Abstract
The most exciting advances in biohydrometallurgy are occurring in the field of microbiology. The two main technologies employed in biohydrometallurgy, agitated tanks for the processing of refractory concentrates and heaps and dumps for the processing of low-grade ores, are technologically sound and widely practised at commercial scale, but their development began at a time when very little was known of the microorganisms that assisted metals extraction from sulfide ores. During and subsequent to those developments it has been shown that microbial communities in metals extraction are more diverse than originally thought, and extremely robust and adaptable to different and variable environments. Recent advances in genomics and proteomics, exploiting hugely increased computing power and speed, have made it possible to describe not only which microorganisms are present in bioleaching systems, but also what physiological functions are being exercised. The body of knowledge being acquired through the application of molecular biology methods will be used increasingly to monitor microbial behaviour, optimise conditions for more appropriate microbiological activity and/or infer the “microbiological health” of bioreactors (tanks and heaps).
1. Introduction
Biohydrometallurgy (biomining or mineral bioprocessing) is a sub-discipline of hydrometallurgy that exploits some attributes of micro-organisms (bio-catalysts) to facilitate and/or enhance the separation of elements from their ores or other materials. Traditional biohydrometallurgy involves the aqueous, inorganic chemistry of acidic sulfate solutions contacted with sulfide concentrates or ores containing valuable metals. As a method of extracting elements, biohydrometallurgy is an alternative to some intense chemical leaching methods [1,2,3,4]. However, its application to the processing of mineral concentrates compares unfavourably with pyrometallurgical processing because it is slower and because, usually, the infrastructure required for pyrometallurgical processing is already in place. According to Holmes and Debus [5], biological processing (of concentrates) would need to demonstrate a greater than 20% advantage over conventional pyrometallurgical processing to interest the mining industry. Similarly, Poulter et al. [6] remarked that industry reluctance to embrace the technology is partly due to the “inherent process and economic advantage of modern smelting technologies” and the “real and perceived risks associated with the introduction of a novel technology”. On the other hand, biohydrometallurgy has achieved acceptance for the processing of low-grade, secondary copper sulfide ores on a very large scale, particularly in Chile, and accounts for approximately 20% of annual global copper production [7,8]. Likewise, pre-treatment of refractory gold concentrates in agitated tanks at commercial scale has a long and successful history from Fairview, South Africa [9] to Runro, the Philippines [10]. Looking to the future, this suite of well-developed and reliable biohydrometallurgical technologies can be applied, for example, to small deposits in remote locations perhaps with unfavourable terrain, to complex ores that are difficult to process, or to concentrates containing impurities that attract smelter penalties.
Man has benefited from the bio-assisted extraction of metals for much longer than the underlying microbiological foundation of the technology has been understood. Accounts from almost 2000 years ago describe the recovery of copper from mine water by cementation (Table 1) but the ‘technology’ could pre-date these accounts by many centuries, possibly since man started to mine ores and extract metals.

Table 1.
Historical biohydrometallurgy: Approximately 2000 years of anecdotes and evidence up to the discovery of Acidithiobacillus (At.) ferrooxidans (collated from [11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28]).
Not surprisingly, metal extraction dominated commercial developments before the discovery that microorganisms were involved in the process. However, that discovery created a broad new research field within which the biogeochemical roles of acidophilic microorganisms were explored in relation to acidic drainage at mine sites and geothermally-heated sulfur-rich environments. Naturally, the greater understanding of microbial attributes was incorporated into established or new technologies. For the purposes of this targeted review of microbiological advances in biohydrometallurgy, it is convenient to discuss separately those advances relating to discoveries about the microorganisms, responses to processing environments, and diversity in managed heaps, columns and agitated tanks.
2. Microbiological Discovery
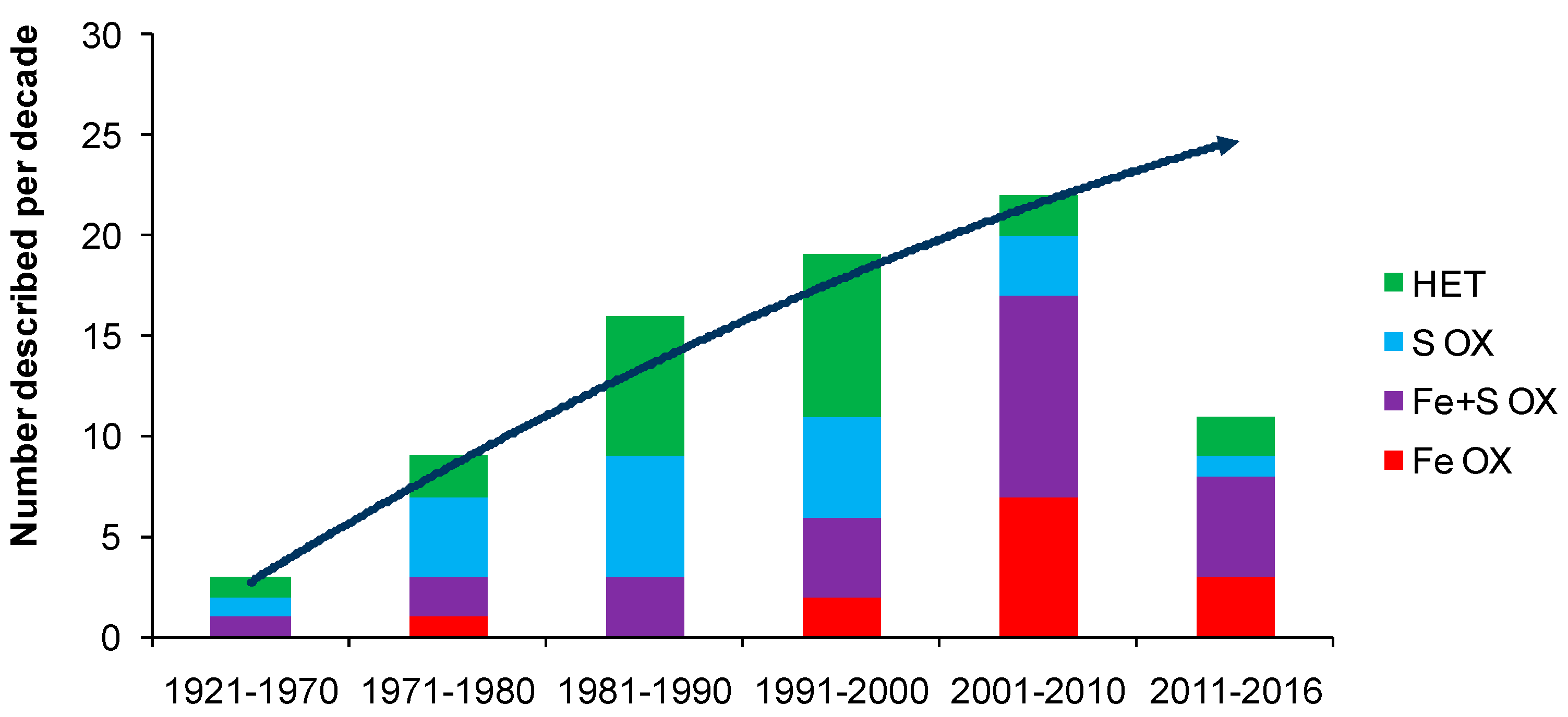
The discovery and description of At. ferrooxidans prompted a rapid increase in microbiological studies related to the oxidation of sulfide minerals. Arguably, At. ferrooxidans is the most studied acidophilic bacterium known, with more than 9000 papers in Web of Science, far more than any other acidophile. However, the perceived importance of At. ferrooxidans in bioleaching environments was largely a consequence of the relative ease of its enrichment and isolation, and its rapid growth in acidic iron(II) media, by which it out-competed other members of its microbial community. In the early years (post-1950), source materials of microorganisms tended to be spoils dumps at coal mines [29] or waste dumps at base metals mines [30]. Since then, many acidophiles have been isolated and described and many more are anticipated (Figure 1), constrained only by the number of research groups interested in acidophiles. Nevertheless, iron(II)- and sulfur-oxidising acidophiles are relatively few in number, especially those isolated from, or detected in, heap- or agitated-tank bioleaching reactors for the processing of sulfide minerals (refer to discussions and collations of microorganisms in [8,31,32,33,34,35].
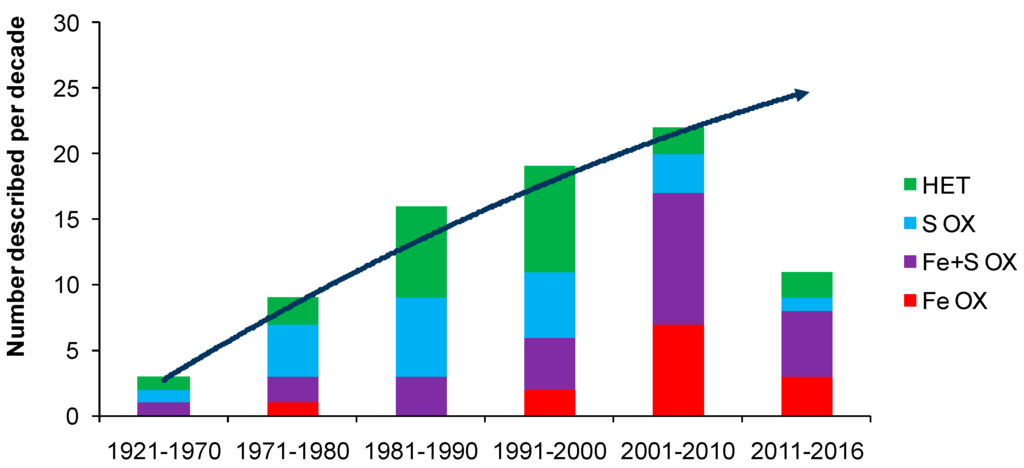
Figure 1.
Trend with time of the numbers of acidophilic microorganisms (Bacteria and Archaea) isolated from environments relevant to biohydrometallurgy since the discovery of At. thiooxidans. They are grouped according to whether they are chemolithotrophs or mixotrophs that utilise iron(II) (Fe OX), both iron(II) and sulfur (Fe + S OX), sulfur but not iron(II) (S OX) or are heterotrophs (HET). Data to February 2016.
More general overviews have been focused on the broader range of microorganisms and microbial interventions associated with acid mine drainage (AMD) environments including collations of acidophilic prokaryotic micro-organisms on the basis of their physiological characteristics [36,37]. These microorganisms are of direct interest in the context of the microbial communities that can inhabit mineral sulfide heaps. Another group of acidophiles of strong interest are the hyperthermophilic archaea found in sulfur-rich geothermal regions (solfataras, volcanoes, hot springs or combusting coal dumps) [38,39,40,41] that might be used to enhance metal extraction from mineral sulfide concentrates in higher-temperature processes.
Before leaving microbiological discovery, it is worth recapping how biomining organisms assist metals extraction. The microbial capabilities of ferrous ion oxidation and RISC oxidation are the two key functions in the most commonly practised form of biohydrometallurgy, ferric sulfate—sulfuric acid leaching in agitated tanks and heaps/dumps. Microbial ferrous-ion oxidation regenerates the oxidant (ferric ion, Reaction (1)) required for sulfide mineral dissolution (e.g., covellite, Reaction (2)). At pH < 4, microbial ferrous ion oxidation is faster than chemical oxidation (e.g., Johnson [36] and references therein). RISC oxidation to sulfuric acid contributes to the acidic environment required for leaching (Reaction (3)).
4Fe2+ + 4H+ + O2 + microbial catalysts → 4Fe3+ + 2H2O
CuS + 2Fe3+ → Cu2+ + 2Fe2+ + S0
2S0 + 3O2 + 2H2O + microbial catalysts → 2H2SO4
However, the geochemistry and biochemistry of sulfide mineral dissolution is more complex than is conveyed by Reactions (1) to (3). It encompasses the electrochemical reactions that take place at the mineral surface during sulfide dissolution as well as physiological processes that control the passage of compounds or ions across the microbial cell membrane and within the cell during growth. These multi-faceted topics are beyond the scope of the review but are discussed comprehensively [42] as well as in some focused reviews on individual species (e.g., At. ferrooxidans [43]), genera (e.g., Sulfolobus spp. [44]), or particular extreme conditions (e.g., microbial mechanisms of coping with high copper concentrations [34], living in acidic environments [39], or the adaptability required to thrive in biomining environments [45]).
3. Microbial Responses to Extreme and Variable Habitats
The two main processes employed in bio-assisted extraction of metals, heap leaching of low-grade sulfide ores and agitated-tank leaching of sulfide concentrates, present very different habitats for the microbial communities that colonise them. Heaps of low-grade ore particles with a broad size distribution (up to 25 mm) are constructed on a large scale and are essentially dynamic, heterogeneous systems in terms of ore mineralogy and grade. Heat generation is a consequence of microbially-catalysed oxidation of sulfide minerals and heat management requires a suitable balance between heap aeration and irrigation [46]. In contrast, agitated tanks are closer to homogeneous in terms of concentrate grade, solution acidity, provision of O2 and CO2 and a selected and controllable temperature for the process. In tanks, there is an abundance of substrate (the sulfide minerals, ferrous ions and sulfur) that diminishes with passage through the train of tanks. Ideally, microorganisms are not exposed to severe fluctuations in conditions.
Much of our understanding of biomining microorganisms, their functions and capabilities, rests upon the collection of acidophiles from mine-impacted sites or geothermal regions that have been isolated and characterised. They form the foundation upon which subsequent studies of the microbial ecology of new sites can be described, monitored and expanded. In particular, the availability of “type strains” deposited in commercial culture collections has facilitated numerous research studies focused on microbial responses to different conditions relevant to bioleaching heap or agitated-tank processes. The four main parameters affecting microbial growth and activity are temperature, acidity, and cations and anions in process water, assuming a substrate is available. In the discussion that follows, unless otherwise noted, it can be assumed that studies are focused on the impact of a particular condition or contaminant on microbial ferrous-ion oxidation rates, this being the key reaction for successful sulfide dissolution and the concomitant extraction of the target metals.
3.1. Temperature
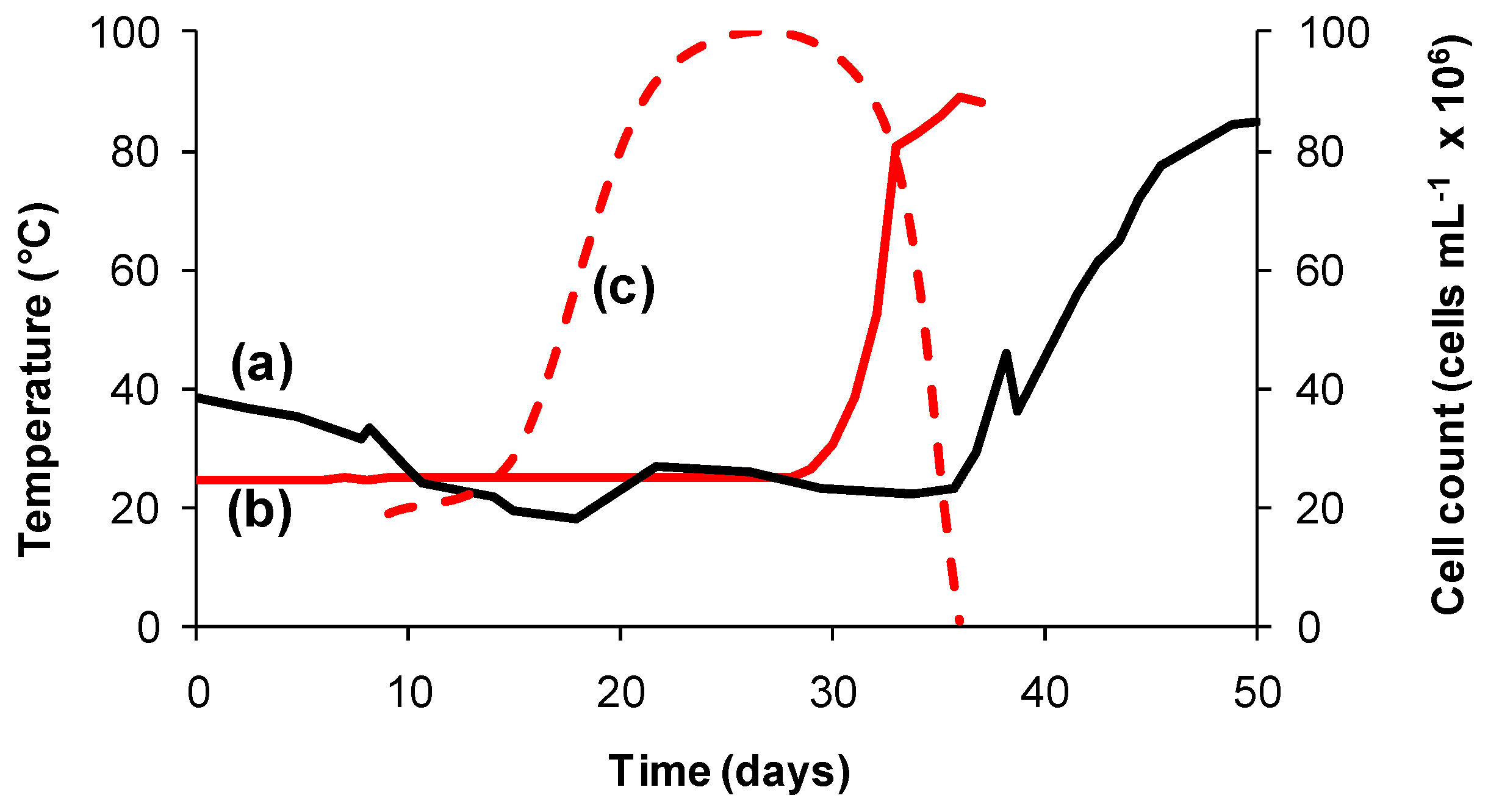
Primarily, microorganisms whether or not they are acidophiles tend to be “classified” according to the temperature range within which they grow, for example mesophiles with temperature optima of 20–40 °C; moderate thermophiles (40–60 °C) or extreme thermophiles (60–80 °C) [36]. Variable temperature is more likely to impact microorganisms in heaps than in agitated tanks. A number of temperature-related examples of advances in understanding taken from laboratory-, pilot- and commercial-scale studies of columns and/or heaps are reported in the literature. Rapid temperature increases and their impacts on microbial communities can be problematic in heap leaching, especially at start up when the sulfide content is at its greatest [47,48,49]. For example, the heat generated during the bioleaching of an ore in a dynamically-controlled, insulated column simulated heat generation in a 5000 tonne test heap of the same ore [47,50]. The catastrophic effect on the microbial population in that simulation as a consequence of the rapid heating to more than 70 °C is of particular interest (Figure 2). The difference in the rates of heat generation is attributed to the initial cell density in the column (5 × 109 cells per kg ore) being higher than that in the test heap (estimated to be 5 × 105 cells per kg ore).
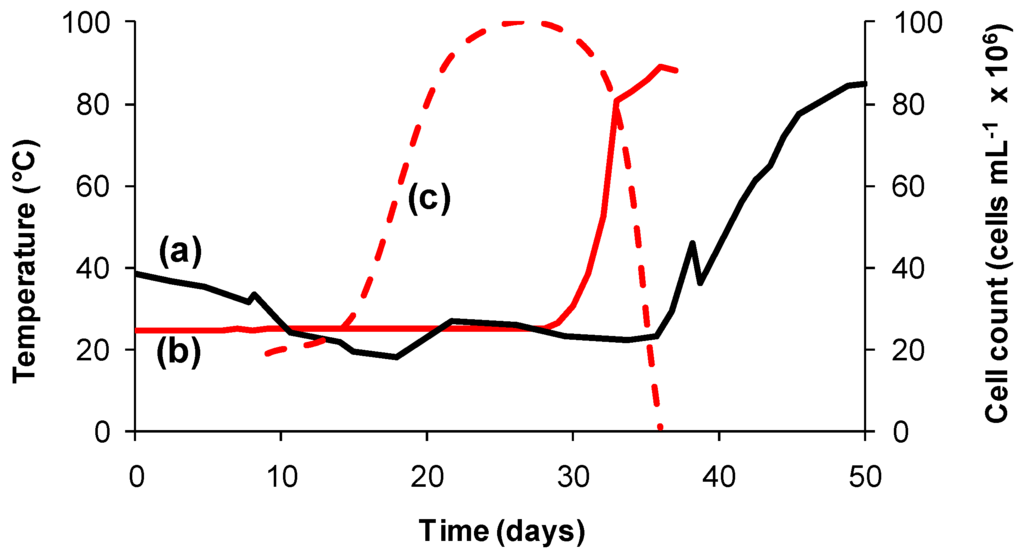
Figure 2.
Heat generation profiles (solid lines) for (a) a 5000 tonne test heap of Ni-Cu-FeS ore and (b) the same ore, acid-agglomerated, inoculated and leached in an aerated, dynamically-controlled, insulated column; (c) estimates of cell numbers (broken line) in column effluent representing the microbial response to increased ore temperature.
The determination of preferred temperatures for growth (ranges and optima) of some single strains of acidophiles, based on their iron(II)- or RISC-oxidation rates, illustrates the rapid decline in the activities of microorganisms at temperatures greater than their respective optima [51,52]. It is generally held that hyperthermophiles, which grow well in sulfur-rich, persistently high-temperature environments such as solfataras and hot springs, would not naturally colonise heaped ores of low-sulfide content [35,51]. The results from a series of columns subjected to different temperature regimes and inoculated with mesophilic, moderately thermophilic and hyperthermophilic microorganisms as appropriate, showed a small benefit from inoculation with hyperthermophiles and led to the subsequent inoculation of a biooxidation heap for refractory gold ore with hyperthermophiles [53,54]. Recently “patches” of Sulfolobus spp. were found in the extremely large Escondida run-of-mine dump, indicative of the existence of persistent high-temperature regions in the dump [55], suggesting that thermophiles may colonise large sulfide dumps and heaps that generate and sustain high temperatures over a long period, removing the need for an inoculation strategy.
3.2. Acidity
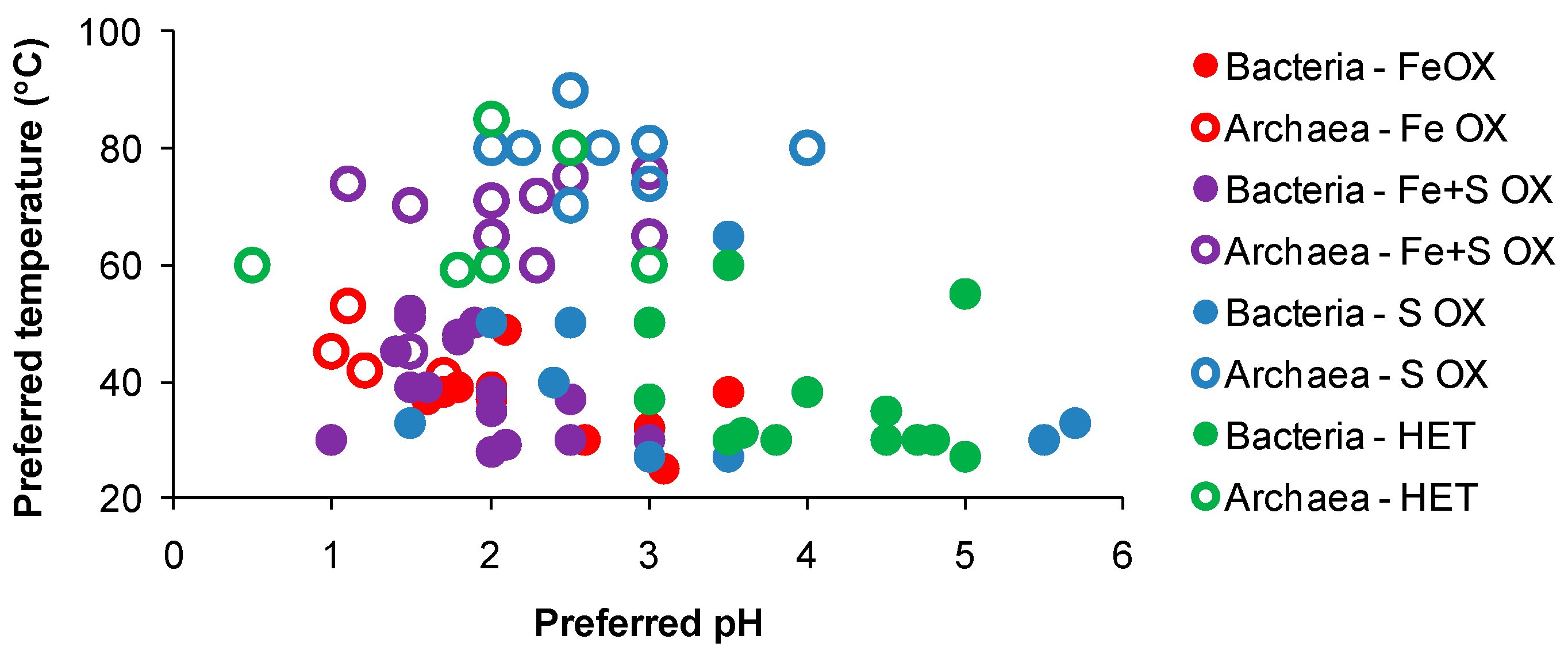
In general, known acidophiles associated with AMD, solfataras and/or managed bioleaching reactors, while widely distributed across the pH range at temperatures favourable to their growth (Figure 3), tend to have a relatively narrow ‘pH window’ below which their activity (iron(II)- or RISC-oxidation) is greatly reduced [56]. However, differences in acid tolerance occur between species/strains. For example, in laboratory flask and reactor tests, a sudden drop in acidity from pH 1.5 to pH 1 caused the cessation of pyrite oxidation by At. ferrooxidans but did not affect the activity of a Sulfobacillus-like strain [57]. Very few acidophiles prefer or are active in habitats poised below pH 1. This was demonstrated in column tests where solution ORP fell, indicative of lower iron(II)-biooxidation activity, when the recycled, pH 1.7 solution, was acidified further to pH < 1 [58]. Similarly, the addition of acid to raffinate prior to recycle to a heap also caused decreased iron(II)- and RISC-oxidising activity in a heap microbial population [59]. Nevertheless, some acidophiles can adapt to very low-pH conditions; Acidiplasma (Ap.) cupricumulans (formerly Ferroplasma cupricumulans) grows in the range pH 0.4–1.8, Ap. aeolicum in the range pH 0–4 and Picrophilus (P.) oshimae and P. torridus can both grow at about pH 0 [60,61,62]. Acidianus sulfidivorans is another extremely acidophilic archaeon with preferred pH range 0.8–1.4 for growth [63].
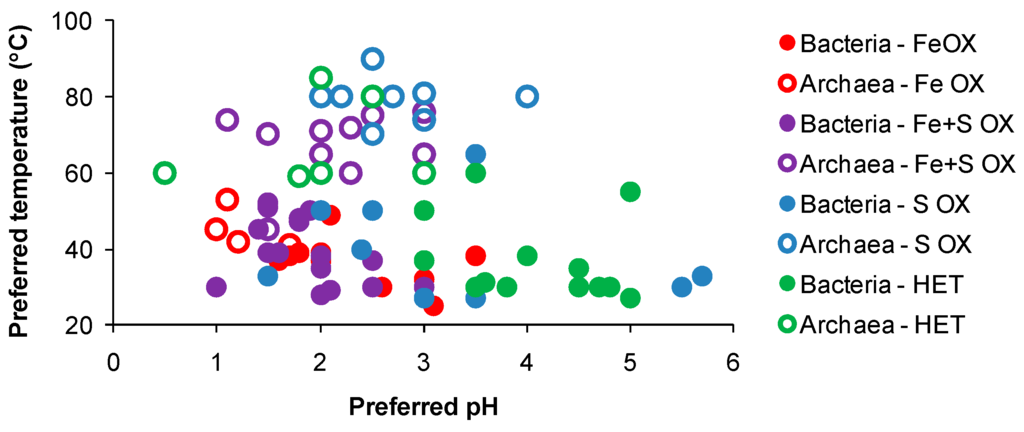
Figure 3.
Preferred solution pH of microorganisms associated with AMD sites, geothermal regions and managed leaching heap or tank bioreactors, referenced against their preferred growth temperatures. Data obtained from their descriptions or from the recommended growth conditions obtained from commercial culture-collection database.
3.3. Cations
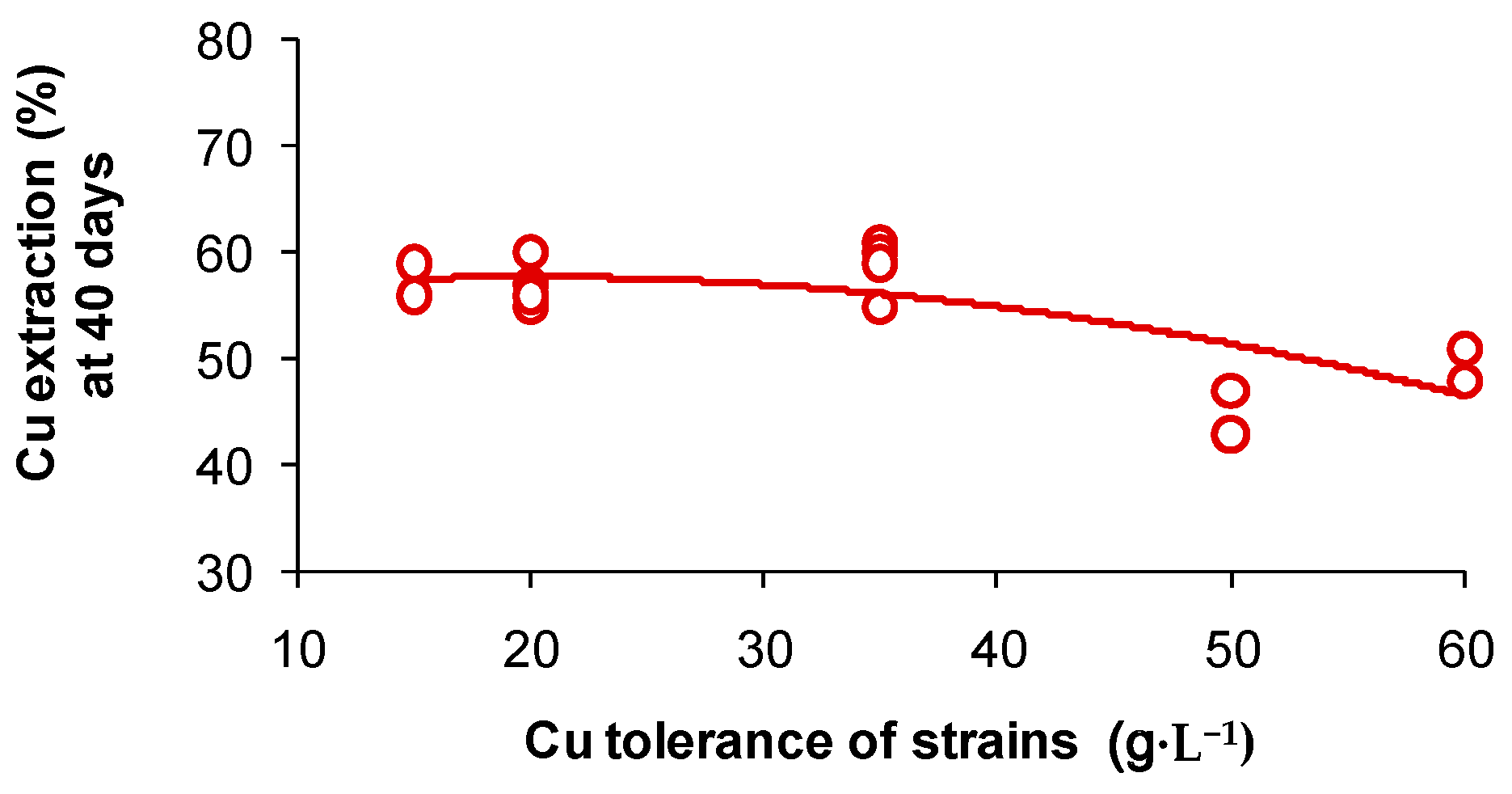
A series of laboratory studies have been conducted in which mainly mesophilic and moderately thermophilic microorganisms were adapted by successive culture to different metal cations, up to (g·L−1): Co, 15; Cu or Ni, 50; Zn, 98 but, overall, there are limited data on natural adaptation [64]. However, an important point to note is that, while tolerance to particular metals concentrations may increase the viability of a microbial population in a leaching operation, it does not necessarily result in increased metals extraction [65]. Indeed, slightly lower copper extractions were achieved in 40 days by the more Cu-tolerant strains of S. thermosulfidooxidans isolated from a commercial copper bioleaching heap (Figure 4).
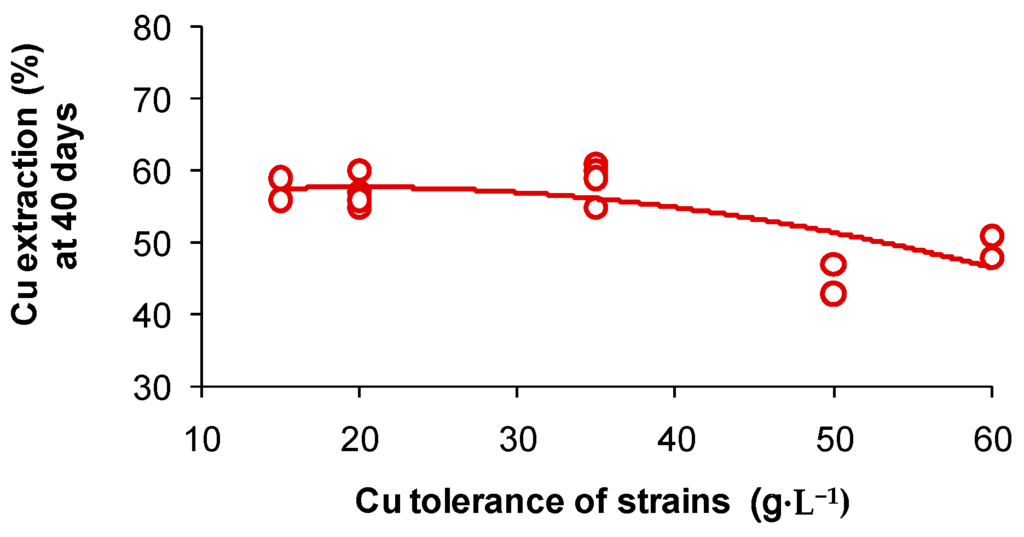
Figure 4.
Bioleaching of chalcopyrite concentrate by isolates of S. thermosulfidooxidans exhibiting different copper tolerances. Tests were conducted with 3 wt % concentrate in basal salts medium (initial pH 1.8) in a shaking incubator (45 °C, 165 rpm). Initial cell density 5 × 106 cells·mL−1.
While data from laboratory studies provide a helpful guide to microbial metals tolerances, more robust data can be obtained from long-term biological processing such as continuous agitated-tank or heap-leach processing. For example, two-years into the continuous pilot-scale operation of a biooxidation plant for the treatment of arsenopyrite gold concentrates, the retention time had reduced from 12 to 3.5 days, the mesophilic microbial population being active in solutions of 13 g·L−1 As [9]. Microbial adaptation to base metals in pilot- and demonstration agitated-tank plants is less well documented but microbial populations have been developed that tolerate >5 g·L−1 Co, 23 g·L−1 Ni with 38 g·L−1 Fe, or 36 g·L−1 Cu in cobalt, nickel and copper continuous plants, respectively [66,67,68,69]. From these data it can be inferred that adaptation has taken place, given that the concentrations are much higher than those encountered in most metal-rich acidic environments.
3.4. Anions
High concentrations of anions can be present in process waters associated with both heap and agitated-tank bioleaching, but in tank leaching sulfate will dominate. Ore heaps are more likely to contain mineral species enriched in chloride, nitrate or fluoride, which are known to inhibit microbial activity.
In bioleaching heaps, especially in Chile, the mineral atacamite (Cu2Cl(OH)3) presents a problem because of the ease with which it dissolves in acidic solutions and releases chloride ion to the circulating process water. In addition, Chile is an arid region with a scarcity of freshwater, leading to the use of seawater at some mines, a factor that has led to strong interest in the development of bacterial cultures active in seawater media. However, efforts to discover or develop acidophilic iron-oxidising strains or adapted cultures that tolerate chloride concentrations equivalent to seawater have had limited success. A survey of microorganisms from saline and acidic drains, lakes and sediments of Western Australia (52 samples) resulted in only five enrichment cultures that could tolerate between 26–41 g·L−1 Cl− in growth media (pH 2) [70]. The authors commented that cultures that oxidised sulfur generally tolerated higher concentrations of salts than those that oxidised iron(II), consistent with previous data [71,72,73].
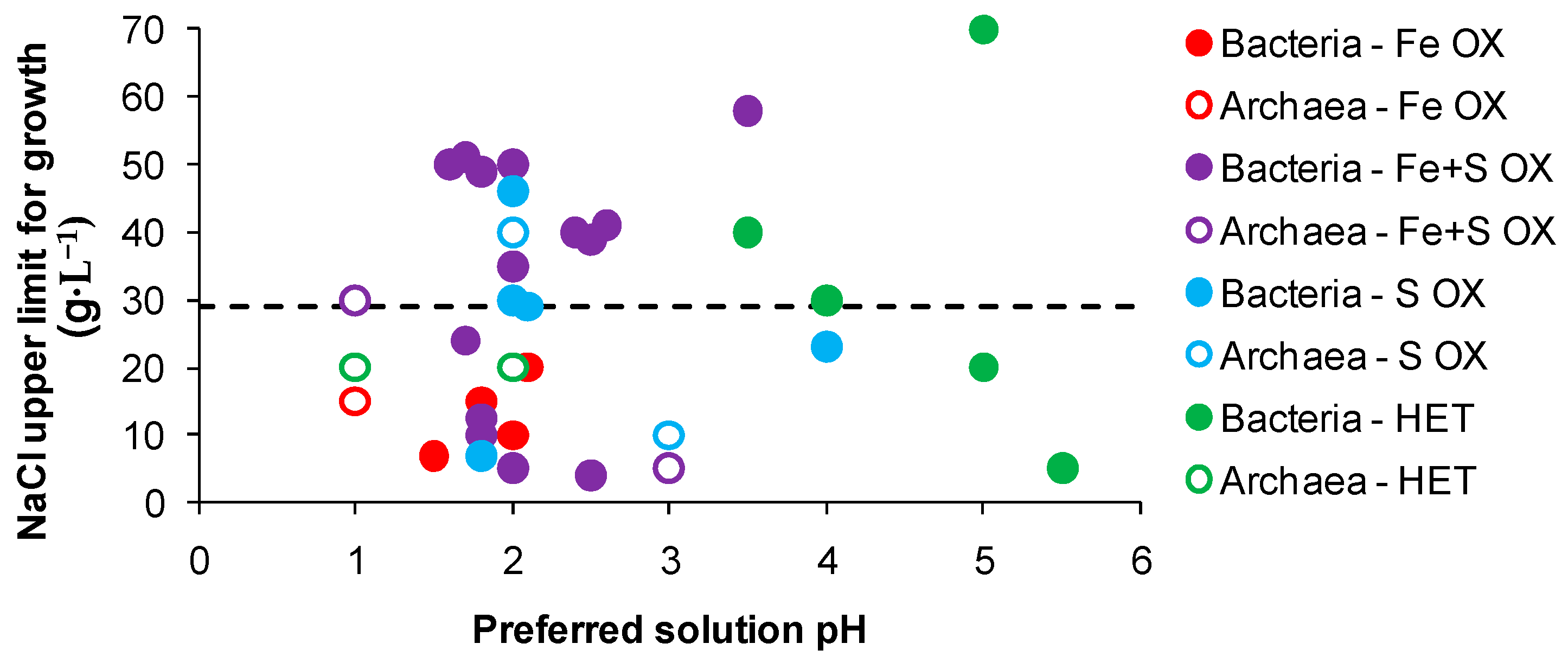
Overall, known biomining acidophiles do not tolerate chloride concentrations equivalent to seawater (approximately equivalent to 29 g·L−1 chloride salts) regardless of the conditions of growth. In several studies, mesophiles tolerated 2–20 g·L−1 NaCl, moderate thermophiles tolerated 10–30 g·L−1 NaCl and a mixed culture of thermophilic archaea tolerated up to 15 g·L−1 NaCl [70,71,72,74]. Data from descriptions of bacteria and archaea for salt tolerance referenced against their preferred pH for growth (Figure 5) indicated that acidophilic iron-oxidising strains with preferred pH 1–2 have limited tolerance to chloride. Strains with the dual capability of iron- and RISC-oxidation, some within slightly less acidic environments, exhibited greater chloride tolerance, particularly Acidihalobacter prosperus (formerly Thiobacillus prosperus [75]) and some Alicyclobacillus-like strains obtained from marine environments. In Figure 5, known biomining microorganisms mainly lie below the dotted line (equivalent chloride content to seawater). Halophilic bacteria, including the RISC-oxidising Halothiobacillus spp [76], and archaea that grow in environments at least 1.5 M NaCl, well above the chloride content of seawater, generally prefer to grow in environments with neutral pH or higher, above the extended acidophile or acid-tolerant range shown in Figure 5 and of lower acidity than would be useful in bio-assisted metals extraction. The discovery or development through adaptation of halo-tolerant acidophiles with iron(II)- and/or RISC-oxidising capability appears to be a major challenge that must be overcome if seawater is to become widely used in heap (bio)leaching.
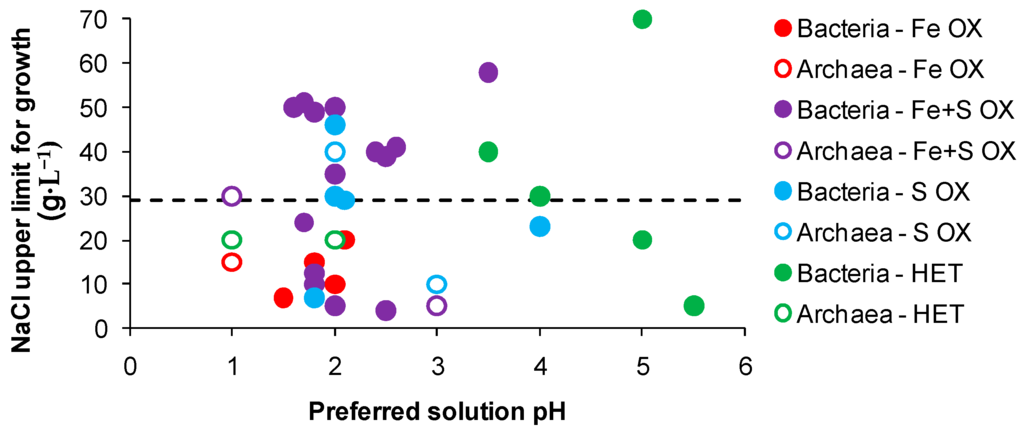
Figure 5.
Maximum salt tolerance of acidophiles referenced against their preferred solution pH. The dotted line represents the concentration of chloride salts in seawater. (Data obtained from published descriptions of species or from the recommended growth conditions obtained from the DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH database).
Concentrations of fluoride higher than 0.5 g·L−1 are detrimental to microbial growth [69], potentially making fluoride in heap process water a greater challenge than chloride. A case in point was a chalcocite heap for which the failure to reach design recoveries was partly attributed to the failure to test the solubility of fluoride-containing minerals in the ore [77]. In that study, concentrations of iron-oxidizing microorganisms were lower than is typical for sulfide heap leaching, diminishing from the critical concentration of 105 cells·mL−1 [78] to 103 cells·mL−1 within a year. In an earlier but related study, finely ground particles of a variety of minerals inhibited iron(II) oxidation by At. ferrooxidans but fluorapatite (Ca5(PO4)3F) particles (<74 µm) caused an anomalously severe response in suspensions < 2 wt % [79].
In a study of sulfur oxidation using At. thiooxidans, the toxicity of fluoride was shown to be pH dependent. It was strongly inhibitory at pH 2.3, where undissociated HF dominates the speciation, and is known to penetrate cell membranes [80,81]. Fluoride was less inhibitory at pH 4.5, where only 10% is present as undissociated HF, and not toxic at pH 7 where the undissociated MF molecule is dominant (M = Li, Na or K) [80]. Fluoride toxicity differs between bacterial species. In a recent comparison of iron(II)- and/or RISC-oxidation by five known bioleaching bacteria, the rank order of fluoride tolerance in media pH 1.5–2 was At. ferrooxidans > At. thiooxidans > Leptospirillum ferrooxidans > At. caldus > Sulfobacillus (S.) thermosulfidooxidans [82]. The fluoride tolerance of bacterial cultures can be increased substantially, in one case from 100 to 850 mg·L−1 through adaptive protocols [81,83].
The impact of the bioleach solution composition on the concentration of the HF species was investigated at 65 °C using Sulfolobus metallicus growing on pyrite (initial solution pH 1.5) and monitoring changes in ORP as a surrogate measure of microbial activity and growth [84]. The tests varied in the concentration of aluminium added to the growth medium, in the knowledge that Al(III) (and Fe(III)) form Al- or Fe-fluoride complexes [85,86,87]. A similar strategy of aluminium addition counteracted fluoride toxicity during the bioleaching of a low-grade, fluoride-containing ore using S. thermosulfidooxidans (50 °C, growth media in the range pH 1.4–2.1) [88], demonstrating that, if fluoride solubilisation is accompanied by strong aluminium or iron(III) solubilisation from gangue minerals, then the effects on the microorganisms may be substantially mitigated due to the formation of aluminium- or iron(III)-fluoro complexes.
As well as being a copper province, Chile is also a nitrate province. The very large Chilean deposits of salitre (saltpetre), mixtures of sodium and potassium nitrate, are a valuable export commodity. It is not surprising therefore that nitrate minerals are sometimes components of sulfide ores and that nitrate in process water may impact on microbial activity. Studies using At. ferrooxidans as the test species showed that nitrate was more inhibitory to iron(II) oxidation than to RISC oxidation [89,90]. However, in a recent comparison of the effects of nitrate ion on substrate utilisation by bacteria and archaea, it was found that the bacteria adapted to the presence of nitrate in growth media and resumed iron(II)-oxidation within a nine-week period in bioleaching tests [91]. Thus nitrate in ores may have only a transient impact on metals extraction in heaps. In contrast, in the same study, it was found that archaea did not adapt to the presence of nitrate. Based on this result, it was proposed that nitrate may be a useful means of controlling ORP during the higher-temperature oxidation of chalcopyrite in an agitated-tank process [91].
Surprisingly, the concentrations of sulfate ion, by far the predominant anion in most bioleaching process waters, are seldom reported and the effect of sulfate on microbial activity seems not to be of great concern. Sulfate concentrations can be more than 100 g·L−1 in agitated tanks [68,69] or up to 150 g·L−1 in the process water circulating through heaps [92,93], concentrations which are much higher than the 40 g·L−1 sulfate that caused an adapted mesophilic culture in iron(II) growth medium to halve its replication rate [94] or At. caldus in tetrathionate growth medium to increase the lag time two- to three-fold [74]. During a column bioleaching study (low-grade copper ore), planktonic cells in process water exiting a column declined rapidly by two orders of magnitude when concentrations of ferric and sulfate ions exceeded 30 and 75 g·L−1, respectively [95], and sulfur-oxidising bioactivity in the Escondida heap was reduced by 50% when sulfate concentrations in process water exceeded 120 g·L−1 [96].
4. Biodiversity in Bioleaching Environments
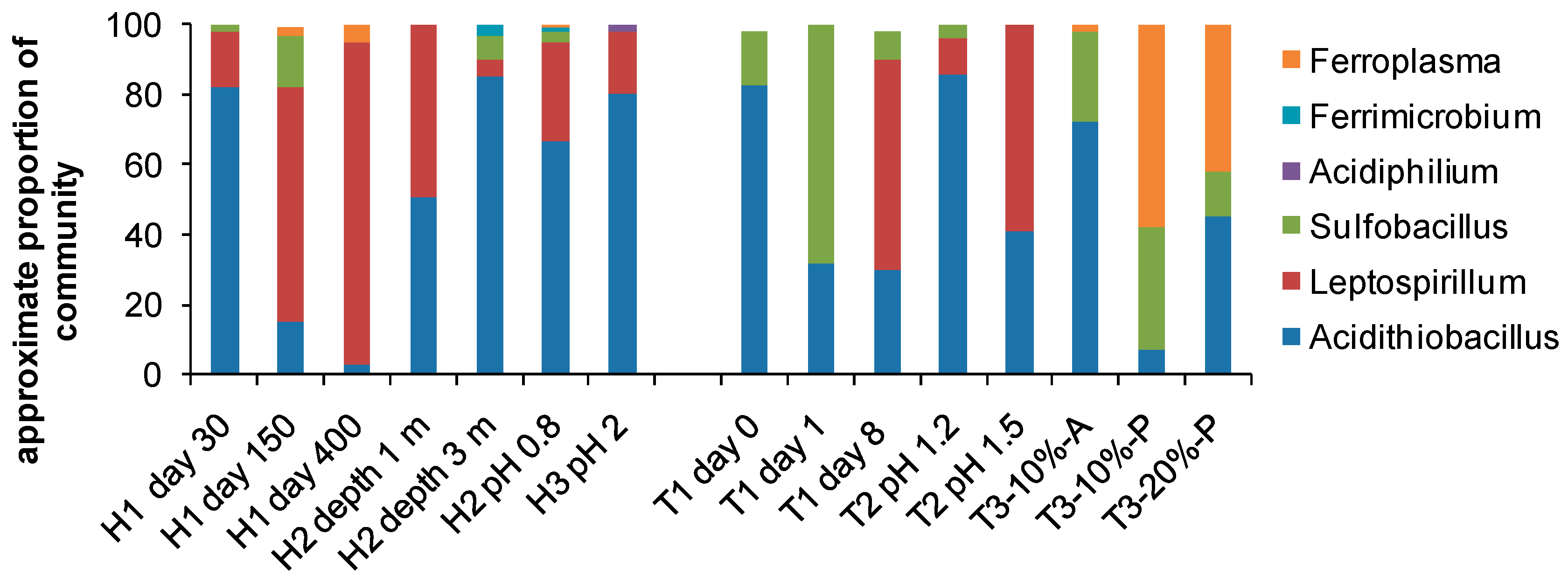
The ease with which the microbial diversity of environmental samples can be examined has led to many recent descriptions of the orders, genera and sometimes species inhabiting bioleaching reactors (heaps, dumps, tanks) and/or AMD impacted environments. The collation of example data in Figure 6 illustrates the increased knowledge of bioreactor diversity as well as some effects of variable environments on that diversity, for example changes in community structure with time (H1, days 30 to 400; and T1, days 0 to 8), with depth in the heap (H2, depth 1 cf. 3 m) and with operating pH of the process water (H2 and H3, T2). In agitated tanks, the population of attached microorganisms differs from the planktonic community (T3, attached vs. planktonic) and the community structure differs with different solids loading (T3, 10% cf. 20%).
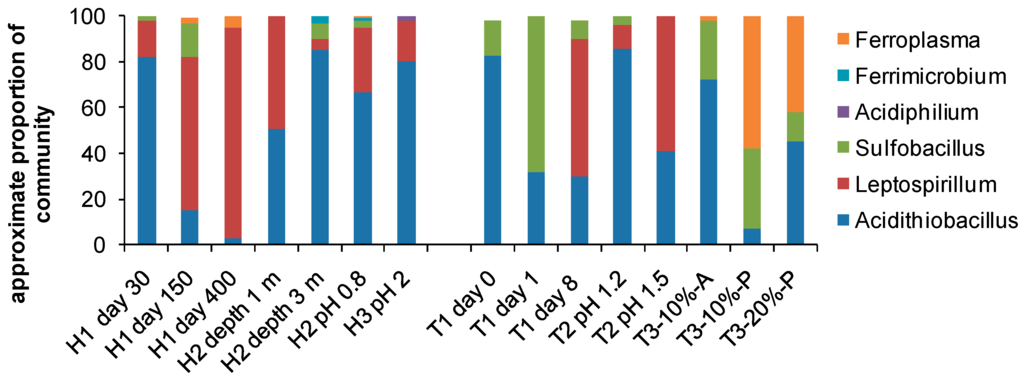
Figure 6.
Microbiological diversity to genus level in heaps (H), and agitated tanks (T); A, attached; P, planktonic cells; % solids loading. Data collated from [97,98,99,100,101,102].
In general, abundances of Acidithiobacillus spp. in heaps including At. ferrooxidans and the sulfur-oxidising species At. caldus, At. thiooxidans and At. albertensis are greater early in leaching when there is a greater sulfur content [99,103]. Leptospirillum and Ferroplasma are more abundant later in the leach when the iron concentrations are higher and possibly inhibitory to Acidithiobacillus and other species [99,104,105]. Microbial communities change with time in both heaps and tanks, influenced by parameters such as substrate content and availability, solution composition and acidity, solids loadings in tanks, and temperature [92,102,103,104,106,107,108]. However, comparisons between studies tend to reveal a variety of effects as the communities respond to a combination of environmental parameters and, in many instances, to adapt to them with time [64].
It is in the nature of sulfide heaps that sulfur contents tend to be low and the heat generated within the bed during sulfide oxidation is transient [49], essentially because heap-management strategies are employed to maintain temperatures in the range suited to bacterial growth. In general, extreme thermoacidophiles with iron(II)- and sulfur-oxidising capabilities colonise habitats that are persistently hot and sulfurous [40,109,110]. As a consequence, it has been assumed that thermophiles such as Acidianus, Metallosphaera and Sulfolobus spp [111,112,113,114] would not naturally colonise sulfide heap operations and that thermophiles would need to be introduced to managed “hot heaps” to exploit increased metal extraction kinetics [49,54]. However, some recent studies on the biodiversity of sulfide heaps and mine-waste dumps included the putative identification of Archaea and some thermophiles have been detected, including Acidianus, Acidiplasma, Ferroplasma, Metallosphaera, Sulfolobus and Thermoplasma spp. [49,115,116,117,118,119].
The advances in both DNA sequencing and protein extraction, accompanied by huge advances in computing power and speed, launched the age of genomics and metagenomics, proteomics and meta-proteomics and metabolomics (Table 2).

Table 2.
Molecular biology terminology.
Not surprisingly, the first genome sequence of a bioleaching microorganism was that of At. ferrooxidans [120]. Since then, the number of genomes of microorganisms of direct interest in bioleaching has increased significantly, largely due to a general interest in acidophiles and their activities in AMD environments. The strong overlap with bioleaching is understandable if bioleaching is viewed as an exemplar of extreme but controlled AMD. In a 2010 review [121], it was reported that there were 30 bacterial genomes and 26 archaeal genomes for extremely acidophilic microorganisms, eight metagenome projects of extremely acidic environments (four associated with the AMD at Iron Mountain, CA) and complete sequences for 38 plasmids and 29 viruses from acidic environments. At the same time, it was concluded that these data were not sufficient “to provide a reasonably complete description of the genomic complexity and, by inference, of the full metabolic potential present in bioleaching operations” because (i) there are many gaps regarding the metabolic data; (ii) the microbial diversity of bioleaching and AMD environments is considerably greater than was previously appreciated; and (iii) often genomes were obtained using laboratory-maintained strains that may have accumulated genetic modifications.
Nevertheless, despite the body of research required to address the gaps and the greater diversity in natural microbial communities, the applications of the complementary techniques (Table 2) are already augmenting our understanding of the microbial processes involved in extracting metals from minerals. Numerous data on microbial functions and the conditions under which they are exercised best are being gathered. For example, their applications to AMD and bioleaching systems are resulting in better understanding of microbial physiology for individual organisms [40,122,123,124,125]. Bacterial, archaeal and eukaryotic diversity and their metabolic networks in AMD habitats have been described [126,127,128] and the ways in which metabolic activity levels and the partitioning of function are affected by community composition and the physico-chemical environment have been explored [129,130]. The extent to which each of the individual participants contributes to the process and how they evolve in time to keep the conglomerate healthy and therefore efficient during the entire process of bioleaching is being investigated [131]. Finally, the hypothesis that advanced knowledge of microbial processes can lead to better process design, control and optimization for metals extraction from minerals is supported by the interest shown by some mining companies in transcriptomic analysis of the genomic sequences of newly-isolated (commercially-sensitive) microorganisms with superior ability to extract copper from sulfides [132].
A valuable output from genomic studies is the generation of genomic sequences for microarray analysis [132]. The development of microarray technology over a number of years [133,134] has the potential to revolutionise the study of biodiversity in environmental samples. A review on the development of functional gene arrays was introduced as follows: “the recent advance of metagenomic technologies such as high throughput sequencing and functional gene arrays provides powerful high throughput tools for analysing microbial communities” [135]. A variety of microarrays have been constructed to contain probes for the genes involved in key microbial functions for specific applications [135]. For example, a microarray was developed based on most of the genes associated with acidophiles, including genes for carbon, nitrogen, sulfur and iron metabolism, DNA replication and repair, and metal resistance [136]. A suite of GeoChip microarrays, of which GeoChip 4 is the most comprehensive [137], have been used to characterize microbial communities in terms of functional diversity, composition, structure and metabolic activity/capability from a variety of habitats (e.g., soils, aquatic, contaminated sites, bioreactors).
Two modes of microarray application have been described. The use of microarrays based on the complete genomes of the members of a microbial community enables a nearly complete view of gene expression under different bioleaching conditions but it is time-consuming and expensive for the study of organisms isolated from the environment. Examples of specific studies based on complete genome sequences in combination with physiological and biochemical studies include for bacteria Acidithiobacillus spp [138,139,140] and for archaea, Metallosphaera sedula [141,142]. The shotgun DNA microarray, is a general approach by which gene expression can be studied in environmental isolates and can be applied to any organism regardless of how much of its genome is sequenced [132,143,144]. Applications for acidophiles include bacteria, Leptospirillum spp. [145,146] and Acidithiobacillus spp. [147] and archaea, Acidianus [148] and Ferroplasma [149]. Applications of microarray analysis of bioleaching and/or AMD environments include the study of bacterial population dynamics at a uranium-contaminated site [150], the monitoring of acidophile activity in AMD systems [151,152] and in the Escondida copper heap [99,153]. As no public domain research on heap or tank microbial populations funded by industry was found in this survey, it is assumed that such applications of advanced molecular-microbiological techniques for the discovery of superior bioleaching microorganisms are commercially sensitive.
5. Summary
Until recently, the success of the two, widely-practised technologies employed in biohydrometallurgy, the pre-treatment of refractory gold concentrates in agitated tanks and the heap and dump leaching of low-grade, copper sulfide ores, was thought to depend on a few acidophilic microorganisms that oxidised iron(II) or sulfur compounds (chemolithotrophs). The technological concern was to optimise processing conditions to suit the microorganisms in the hope of faster and greater metals production.
In the late 20th century, the research emphasis was focused in two areas, discovering microorganisms with ‘superior’ metal-extraction capabilities and gaining a better understanding of how adaptable and resilient they were to extreme and variable mineral processing environments. The numbers of new species from natural and mine-impacted acidic environments increased steadily, with particular value being placed on extremely acidophilic, thermophilic and/or halophilic species with the requisite iron(II) and sulfur oxidising capabilities. Once characterised, their ready availability from commercial culture collections promoted an impressive number of studies on their responses to acid, metals and other soluble components, high ionic strength, high temperature and other processing parameters, depending on whether the research interest was rapid metals extraction from ores/concentrates or remediation of acid-drainage systems. The result is that what were once thought to be environments with low biodiversity have been shown to host diverse, robust, adaptable communities of acidophiles.
Now, in the 21st century, more than 50 genomes of acidophilic microorganisms have been published and analysed, leading to greatly increased knowledge of their physiological capabilities. Functional-gene microarrays are providing snapshots of microbial diversity in AMD environments and managed bioleaching reactors in terms of community structure and metabolic activity. The increasing capability of microarrays to describe not only what acidophiles are present but also what active contributions the acidophiles are making to metals extraction, is set to revolutionise both biodiversity studies of operating heaps and bioreactors as well as laboratory-based studies of the impacts of changing growth conditions on microbial activities. The body of knowledge being acquired through such studies can be used to predict microbial behaviour under varied conditions and from those data, to modify conditions for better metal extraction or for more appropriate microbial activity. Conversely, it should become possible to infer “microbial health” particularly in heaps based on measured physicochemical parameters and microbial diversity in process water, without having to disturb a production heap by sampling the solids.
Acknowledgments
The Australian Government is thanked for funding through CSIRO Mineral Resources.
Conflicts of Interest
The author declares no conflict of interests.
References
- McDonald, R.G.; Whittington, B.I. Atmospheric leaching of nickel laterites review. Part 1. Sulphuric acid technologies. Hydrometallurgy 2008, 91, 35–55. [Google Scholar] [CrossRef]
- McDonald, R.G.; Whittington, B.I. Atmospheric leaching of nickel laterites review. Part 2. Chloride and bio-technologies. Hydrometallurgy 2008, 91, 56–69. [Google Scholar] [CrossRef]
- McDonald, R.G.; Muir, D.M. Pressure oxidation leaching of chalcopyrite. Part I. Comparison of high and low temperature reaction kinetics and products. Hydrometallurgy 2007, 86, 191–205. [Google Scholar] [CrossRef]
- McDonald, R.G.; Muir, D.M. Pressure oxidation leaching of chalcopyrite. Part II. Comparison of medium temperature kinetics and products and effect of chloride ion. Hydrometallurgy 2007, 86, 206–220. [Google Scholar] [CrossRef]
- Holmes, D.S.; Debus, K.A. Biological opportunities for metal recovery. In Proceedings of the International Symposium on Biotechnology for Energy, Faisalabad, Pakistan, 16–21 December 1989; Malik, K.A., Naqvi, S.H.M., Aleem, N.I.H., Eds.; Nuclear Institute for Agriculture and Biology: Faisalabad, Pakistan, 1991; pp. 341–358. [Google Scholar]
- Poulter, S.; Canterford, J.; Lunt, D. Biotechnology for sulphide minerals. A comparison with other leaching processes. In Proceedings of the Biomine 99 and Waste Management in Metallurgical Operations Conference, Perth, Australia, 23–24 August 1999; Australian Mineral Foundation: Glenside, Australia, 1999; pp. 1–14. [Google Scholar]
- Domic, E. A review of the development and current status of copper bioleaching operations in Chile: 25 years of successful commercial implementation. In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer: Berlin, Germany, 2007; pp. 81–95. [Google Scholar]
- Watling, H.R. The bioleaching of sulphide minerals with emphasis on copper sulphides—A review. Hydrometallurgy 2006, 84, 81–108. [Google Scholar] [CrossRef]
- Van Aswegen, P.C.; Haines, A.K.; Marais, H.J. Design and operation of a commercial bacterial oxidation plant at Fairview. In Proceedings of the Randol Gold Conference, Perth, Australia, 14–17 March 1995; Randol International: Golden, CO, USA, 1988; pp. 144–147. [Google Scholar]
- Van Niekerk, J. Factors affecting the selection of BIOX® as the preferred technology for the treatment of a refractory gold concentrate. Adv. Mater. Res. 2015, 1130, 191–196. [Google Scholar] [CrossRef]
- Hiskey, J.B. In situ leaching of copper: What’s next? In Proceedings of the Hydrometallurgy ’94, Cambridge, UK, 11–15 July 1994; Chapman & Hall: London, UK, 1994; pp. 43–67. [Google Scholar]
- Pliny the Elder: the Natural History. Volume 37. Available online: http://penelope.uchicago.edu/Thayer/E/Roman/Texts/Pliny_the_Elder/home.html (accessed on 20 May 2016).
- Walsh, J. Galen visits the Dead Sea and the copper mines of Cyprus. Bull. Geogr. Soc. Phila. 1927, 25, 93–110. [Google Scholar]
- Lung, T.N. The history of copper cementation on iron—World’s first hydrometallurgical process from medieval China. Hydrometallurgy 1986, 17, 113–129. [Google Scholar] [CrossRef]
- Salkield, L.U. A Technical History of the Rio Tinto Mines: Some Notes on Exploitation from Pre-Phoenician Times to the 1950s; Institution of Mining and Metallurgy: London, UK, 1987. [Google Scholar]
- Agricola, G. De re Metallica; Froben: Basel, Switzerland, 1556; Available online: www.gutenberg.org/ebooks/38015 (accessed on 20 May 2016).
- Lipman, J.G.; Waksman, S.A.; Joffe, J.S. The oxidation of sulfur by soil microorganisms. Soil Sci. 1921, 12, 475–489. [Google Scholar] [CrossRef]
- Waksman, S.A.; Joffe, J.S. Microorganisms concerned in the oxidation of sulfur in the soil. II. Thiobacillus thiooxidans, a new sulfur-oxidizing organism isolated from the soil. J. Bacteriol. 1922, 7, 239–256. [Google Scholar] [PubMed]
- Rudolfs, W. Oxidation of iron pyrites by sulfur-oxidizing organisms and their use for making mineral phosphates available. Soil Sci. 1922, 14, 135–147. [Google Scholar] [CrossRef]
- Rudolfs, W.; Helbronner, A. Oxidation of zinc sulfide by microorganisms. Soil Sci. 1922, 14, 459–464. [Google Scholar] [CrossRef]
- Falck, R.; Kingma, V.T. The methodological and principles of production of organic acids by biological methods with help of aphanomyces. Ber. Deutsch. Chem. Ges. 1924, 57, 915–920. [Google Scholar] [CrossRef]
- Bernhauer, K. The problem of the formation of acids by Aspergillus niger. (Introductory announcement). Biochem. Z. 1924, 153, 517–521. [Google Scholar]
- Colmer, A.R.; Hinkle, M.E. The role of microorganisms in acid mine drainage. Science 1947, 106, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Colmer, A.R.; Temple, K.L.; Hinkle, H.E. An iron-oxidizing bacterium from the acid mine drainage of some bituminous coal mines. J. Bacteriol. 1950, 59, 317–328. [Google Scholar] [PubMed]
- Temple, K.L.; Colmer, A.R. The autotrophic oxidation of iron by a new bacterium: Thiobacillus ferrooxidans. J. Bacteriol. 1951, 62, 605–611. [Google Scholar] [PubMed]
- Oldfield, J.D.; Shaw, D.J.B. VI Vernadskii and the development of biogeochemical understandings of the biosphere, c. 1880s–1968. Brit. J. Hist. Sci. 2013, 46, 287–310. [Google Scholar] [CrossRef]
- Krasilnikov, N.A. The role of micro-organisms in rock weathering. I. Microflora of rock stratum surfaces. Mikrobiologiya 1949, 18, 318–323. [Google Scholar]
- Krasilnikov, N.A. The role of micro-organisms in rock weathering. II. The spread of micro-organisms from foci on rock surfaces. Mikrobiologiya 1949, 18, 492–497. [Google Scholar]
- Leathen, W.W.; Braley, S.S., Sr.; McIntyre, L.D. The role of bacteria in the formation of acid from sulfuritic constituents associated with bituminous coal: II. Ferrous iron oxidizing bacteria. Appl. Microbiol. 1953, 1, 65–68. [Google Scholar] [PubMed]
- Bryner, L.C.; Jameson, A.K. Microorganisms in leaching sulfide minerals. Appl. Microbiol. 1958, 6, 281–287. [Google Scholar] [PubMed]
- Rawlings, D.E. Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Factories 2005, 4. [Google Scholar] [CrossRef] [PubMed]
- Norris, P.R. Acidophile diversity in mineral sulfide oxidation. In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer: Berlin, Germany, 2007; pp. 199–216. [Google Scholar]
- Johnson, D.B. Biodiversity and interactions of acidophiles: Key to understanding and optimizing microbial processing of ores and concentrates. Trans. Nonferr. Met. Soc. China 2008, 18, 1367–1373. [Google Scholar] [CrossRef]
- Orell, A.; Navarro, C.A.; Aranciba, R.; Mobarec, J.C.; Jerez, C.A. Life in blue: Copper resistance mechanisms of Bacteria and Archaea used in industrial biomining of minerals. Biotechnol. Adv. 2010, 28, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Watling, H.R.; Watkin, E.L.J.; Ralph, D.E. The resilience and versatility of acidophiles that contribute to the bioassisted extraction of metals from mineral sulphides. Environ. Technol. 2010, 31, 915–933. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B. Extremophiles: Acidic Environments. In Encyclopedia of Microbiology; Schaechter, M., Ed.; Elsevier: Oxford, UK, 2009; pp. 107–126. [Google Scholar]
- Johnson, D.B.; Hallberg, K.B. Carbon, iron and sulfur metabolism in acidophilic micro-organisms. Adv. Microb. Physiol. 2009, 54, 201–255. [Google Scholar]
- Norris, P.R.; Burton, N.P.; Foulis, N.A.M. Acidophiles in bioreactor mineral processing. Extremophiles 2000, 4, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Dopson, M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007, 15, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Auernik, K.S.; Cooper, C.R.; Kelly, R.M. Life in hot acid: Pathway analyses in extremely thermoacidophilic archaea. Curr. Opin. Biotechnol. 2008, 19, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Itoh, T.; Yamagishi, A. Archaeal diversity in a terrestrial acidic spring field revealed by PCR primer targeting archaeal 16S rRNA genes. FEMS Microbiol. Lett. 2011, 319, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, H.L.; Newman, D.K.; Kappler, A. Geomicrobiology, 6th ed.; CRC Press: Boca Raton, FA, USA, 2015; p. 635, (and references therein). [Google Scholar]
- Alexander, B.; Leach, S.; Ingledew, W.J. The relationship between chemiosmotic parameters and sensitivity to anions and organic acids in the acidophile Thiobacillus ferrooxidans. J. Gen. Microbiol. 1987, 133, 1171–1179. [Google Scholar] [CrossRef]
- Wheaton, G.; Counts, J.; Mukherjee, A.; Kruh, J.; Kelly, R. The confluence of heavy metal biooxidation and heavy metal resistance: Implications for bioleaching by extreme thermoacidophiles. Minerals 2015, 5, 397–451. [Google Scholar] [CrossRef]
- Rawlings, D.E. Relevance of cell physiology and genetic adaptability of biomining microorganisms to industrial processes. In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer-Verlag: Heidelberg, Germany, 2007; pp. 177–198. [Google Scholar]
- Dixon, D.G. Analysis of heat conservation during copper sulphide heap leaching. Hydrometallurgy 2000, 58, 27–41. [Google Scholar] [CrossRef]
- Hunter, C. BioHeap™ leaching of a primary nickel-copper sulphide ore. In Proceedings of the Nickel-Cobalt-8 Technical Proceedings, Perth, Australia, 20–22 May 2002; ALTA Metallurgical Services: Melbourne, Australia, 2002. [Google Scholar]
- Readett, D.; Sylwestrzak, L.; Franzmann, P.D.; Plumb, J.J.; Robertson, W.R.; Gibson, J.A.E.; Watling, H. The life cycle of a chalcocite heap bioleach system. In Hydrometallurgy 2003; Young, C.A., Alfantazi, A.M., Anderson, C.G., Dreisinger, D.B., Harris, B., James, A., Eds.; TMS: Warrendale, PA, USA, 2003; pp. 365–374. [Google Scholar]
- Plumb, J.J.; Hawkes, R.B.; Franzmann, P.D. The microbiology of moderately thermophilic and transiently thermophilic ore heaps. In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer-Verlag: Berlin, Germany, 2007; pp. 217–235. [Google Scholar]
- Shiers, D.W.; Maree, M.D.; Collinson, D.M.; Watling, H.R.; Hosken, T.; Ingram, G.D. Use of a dynamically controlled column to assess the impact of temperature on copper extraction and microbial activity during copper sulfide bioleaching. In Proceeding of the Goldschmidt 2015 25th Anniversary, Praguz, Czech Republic, 16–21 August 2015.
- Franzmann, P.D.; Haddad, C.M.; Hawkes, R.B.; Robertson, W.J.; Plumb, J.J. Effects of temperature on the rates of iron and sulfur oxidation by selected bioleaching Bacteria and Archaea: Application of the Ratkowsky equation. Min. Eng. 2005, 18, 1304–1314. [Google Scholar] [CrossRef]
- Watling, H.R.; Perrot, F.A.; Shiers, D.W. Comparison of selected characteristics of Sulfobacillus species and review of their occurrence in acidic and bioleaching environments. Hydrometallurgy 2008, 93, 57–65. [Google Scholar] [CrossRef]
- Brierley, J.A. Response of microbial systems to thermal stress in biooxidation-heap pretreatment of refractory gold ores. Hydrometallurgy 2003, 71, 13–19. [Google Scholar] [CrossRef]
- Logan, T.C.; Seal, T.; Brierley, J.A. Whole ore heap biooxidation of sulfidic gold-bearing ores. In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer: Berlin, Germany, 2007; pp. 113–138. [Google Scholar]
- Soto, P.; Acosta, M.; Tapia, P.; Contador, Y.; Velásquez, A.; Espoz, C.; Pinilla, C.; Galleguillos, P.; Demergasso, C. From mesophilic to moderate thermophilic populations in an industrial heap bioleaching process. Adv. Mater. Res. 2013, 825, 376–379. [Google Scholar] [CrossRef]
- Plumb, J.J.; Muddle, R.; Franzmann, P.D. Effect of pH on rates of iron and sulfur oxidation by bioleaching microorganisms. Min. Eng. 2008, 21, 76–82. [Google Scholar] [CrossRef]
- Yahya, A.; Johnson, D.B. Bioleaching of pyrite at low pH and low redox potentials by novel mesophilic gram-positive bacteria. Hydrometallurgy 2002, 63, 181–188. [Google Scholar] [CrossRef]
- Tupikina, O.V.; Ngoma, I.E.; Minnaar, S.; Harrison, S.T.L. Some aspects of the effect of pH and acid stress in heap bioleaching. Min. Eng. 2011, 24, 1209–1214. [Google Scholar] [CrossRef]
- Zepeda, V.J.; Cautivo, D.; Galleguillos, P.A.; Salazar, C.N.; Velasquez, A.; Pinilla, C.; Demergasso, C.S. Effect of increased acid concentration on the microbial population inhabiting an industrial heap bioleaching plant. Adv. Mater. Res. 2013, 825, 348–351. [Google Scholar] [CrossRef]
- Hawkes, R.B.; Franzmann, P.D.; O’Hara, G.; Plumb, J.J. Ferroplasma cupricumulans sp. nov. a novel moderately thermophilic acidophilic archaeon isolated from an industrial-scale chalcocite bioleach heap. Extremophiles 2008, 10, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Golyshina, O.V.; Yakimov, M.M.; Lunsdorf, H.; Ferrer, M.; Nimtz, M.; Timmis, K.N.; Wray, V.; Tindall, B.J.; Golyshin, P.N. Acidiplasma aeolicum gen. nov., sp. nov., a euryarchaeon of the family Ferroplasmaceae isolated from a hydrothermal pool and transfer of Ferroplasma cupricumulans to Acidiplasma cupricumulans comb. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2815–2823. [Google Scholar] [CrossRef] [PubMed]
- Schleper, C.; Pühler, G.; Klenk, H.-P.; Zillig, W. Picrophilus oshimae and Picrophilus torridus fam. nov., gen. nov., sp. nov., two species of hyperacidophilic, thermophilic, heterotrophic, aerobic archaea. Int. J. Syst. Bacteriol. 1996, 46, 814–816. [Google Scholar] [CrossRef]
- Plumb, J.J.; Haddad, C.M.; Gibson, J.A.E.; Franzmann, P.D. Acidianus sulfidivorans sp. nov., an extremely acidophilic, thermophilic archaeon isolated from a solfatara on Lihir Island, Papua New Guinea, and emendation of the genus description. Int. J. Syst. Evol. Microbiol. 2007, 57, 1418–1423. [Google Scholar] [CrossRef] [PubMed]
- Watling, H.R. Adaptability of biomining organisms in hydrometallurgical processes. In Biohydrometallurgical Processes: A Practical Approach; Santos Sobral, L.G., Monteiro de Oliveira, D., Gomes de Souza, C.E., Eds.; CETEM/MCTI: Rio de Janeiro, Brazil, 2011; pp. 39–70. [Google Scholar]
- Watkin, E.L.J.; Keeling, S.E.; Perrot, F.A.; Shiers, D.W.; Palmer, M.-L.; Watling, H.R. Metals tolerance in moderately thermophilic isolates from a spent copper sulfide heap, closely related to Acidithiobacillus caldus, Acidimicrobium ferrooxidans and Sulfobacillus thermosulfidooxidans. J. Ind. Microbiol. Biotechnol. 2009, 36, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, F.; Morin, D.; Ollivier, P. Dissolution of cobaltiferous pyrite by Thiobacillus ferrooxidans and Thiobacillus thiooxidans: Factors influencing bacterial leaching efficiency. J. Biotechnol. 1994, 32, 11–16. [Google Scholar] [CrossRef]
- D’Hughes, P.; Cezac, P.; Cabral, T.; Battaglia, F.; Truong-Meyer, X.M.; Morin, D. Bioleaching of a cobaltiferous pyrite: A continuous laboratory-scale study at high solids concentration. Min. Eng. 1997, 10, 507–527. [Google Scholar]
- Heinzle, T.; Miller, D.; Nagel, V. Results of an integrated pilot plant operation using the BioNIC® process to produce nickel metal. In Proceedings of the Biomine 99 and Water Management in Metallurgical Operations 99, Perth, Australia, 24–25 August 1999; AusIMM: Melbourne, Australia, 1999; pp. 16–25. [Google Scholar]
- Du Plessis, C.A.; Batty, J.D.; Dew, D.W. Commercial applications of thermophile bioleaching. In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer-Verlag: Berlin, Germany, 2007; pp. 57–80. [Google Scholar]
- Rea, S.M.; McSweeney, N.J.; Degens, B.P.; Morris, C.; Siebert, H.M.; Kaksonen, A.H. Salt-tolerant microorganisms potentially useful for bioleaching operations where fresh water is scarce. Min. Eng. 2015, 75, 126–132. [Google Scholar] [CrossRef]
- Harahuc, L.; Lizama, H.M.; Suzuki, I. Selective inhibition of the oxidation of ferrous iron or sulfur in Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 2000, 66, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Zammit, C.M.; Mangold, S.; rao Jonna, V.; Mutch, L.A.; Watling, H.R.; Dopson, M.; Watkin, E.L.J. Bioleaching in brackish waters—Effect of chloride ions on the acidophilic population and proteomes of model species. Appl. Microbiol. Biotechnol. 2012, 93, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Grail, B.M.; Bonnefoy, V. New insights into salt-tolerance in acidophilic iron-oxidising bacteria. Adv. Mater. Res. 2015, 1130, 3–6. [Google Scholar] [CrossRef]
- Watling, H.R.; Shiers, D.W.; Zhang, G.J. Microbial behaviour under conditions relevant to heap leaching: Studies using the sulfur-oxidising, moderate thermophile Acidithiobacillus caldus. Hydrometallurgy 2012, 127–128, 104–111. [Google Scholar] [CrossRef]
- Cardenas, J.P.; Ortiz, R.; Norris, P.R.; Watkin, E.; Holmes, D.S. Reclassification of ‘Thiobacillus prosperus’ Huber and Stetter 1989 as Acidihalobacter prosperus gen. nov., sp. nov., a member of the family Ectothiorhodospiraceae. Int. J. Syst. Evol. Microbiol. 2015, 65, 3641–3644. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.P.; Wood, A.P. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov., and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Brierley, J.A.; Kuhn, M.C. Fluoride toxicity in a chalcocite bioleach heap process. Hydrometallurgy 2010, 104, 410–413. [Google Scholar] [CrossRef]
- Murr, L.E.; Brierley, J.A. The use of large-scale test facilities in commercial leaching operations. In Metallurgical Applications of Bacterial Leaching and Related Microbiological Phenomena; Murr, L., Torma, A., Brierley, J., Eds.; Academic Press: New York, NY, USA, 1978; pp. 491–520. [Google Scholar]
- Soljanto, P.; Rehtijarvi, P.; Tuovinen, O.H. Ferrous iron oxidation by Thiobacillus ferrooxidans: Inhibition by finely ground particles. Geomicrobiol. J. 1980, 2, 1–12. [Google Scholar] [CrossRef]
- Suzuki, I.; Lee, D.; McKay, B.; Harahuc, L.; Oh, J.K. Effect of various anions, pH, and osmotic pressure on oxidation of elemental sulfur by Thiobacillus thiooxidans. Appl. Environ. Microbiol. 1999, 65, 5163–5168. [Google Scholar] [PubMed]
- Peng, Z.; Yu, R.; Qiu, G.; Qin, W.; Gu, G.; Wang, Q.; Li, Q.; Liu, X. Really active form of fluorine toxicity affecting Acidithiobacillus ferrooxidans activity in bioleaching uranium. Trans. Nonferr. Met. Soc. China 2013, 23, 812–817. [Google Scholar] [CrossRef]
- Ma, L.; Li, Q.; Xiao, Y.; Wang, Q.; Yin, H.; Liang, Y.; Qiu, G.; Liu, X. Comparative study of fluoride tolerance of five typical bioleaching microorganisms. Adv. Mater. Res. 2013, 825, 214–218. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Y.; Hu, K.; Hu, E. Experimental study on acclimatization and culture of acid-tolerant and fluoride-tolerant bacteria. Min. Res. Dev. 2005, 25, 35–36. [Google Scholar]
- Sundkvist, J.-E.; Sandström, Å.; Gunneriusson, L.; Lindstrom, E.B. Fluorine toxicity in bioleaching systems. In Proceedings of the 16th International Biohydrometallurgy Symposium, Cape Town, South Africa, 25–29 September 2005; Harrison, S.T.L., Rawlings, D.E., Petersen, J., Eds.; IBS: Cape Town, South Africa, 2005; pp. 19–28. [Google Scholar]
- Corbillon, M.S.; Olazabal, M.A.; Madariaga, J.M. Potentiometric study of aluminium fluoride complexation equilibria and definition of the thermodynamic model. J. Solut. Chem. 2008, 37, 567–579. [Google Scholar] [CrossRef]
- Radic, N.; Bralic, M. Aluminium fluoride complexation and its ecological importance in the aquatic environment. Sci. Tot. Environ. 1995, 172, 237–243. [Google Scholar] [CrossRef]
- Soli, A.L.; Byrne, R.H. The hydrolysis and fluoride complexation behaviour of Fe(III) at 25 °C and 0.68 molal ionic strength. J. Solut. Chem. 1996, 25, 773–785. [Google Scholar] [CrossRef]
- Sicupira, L.; Veloso, T.; Reis, F.; Leao, V. Assessing metal recovery from low-grade copper ores containing fluoride. Hydrometallurgy 2011, 109, 202–210. [Google Scholar] [CrossRef]
- Razzell, W.E.; Trussell, P.C. Isolation and properties of an iron-oxidizing Thiobacillus. J. Bacteriol. 1963, 85, 595–603. [Google Scholar] [PubMed]
- Harahuc, L.; Lizama, H.; Suzuki, I. Effect of anions on selective solubilisation of zinc and copper in bacterial leaching of sulfide ores. Biotechnol. Bioeng. 2000, 69, 196–203. [Google Scholar] [CrossRef]
- Shiers, D.W.; Ralph, D.E.; Watling, H.R. The effects of nitrate on substrate utilisation by some iron(II)- and sulfur-oxidising Bacteria and Archaea. Hydrometallurgy 2014, 150, 259–268. [Google Scholar] [CrossRef]
- Romero, J.; Yañez, C.; Vásquez, M.; Moore, E.R.B.; Espejo, R.T. Characterization and identification of an iron-oxidizing, Leptospirillum-like bacterium present in the high-sulfate leaching solution of a commercial bioleaching plant. Res. Microbiol. 2003, 154, 353–359. [Google Scholar] [CrossRef]
- Ruan, R.; Liu, X.; Zou, G.; Chen, J.; Wen, J.; Wang, D. Industrial practice of a distinct bioleaching system operated at low pH, high ferric concentration, elevated temperature and low redox potential for secondary copper sulfide. Hydrometallurgy 2011, 108, 130–135. [Google Scholar] [CrossRef]
- Shiers, D.W.; Blight, K.R.; Ralph, D.E. Sodium sulphate and sodium chloride effects on batch culture of iron oxidising bacteria. Hydrometallurgy 2005, 80, 75–82. [Google Scholar] [CrossRef]
- Watling, H.R.; Collinson, D.M.; Li, J.; Mutch, L.A.; Perrot, F.A.; Rea, S.M.; Reith, F.; Watkin, E.L.J. Bioleaching of a low-grade copper ore linking leach chemistry and microbiology. Min. Eng. 2014, 56, 35–44. [Google Scholar] [CrossRef]
- Cautivo, D.; Soto, P.; Zepeda, V.J.; Galleguillos, P.A.; Velasquez, A.; Pinilla, C.; Demergasso, C.S. Estimation of ionic load effect on the oxidizing activity of the microbial population in the heap bioleaching process at Escondida Mine. Adv. Mater. Res. 2013, 825, 219–222. [Google Scholar] [CrossRef]
- He, Z.; Xiao, S.; Xie, X.; Hu, Y. Microbial diversity in acid mineral bioleaching systems of Dongxiang copper mine and Yinshan lead-zinc mine. Extremophiles 2008, 12, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Wang, L.; Dong, H.; Zhang, H. Succession of acidophilic bacterial community during bio-oxidation of refractory gold-containing sulfides. Geomicrobiol. J. 2010, 27, 683–691. [Google Scholar] [CrossRef]
- Remonsellez, F.; Galleguillos, F.; Moreno-Paz, M.; Parro, V.; Acosta, M.; Demeragasso, C. Dynamic of active microorganisms inhabiting a bioleaching industrial heap of low-grade copper sulfide ore monitored by real-time PCR and olignucleotide prokaryotic acidophile microarray. Microbiol. Biotechnol. 2009, 2, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, B.; Wen, J.; Ruan, R. Leptospirillum forms a minor portion of the population in Zijinshan commercial non-aeration copper bioleaching heap identified by 16S rRNA clone libraries and real-time PCR. Hydrometallurgy 2010, 104, 399–403. [Google Scholar]
- Liu, X.; Wu, B.; Chen, B.; Wen, J.; Ruan, R.; Yao, G.; Wang, D. Bioleaching of chalcocite started at different pH: Response of the microbial community to environmental stress and leaching kinetics. Hydrometallurgy 2010, 103, 1–6. [Google Scholar]
- Wang, Y.; Su, L.; Zeng, W.; Wan, L.; Chen, Z.; Zhang, L.; Qiu, G.; Chen, X.; Zhou, H. Effect of pulp density on planktonic and attached community dynamics during bioleaching of chalcopyrite by a moderately thermophilic microbial culture under uncontrolled conditions. Min. Eng. 2014, 61, 66–72. [Google Scholar] [CrossRef]
- Demergasso, C.; Galleguillos, F.; Soto, P.; Serón, M.; Iturriaga, V. Microbial succession during heap bioleaching cycle of low grade copper sulfides. Does this knowledge mean a real input for industrial process design and control? Hydrometallurgy 2010, 104, 382–390. [Google Scholar] [CrossRef]
- Okibe, N.; Gericke, M.; Hallberg, K.B.; Johnson, D.B. Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred-tank bioleaching operation. Appl. Environ. Microbiol. 2003, 69, 1936–1943. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, Y.; Inoue, C.; Suto, K.; Chida, T. Inhibitory effect of high concentrations of ferric ions on the activity of Acidithiobacillus ferrooxidans. J. Biosci. Bioeng. 2003, 96, 375–379. [Google Scholar] [CrossRef]
- Cameron, R.A.; Yeung, C.W.; Greer, C.W.; Gould, W.D.; Mortazavi, S.; Bedard, P.L.; Morin, L.; Lortie, L.; Dinardo, O.; Kennedy, K.J. The bacterial community structure during bioleaching of a low-grade nickel sulphide ore in stirred tank reactors at different combinations of temperature and pH. Hydrometallurgy 2010, 104, 207–215. [Google Scholar] [CrossRef]
- Mutch, L.A.; Watling, H.R.; Watkin, E.L.J. Microbial population dynamics of inoculated low-grade chalcopyrite bioleaching columns. Hydrometallurgy 2010, 104, 391–398. [Google Scholar] [CrossRef]
- Yu, R.; Shi, L.; Gu, G.; Zhou, D.; You, L.; Chen, M.; Qiu, G.; Zeng, W. The shift in microbial community under the adjustment of initial and processing pH during bioleaching of chalcopyrite concentrate by moderate thermophiles. Bioresour. Technol. 2014, 162, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Segerer, A.H.; Burggraf, S.; Fiala, G.; Huber, G.; Huber, R.; Pley, U.; Stetter, K.O. Life in hot springs and hydrothermal vents. Orig. Life Evolut. Biosph. 1993, 23, 77–90. [Google Scholar] [CrossRef]
- Childs, A.M.; Mountain, B.W.; O’Toole, R.; Stott, M.B. Relating microbial community and physicochemical parameters of a hot spring: Champagne Pool, Wai-o-tapu, New Zealand. Geomicrobiol. J. 2008, 25, 441–453. [Google Scholar] [CrossRef]
- Stott, M.B.; Sutton, D.C.; Watling, H.R.; Franzmann, P.D. Comparative leaching of chalcopyrite by selected acidophilic Bacteria and Archaea. Geomicrobiol. J. 2003, 20, 215–230. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, J.; Wei, M.; Ding, J.; Zhou, H. Bioleaching of chalcopyrite with Acidianus manzaensis YN25 under contact and non-contact conditions. Trans. Nonferr. Met. Soc. China 2010, 20, 1981–1986. [Google Scholar] [CrossRef]
- Zhu, W.; Xia, J.; Yang, Y.; Nie, Z.; Peng, A.; Liu, H.; Qiu, G. Thermophilic archaeal community succession and function change associated with the leaching rate in bioleaching of chalcopyrite. Bioresour. Technol. 2013, 133, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Shiers, D.W.; Ralph, D.E.; Bryan, C.G.; Watling, H.R. Substrate utilisation by Acidianus brierleyi, Metallospaera hakonensis and Sulfolobus metallicus in mixed ferrous ion and tetrathionate growth media. Min. Eng. 2013, 48, 86–93. [Google Scholar] [CrossRef]
- Demergasso, C.; Galleguillos, P.; Escudero, L.; Zepeda, V.; Castillo, D.; Casamayor, E. Molecular characterization of microbial populations in a low grade copper ore bioleaching test heap. Hydrometallurgy 2005, 80, 241–253. [Google Scholar] [CrossRef]
- Fuchs, T.; Huber, H.; Teiner, K.; Burggraf, S.; Stetter, K.O. Metallosphaera prunae, sp. nov., a novel metal-mobilizing, thermoacidophilic Archaeum isolated from a uranium mine in Germany. Syst. Appl. Microbiol. 1995, 18, 560–566. [Google Scholar] [CrossRef]
- Xiao, S.; Xie, X.; Liu, J.; He, Z.; Hu, Y. Composition and structures of archaeal communities in acid mineral bioleaching system of Dongxiang copper mine and Yinshan lead-Zinc Mine, China. Curr. Microbiol. 2008, 57, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Xiao, S.; He, Z.; Liu, J.; Qiu, G. Microbial populations in acid mineral bioleaching systems of Tong Shankou copper mine, China. J. Appl. Microbiol. 2007, 103, 1227–1238. [Google Scholar] [CrossRef] [PubMed]
- Halinen, A.-K.; Beecroft, N.J.; Määttä, K.; Nurmi, P.; Laukkanen, K.; Kaksonen, A.H.; Riekkola-Vanhanen, M.; Puhakka, J.A. Microbial community dynamics during a demonstration-scale bioheap. Hydrometallurgy 2012, 125–126, 34–41. [Google Scholar] [CrossRef]
- Selkov, E.; Overbeek, R.; Kogan, Y.; Chu, L.; Vonstein, V.; Holmes, D.S.; Silver, S.; Haselkorn, R.; Fonstein, M. Functional analysis of gapped microbial genomes: Amino acid metabolism of Thiobacillus ferrooxidans. PNAS 2000, 97, 3509–3514. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, J.P.; Valdes, J.; Quatrini, R.; Duarte, F.; Holmes, D.S. Lessons from the genomes of extremely acidophilic bacteria and archaea with special emphasis on bioleaching microorganisms. Appl. Microbiol. Biotechnol. 2010, 88, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Justice, N.B.; Norman, A.; Brown, C.T.; Singh, A.; Thomas, B.C.; Banfield, J.F. Comparison of environmental and isolate Sulfobacillus genomes reveals diverse carbon, sulfur, nitrogen, and hydrogen metabolisms. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-L.; Jiang, C.-Y.; Liu, S.-J. Insight into the sulfur metabolism by thermoacidophilic archaeon Metallosphaera cuprina with genomic, proteomic and biochemical tools. Adv. Mater. Res. 2015, 1130, 145–148. [Google Scholar] [CrossRef]
- Ullrich, S.R.; Poehlein, A.; Daniel, R.; Tischler, J.S.; Vogel, S.; Schlömann, M.; Mühling, M. Comparative genomics underlines the functional and taxonomic diversity of novel “Ferrovum” related iron oxidizing bacteria. Adv. Mater. Res. 2015, 1130, 15–18. [Google Scholar] [CrossRef]
- Goltsman, D.S.; Denef, V.J.; Singer, S.W.; VerBerkmoes, N.C.; Lefsrud, M.; Mueller, R.S.; Dick, G.J.; Sun, C.L.; Wheeler, K.E.; Zemla, A.; et al. Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing “Leptospirillum rubarum” (Group II) and “Leptospirillum ferrodiazotrophum” (Group III) bacteria in acid mine drainage biofilms. Appl. Environ. Microbiol. 2009, 75, 4599–4615. [Google Scholar] [CrossRef] [PubMed]
- Hemme, C.L.; Deng, Y.; Gentry, T.J.; Fields, M.W.; Wu, L.; Barua, S.; Barry, K.; Tringe, S.G.; Watson, D.B.; He, Z.; et al. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 2010, 4, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Méndez-García, C.; Peláez, A.I.; Mesa, V.; Sánchez, J.; Golyshina, O.V.; Ferrer, M. Microbial diversity and metabolic networks in acid mine drainage habitats. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Chen, L.; Huang, L.; Méndez-Garcia, C.; Kuang, J.; Hua, Z.; Liu, J.; Shu, W. Microbial communities, processes and functions in acid mine drainage systems. Curr. Opin. Biotechnol. 2016, 38, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Banfield, J.F.; Verboekmoes, N.C.; Hettich, R.L.; Thelen, M.P. Proteogenomic approaches for the molecular characterization of natural microbial communities. OMICS 2005, 9, 301–333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Niu, J.; Liang, Y.; Liu, X.; Yin, H. Metagenome-scale analysis yields insights into the structure and function of microbial communities in a copper bioleaching heap. BMC Genet. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, L.; Chi, A.; Beard, S.; Orell, A.; Guiliani, N.; Shabanowitz, J.; Hunt, D.F.; Jerez, C.A. Genomics, metagenomics and proteomics in biomining microorganisms. Biotechnol. Adv. 2006, 24, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Jerez, C.A. Metal extraction and biomining. In The Desk Encyclopedia of Microbiology; Schaechter, M., Ed.; Elsevier: Oxford, UK, 2009; pp. 762–775. [Google Scholar]
- Stoughton, R.B. Applications of DNA microarrays in biology. Ann. Rev. Biochem. 2005, 74, 53–82. [Google Scholar] [CrossRef] [PubMed]
- Seidel, M.; Niessner, R. Automated analytical microarrays: A critical review. Anal. Bioanal. Chem. 2008, 391, 1521–1544. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Deng, Y.; Zhou, Y. Development of functional gene arrays for microbial community analysis. Curr. Opin. Biotechnol. 2012, 23, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Cao, L.; Qiu, G.; Wang, D.; Kellogg, L.; Zhou, J.; Dai, Z.; Liu, X. Development and evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in acid mine drainages and bioleaching systems. J. Microbiol. Methods 2007, 70, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Tu, Q.; Yu, H.; He, Z.; Deng, Y.; Wu, L.; Van Nostrand, J.D.; Zhou, A.; Voordeckers, J.; Lee, Y.-J.; Qin, Y.; et al. GeoChip 4: A functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol. Ecol. Resour. 2014, 14, 914–928. [Google Scholar] [CrossRef] [PubMed]
- Quatrini, R.; Appia-Ayme, C.; Denis, Y.; Ratouchniak, J.; Veloso, F.; Valdes, J.; Lefimil, C.; Silver, S.; Roberto, F.; Orellana, O.; et al. Insights into the iron and sulfur energetic metabolism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy 2006, 83, 263–272. [Google Scholar] [CrossRef]
- Quatrini, R.; Appia-Ayme, C.; Denis, Y.; Jedlicki, E.; Holmes, D.S.; Bonnefoy, V. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genom. 2009, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Valdés, J.; Pedroso, I.; Quatrini, R.; Holmes, D.S. Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: Insights into their metabolism and ecophysiology. Hydrometallurgy 2008, 94, 180–184. [Google Scholar] [CrossRef]
- Auernik, K.S.; Kelly, R.M. Physiological versatility of the extremely thermoacidophilic archaeon Metallosphaera sedula supported by transcriptomic analysis of heterotrophic, autotrophic and mixotrophic growth. Appl. Environ. Microbiol. 2010, 76, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Auernik, K.S.; Maezato, Y.; Blum, P.H.; Kelly, R.M. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 2008, 74, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Haward, R.E.; Derisi, J.L.; Alfadhli, S.; Kaslow, D.C.; Brown, P.O.; Rathod, P.K. Shotgun DNA microarrays and stage-specific gene expression in Plasmodium falciparum malaria. Mol. Microbiol. 2000, 35, 6–14. [Google Scholar] [CrossRef]
- Parro, V.; Moreno-Paz, M. Gene function analysis in environmental isolates: The nif regulon of the strict iron oxidizing bacterium Leptospirillum ferrooxidans. PNAS 2003, 100, 7883–7888. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.W.; Lo, I.; Baker, B.J.; Allen, E.E.; Hugenholtz, P.; Banfield, J.F. Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. from an acidophilic microbial community. Appl. Environ. Microbiol. 2005, 71, 6319–6324. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Paz, M.; Gómez, M.J.; Arcas, A.; Parro, V. Environmental transcriptome analysis reveals physiological differences between biofilm and planktonic modes of life of the iron oxidizing bacteria Leptospirillum spp. in their natural microbial community. BMC Genomics 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Vera, M.; Krok, B.; Bellenberg, S.; Sand, W.; Poetsch, A. Shotgun proteomics study of early biofilm formation process of Acidithiobacillus ferrooxidans ATCC 23270 on pyrite. Proteomics 2013, 13, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Laska, S.; Lottspeich, F.; Kletzin, A. Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology 2003, 149, 2357–2371. [Google Scholar] [CrossRef] [PubMed]
- Golyshina, O.V. Environmental, biogeographic, and biochemical patterns of archaea of the family Ferroplasmaceae. Appl. Environ. Microbiol. 2011, 77, 5071–5078. [Google Scholar] [CrossRef] [PubMed]
- Brodie, E.L.; DeSantis, T.Z.; Joyner, D.C.; Baek, S.M.; Larsen, J.T.; Andersen, G.L.; Hazen, T.C.; Richardson, P.M.; Herman, D.J.; Tokunaga, T.K.; et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 2006, 72, 6288–6298. [Google Scholar] [CrossRef] [PubMed]
- Garrido, P.; Gonzalez-Toril, E.; Garcia-Moyano, A.; Moreno-Paz, M.; Amils, R.; Parro, V. An oligonucleotide prokaryotic acidophile microarray: Its validation and its use to monitor seasonal variations in extreme acidic environments with total environmental RNA. Environ. Microbiol. 2008, 10, 836–850. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Yin, H.; Hu, Q.; Liu, X.; Qiu, G. Microarray-based characterization of microbial community functional structure and heterogeneity associated with acid mine drainages. Adv. Mater. Res. 2015, 1130, 40–44. [Google Scholar] [CrossRef]
- Acosta, M.; Galleguillos, P.; Ghorbani, Y.; Tapia, P.; Contador, Y.; Velásquez, A.; Espoz, C.; Pinilla, C.; Demergasso, C. Variation in microbial community from predominantly mesophilic to thermotolerant and moderately thermophilic species in an industrial copper heap bioleaching operation. Hydrometallurgy 2014, 150, 281–289. [Google Scholar] [CrossRef]
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).