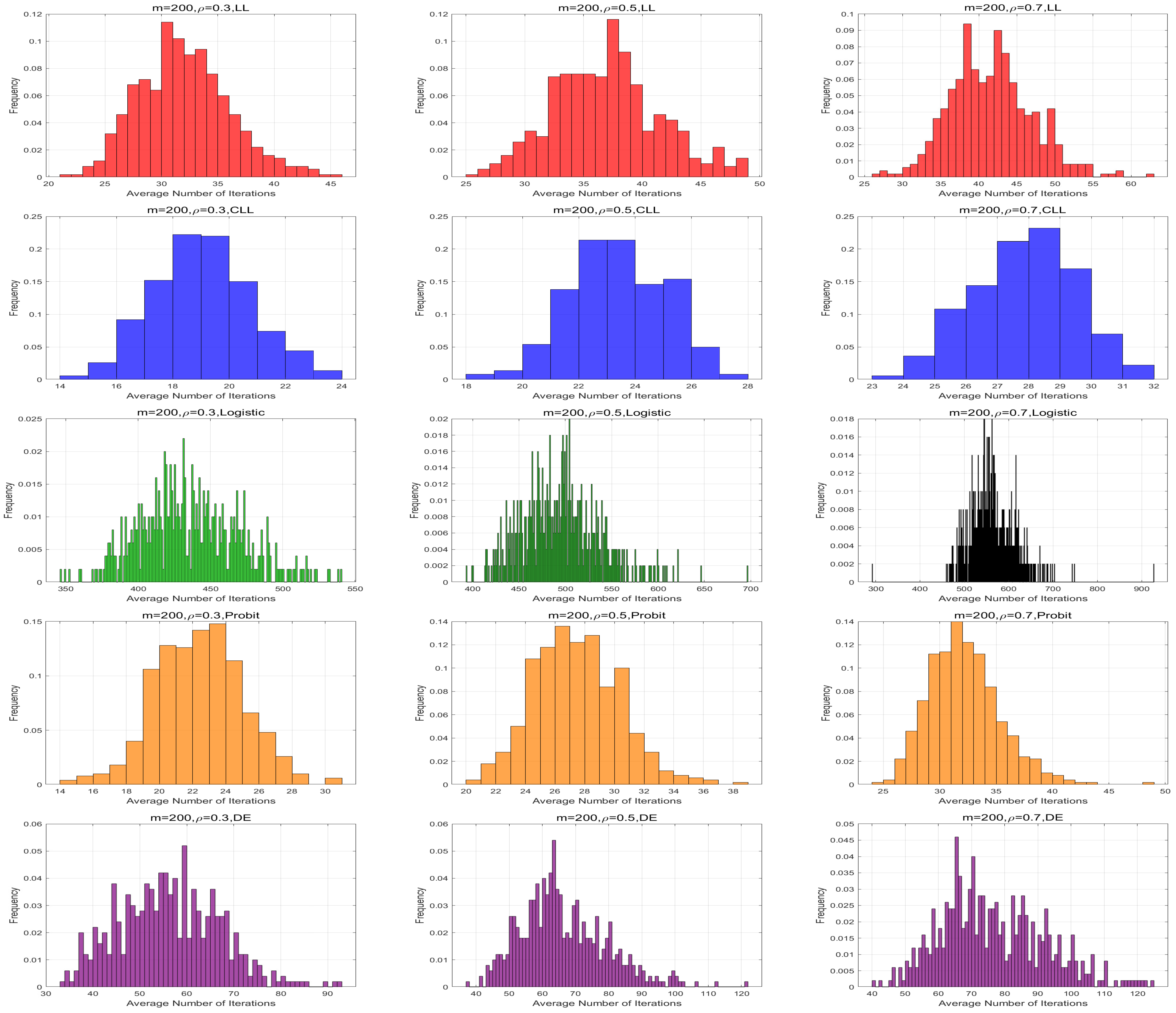
Figure 1 and
Figure 2 provide the frequency distributions of the average iteration numbers under the different link functions. The average number of iterations under the logistic link function is significantly higher than that of the other four link functions across all parameter settings. On the other hand, we fixed the iteration number to 100 and recorded their time cost. The average iteration time for five link functions is shown in
Table 5. Under all parameter settings, the average iteration time for the logistic link function is also notably longer than for the other link functions.
These findings suggest that the computational performance of the Fisher-bound-based NR algorithm is substantially more efficient than the QLB-based NR algorithm. Through comparisons across different link functions, we observe that both the average number of iterations and the average computation time increase with the sample size and the correlation coefficient. This demonstrates that both factors have a significant impact on the computational burden of the algorithms. Moreover, it is worth noting that the Fisher-bound-based NR algorithm with the CLL link function consistently exhibits the best performance across all parameter settings.
3.3. Comparison of Test Statistics
Under the hypothesis test (
5), we evaluate the performance of the three statistics for given link functions with parameter settings. Take the sample size
and the correlation coefficient
. The covariate vector
is defined in Cases (1) and (2). Other parameter settings are based on the two cases: (I)
,
,
,
. (II)
,
,
, and
Similarly, the samples
are generated from the discrete distribution
for 10,000 times, where
. The empirical type I error rate is calculated by dividing the number of rejections of the null hypothesis
by the total number of simulations, which is 10,000. The empirical TIEs are summarized in
Table 6. Power refers to the probability of correctly rejecting the null hypothesis
when it is false. We consider sample sizes
n = 50, 80, 100, 200, 400, and
= 0.3, 0.5, 0.7. The following two cases of regression coefficient vectors are considered: (A)
and (B)
. The other parameter configurations remain consistent with Cases (I) and (II). For each
, we generate
10,000 times, where
. The empirical power of each test is computed as the proportion of rejections at the 0.05 significance level over 10,000 simulations.
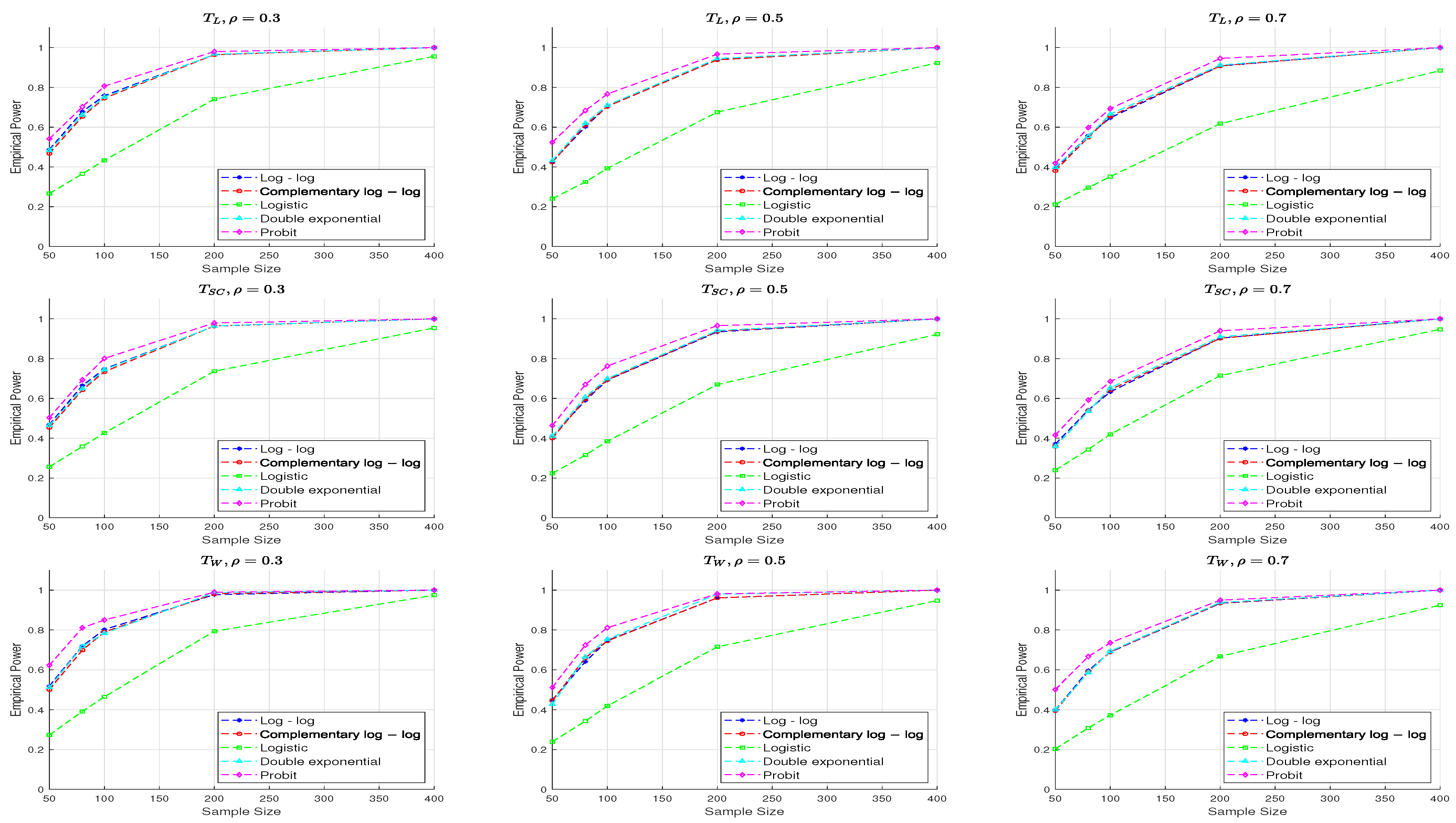
Figure 3 and
Figure 4 display the empirical powers of the three test statistics across five different link functions. A comparison of the results shows that both the
and
tests consistently maintain empirical type I error rates (TIEs) close to the nominal level of 0.05 across all link functions considered (LL, CLL, L, DE, and Pt), indicating robust performance. In contrast, the
test tends to yield lower empirical TIEs under the double exponential link, though its TIEs for other link functions fluctuate around the nominal level. Furthermore, empirical TIEs are generally higher when
compared to
, with this effect being more noticeable for smaller sample sizes.
Figure 3 illustrates that increasing the sample size enhances the power of all three test statistics. Power decreases slightly for a fixed sample size as
increases. The
statistic demonstrates the best performance, while
and
produce comparable results across all parameter settings. Among the link functions, the probit link generally yields the highest power, especially for small sample sizes, whereas the logistic link results in the lowest. The log–log, complementary log–log, and double exponential links exhibit similar power levels.
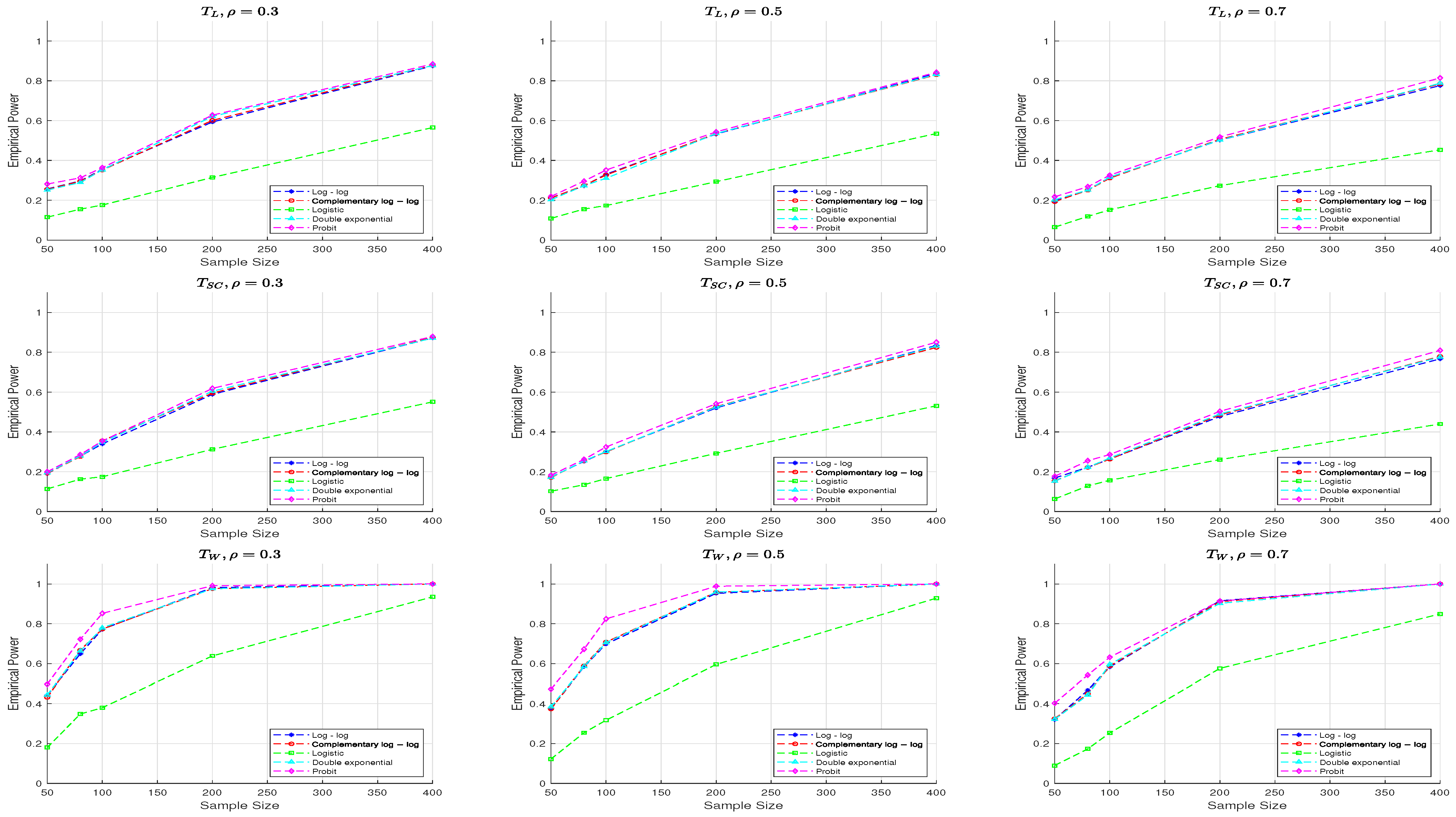
From
Figure 4, we observe that the performance of the three test statistics for
is generally similar to that for
, though there are some differences. Specifically, compared to
, the power of all test statistics decreases when
, with the
and
test statistics showing a more pronounced decline across all parameter settings and link functions.
In addition to the empirical findings, theoretical analysis suggests that the Wald test achieves higher power under the probit link due to its derivative structure, which yields greater Fisher information and a larger non-centrality parameter
[
30].
Moreover, considering that the test statistic under the probit link function shows robust empirical TIEs, it is recommended as the most effective test for assessing the significance of confounding effects on the response rate.