Abstract
Centrioles make up the centrosome and basal bodies in animals and as such play important roles in cell division, signalling and motility. They possess characteristic 9-fold radial symmetry strongly influenced by the protein SAS-6. SAS-6 is essential for canonical centriole assembly as it forms the central core of the organelle, which is then surrounded by microtubules. SAS-6 self-assembles into an oligomer with elongated spokes that emanate towards the outer microtubule wall; in this manner, the symmetry of the SAS-6 oligomer influences centriole architecture and symmetry. Here, we summarise the form and symmetry of SAS-6 oligomers inferred from crystal structures and directly observed in vitro. We discuss how the strict 9-fold symmetry of centrioles may emerge, and how different forms of SAS-6 oligomers may be accommodated in the organelle architecture.
1. Introduction
Centrioles are conserved eukaryotic organelles essential for correct cellular behaviour. In animals, a pair of centrioles together with the pericentriolar material comprise the centrosome, which is the main microtubule organising centre of cells and directs the formation of the mitotic spindle during division [1,2,3], thereby ensuring correct chromosome segregation. Centrioles are also essential for organising cilia and flagella at the surface of cells. As such, they play a crucial role in cellular motility, for example of sperm, cell signalling via the primary cilium, and in the interaction of cells with their surroundings, as seen in tracheal lining cells that sweep mucus and dirt out of the respiratory system. Thus, aberrations in centriole structure or numbers are linked to disease conditions such as ciliopathies, male sterility, ectopic pregnancies, primary microcephaly and possibly cancer [4,5,6,7,8,9,10].
Centrioles are cylinders characterised by 9-fold radial symmetry in the vast majority of organisms [11,12,13] that are approximately 100–250 nm in diameter and 150–400 nm in length, depending on species and cell type. In centrioles, single, double or triple microtubule ‘blades’ form the outer wall of the cylinder and they are arranged around a central scaffold, termed the ‘cartwheel’ [14]. The cartwheel typically comprises a stack of 9-fold symmetric rings consisting of a ‘central hub’ with ‘spokes’ emanating radially. These spokes interact with microtubules at the outer centriole wall via a structure known as the ‘pinhead’, thereby linking the cartwheel radial symmetry to that of the entire organelle. Thus, cartwheels provide an essential building block for centriole assembly that influences organelle symmetry.
Genetic and functional studies in Caenorhabditis elegans aiming to isolate spindle assembly-defective mutants originally identified five proteins, SAS-6, SAS-4, SAS-5, SPD-2 and ZYG-1, essential for centriole formation [15,16,17,18,19,20,21,22]; relatives of these proteins were quickly located across eukaryotic evolution [23,24]. SAS-6 was perceived as a key protein for centriole symmetry, as immunolocalisation electron microscopy and sub-diffraction fluorescence microscopy studies placed this protein in cartwheels [25,26,27]. X-ray crystallography studies showed that SAS-6 comprises a globular N-terminal domain followed by a long coiled coil (Figure 1) and a C-terminal disordered region. SAS-6 strongly dimerises in vitro and in vivo via its coiled-coil domain, and protein dimers further interact via their N-terminal domains to form oligomers with 9-fold radial symmetry on average [28,29,30,31]. The SAS-6 N-terminal domain bears strong structural resemblance to similar domains of XRCC4 and XLF (Figure 2), proteins involved in DNA repair and non-homologous end joining. Further, similar to SAS-6, XRCC4 forms dimers via a coiled-coil domain, and it hetero-oligomerises with XLF via the globular N-terminal domains to form a spiral [32,33,34,35,36].
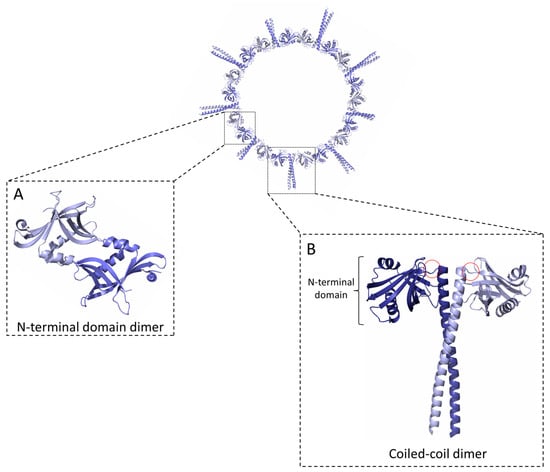
Figure 1.
SAS-6 oligomerisation. A SAS-6 ring containing nine protein dimers is shown. SAS-6 self-assembles into rings via two dimerisation interfaces mediated by (A) an N-terminal globular domain (NN) and (B) a coiled coil (CC) 35 to 50 nm long, depending on species. The full extent of the SAS-6 coiled coil and a disordered C-terminal protein region are not shown. Hinge regions connecting the SAS-6 N-terminal domains with the coiled coil are indicated by red circles in (B).
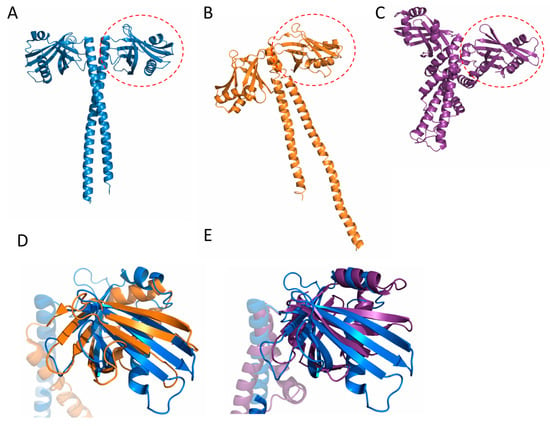
Figure 2.
Comparison of SAS-6 to structurally similar proteins. (A) SAS-6; (B) XRCC4; and (C) XLF proteins display structurally related N-terminal globular domains and helical dimerisation regions, which comprise coiled coils in SAS-6 and XRCC4. Superposition of the N terminal domain of SAS-6 with XLF (D) and XRCC4 (E) (a single chain is shown for clarity).
Most SAS-6 oligomers resolved to date are highly reminiscent of cartwheel rings, with a central hub derived from the N-terminal globular domains and the SAS-6 coiled coils comprising the cartwheel spokes [28,29,31]. Furthermore, disruption of SAS-6 oligomerisation abrogates formation of the central centriole scaffold and strongly hinders organelle assembly [29,31]. Thus, SAS-6 oligomers are thought to comprise the centriole cartwheels, thereby influencing the radial symmetry of this organelle [26,37].
Although the tertiary structure of SAS-6 is highly conserved amongst homologues, there exists both subtle and large variations in oligomer morphologies. For example, Caenorhabditis elegans SAS-6 (CeSAS-6) self-assembles into a spiral [30] rather than the planar rings observed in the Chlamydomonas reinhardtii (CrSAS-6; [31]), Danio rerio (DrSAS-6; [29]) and Leishmania major (LmSAS-6; [28]) variants of the same protein. Furthermore, there is substantial heterogeneity in the intrinsic symmetry of SAS-6 oligomers, both inferred by X-ray crystallography and directly visualised by microscopy. Here, we summarise the structural differences seen across SAS-6 variants, with emphasis on the symmetry of protein oligomers, and discuss how SAS-6 variants may support the formation of a highly conserved organelle architecture with strict symmetry requirements.
2. SAS-6 Rings
Initial insight into the symmetry of SAS-6 oligomers was obtained through in silico models, where oligomer symmetry was inferred by combining separate crystal structures of SAS-6 constructs comprising dimers of the N-terminal globular domain (NN interface) or coiled-coil dimers (CC interface). Such studies suggested that SAS-6 oligomers feature radial symmetries close to the canonical centriolar 9-fold arrangement, although significant symmetry variations were observed that could be attributed to artefacts of crystallisation or protein conformational heterogeneity. For example, there exist three crystallographic models of the CrSAS-6 NN interface and one of the CrSAS-6 CC interface [31]; thus, three different oligomer variants can be built depending on the NN interface model employed resulting in 9.6- to 10.1-fold radial symmetry. Similarly, when DrSAS-6 oligomers are built from available crystal structures a variation in oligomer architecture is observed, with DrSAS-6 forming either an 8-fold symmetric shallow spiral or an 8-fold symmetric ring [29], while LmSAS-6 inferred oligomer symmetry ranges from 7.8- to 11.3-fold [28]. The variation in oligomer symmetry from different SAS-6 crystallographic models is summarised in Figure 3A.
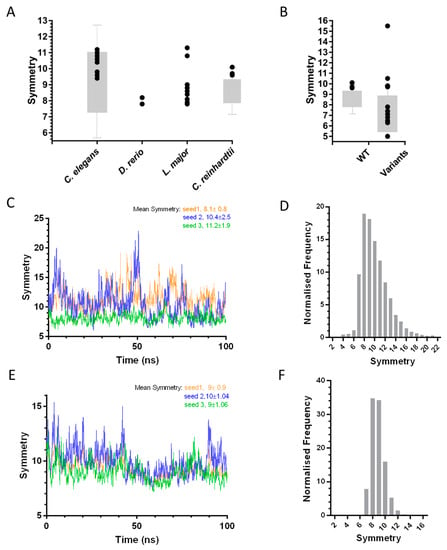
Figure 3.
Analysis of variation in SAS-6 oligomer symmetry. (A) Variation of symmetry in SAS-6 oligomers [28,29,30,31] inferred from crystal structures (black circles) or directly visualised by electron microscopy (grey bars). (B) Variation in oligomer symmetry of wild type [37] and engineered CrSAS-6 [31] (denoted as in panel A). (C,E) Fluctuations in LmSAS-6 (C) and CeSAS-6 (E) oligomer symmetry over a 100-ns molecular dynamics (MD) simulation. Triplicate simulations are shown in blue, green and orange. (D,F) Frequency of LmSAS-6 (D) and CeSAS-6 (F) oligomer symmetry derived from 10,000 conformations where both NN and CC interfaces are varied.
Heterogeneity in symmetry was also observed in in vitro experiments where SAS-6 oligomers were reconstituted and their symmetry directly examined. For example, rotary metal shadowing electron microscopy (EM) of CrSAS-6 oligomers revealed the formation of rings with variable diameters corresponding to 7.8 to 9.4-fold radial symmetry [31], a conclusion also supported by atomic force microscopy (AFM) studies [37,38] of the same oligomers (Figure 4A). These experiments argued that symmetry differences inferred from the SAS-6 crystallographic structures did not represent artefacts, but rather genuine protein flexibility at the SAS-6 NN and CC interaction interfaces which drive oligomerisation. Atomistic molecular dynamics (MD) simulations of CrSAS-6 performed previously [37], and similar work on LmSAS-6 presented here (described in Appendix A), showcase this conformational flexibility. In this type of analysis, SAS-6 oligomers are built using a fixed model of one interface from crystallography and variable models of the second interface derived from snapshots of the interface MD trajectory. The resulting SAS-6 oligomers show substantial fluctuations in symmetry; for example, CrSAS-6 oligomers built using a simulated NN interface range from 7- to 13-fold symmetry [37], while we noted 6- to 23-fold symmetries for LmSAS-6 oligomers built from a simulated CC interface (Figure 3C). Furthermore, we derived 10,000 distinct oligomer models of LmSAS-6 by combining independent MD simulations of the NN and CC dimerization interfaces (Figure 3D), and noted that a substantial fraction (24.5 ± 2.7%) of LmSAS-6 oligomers acquire 12-fold or larger symmetries, with a mean oligomer symmetry at approximately 10-fold.
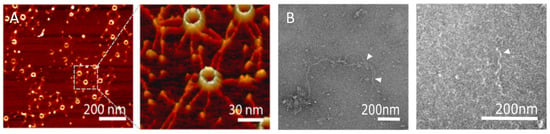
Figure 4.
In vitro visualisation of SAS-6 oligomers. (A) Atomic force microscopy images of CrSAS-6 oligomers forming rings. Left panel presents an overview of the sample; right panel magnifies the indicated region. Reprinted with permission from [38]. Copyright 2014 American Chemical Society. (B) Negative stain transmission electron microscopy images of CeSAS-6 protein spirals [30]. Left panel shows long CeSAS-6 double spirals; white arrowheads denote points at which individual strands of the double spirals separate or come together. Right panel shows a magnified view of isolated CeSAS-6 spiral strands.
3. SAS-6 Spirals
Although C. elegans centrioles show strong structural and functional conservation compared to those of other species [28,29,31], crystallographic and electron microscopy studies of CeSAS-6 suggested that this protein variant forms spiral oligomers rather than rings. Interestingly, despite the limited sequence similarity between CeSAS-6 and its homologues [24,39,40], both the overall SAS-6 architecture of a globular N-terminal domain succeeded by a coiled coil and the tertiary structure of the N-terminal domain itself were highly similar. Rather, the discrepancy of SAS-6 oligomeric architecture between C. elegans and other organisms appears to originate at the protein region connecting the N-terminal domain and the coiled coil, known as the hinge (Figure 1 and Figure 5). There, in ring-forming SAS-6, invariably exists a small amino acid residue, typically glycine or alanine. In contrast, in the spiral forming CeSAS-6 this position is occupied by a valine (Figure 5); a valine is also found at the hinge position of XRCC4, which forms spirals similar to CeSAS-6 [32,34,35,36]. The steric effect imposed by the valine in CeSAS-6 moves the coiled-coil helix relative to the N-terminal domain by approximately 40° compared to other SAS-6 variants [30], thereby driving successive CeSAS-6 dimers out of the horizontal plane and into a steep spiral of ~30 nm pitch. Such CeSAS-6 spirals were observed to intertwine in vitro (Figure 4B); thus, it was proposed that in C. elegans centrioles two CeSAS-6 spirals may form a central scaffold analogous to the cartwheel [30].
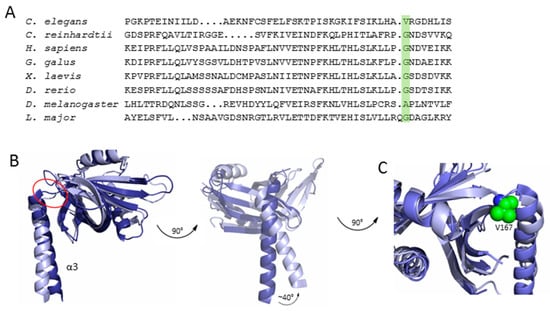
Figure 5.
Comparison of SAS-6 variants. (A) Multiple sequence alignment of SAS-6 variants with the hinge residue highlighted in green. (B) Overlay of LmSAS-6 (dark blue) and CeSAS-6 (light blue) with the hinge indicated by the red circle. A zoomed-in view of the hinge region is shown in panel (C), with the hinge valine of CeSAS-6 shown in the green sphere representation.
Similar to SAS-6 from other species, oligomers of CeSAS-6 inferred from crystallographic structures of the NN and CC interfaces show substantial variation in symmetry ranging from approximately 9- to 11-fold per turn of an intertwined spiral (Figure 3A), thus suggesting the presence of conformational flexibility in the NN and CC interfaces. Indeed, direct visualization of CeSAS-6 spiral oligomers by negative stain EM demonstrated an even broader symmetry distribution from 7- to 11-fold; it is therefore possible that the structural variation seen in crystallographic structures underestimates the true degree of interface flexibility. MD simulations of the CeSAS-6 CC interface resulted in spiral SAS-6 oligomers of 7- to 16-fold symmetry (Figure 3E; [37]); more strikingly, when combining simulations of the NN and CC interfaces, CeSAS-6 oligomer symmetries ranged from 6.4- to 16-fold, with a mean symmetry of 8.8-fold (Figure 3F). This indicates that, despite the steric clash at the hinge region imposed on CeSAS-6, the protein remains flexile and able to accommodate a range of oligomer symmetries.
4. Discussion
Crystallographic, biophysical and computational analysis of the SAS-6 NN and CC interfaces suggest that these exhibit substantial flexibility, which in turn leads to heterogeneity of SAS-6 oligomers. Thus, SAS-6, if acting alone, would have given rise to highly variable cartwheels and centrioles; however, centriole symmetry is strictly controlled in vivo which suggests that additional factors or proteins beyond SAS-6 influence the radial organisation of this organelle. This conclusion was reinforced by a very recent study where CrSAS-6 was subjected to rational engineering [37]. There, amino acid variants of the NN interface produced CrSAS-6 oligomers with symmetries ranging from 5- to 10-fold as judged by crystallography, negative stain EM and AFM (Figure 3B). However, centrioles in C. reinhardtii cells incorporating these CrSAS-6 variants almost always retained 9-fold symmetry; the sole exception was a CrSAS-6 variant giving rise to 5-fold symmetric oligomers in vitro that produced a 2:1 ratio of 9- and 8-fold symmetric centrioles in vivo. Thus, it is clear that while SAS-6 plays a central role in determining centriole symmetry, the organelle architecture must also be dependent on other factors. We note, for example, that null mutants of C. reinhardtii bld10, a gene orthologous to human cep135, generate centrioles with variable numbers of microtubule blades, indicating that Bld10 protein may also contribute to centriolar symmetry [41].
How may SAS-6, Bld10/Cep135 and microtubules work together to determine centriole symmetry? A likely relevant observation is that cartwheel diameter, primarily determined by the length of the SAS-6 coiled coil plus Bld10/Cep135, instructs the diameter and hence the circumference of centrioles. This, in turn, governs the number of microtubule blades that can be accommodated around the perimeter of the centriole central core. On the other hand, the size of microtubule blades and the geometry of the linkers connecting them also define a preferred circumference for centrioles. Thus, strict organelle 9-fold symmetry may emerge as a result of coinciding but independent weak circumference preferences derived by SAS-6 and Bld10/Cep135, and microtubules at the centriole outer wall. In this manner, different SAS-6 and Bld10/Cep135 variants in organisms, or expression of different protein isoforms in different cell types, could lead to different cartwheel diameters and hence shift organelle symmetry towards different modal points. Although speculative, such molecular processes may account for observations in Acerentomon centrioles, which possess 9-fold symmetry in somatic cells but 14-fold symmetry in spermatocytes with a corresponding enlargement of diameter [42]. Alternatively, different SAS-6 and Bld10/Cep135 isoforms could be accommodated in centrioles without varying symmetry by changing the size of microtubule blades at the outer organelle wall in a compensatory manner. We note, for example, that D. melanogaster germ line centrioles possess a triplet of microtubules in each organelle blade, while centrioles in somatic cells have microtubule doublets [43].
Further molecular mechanisms may of course influence centriole symmetry; for example, work in human cells has suggested that pre-existing centrioles may template newly assembled organelles through possible interactions of new SAS-6 cartwheels with the mother centriole lumen [44,45]. However, it is worth noting that the lumen of human centrioles becomes vacant, and hence available to template new cartwheels, only due to SAS-6 degradation during the G1 phase [1]. Other organisms, such as C. reinhardtii and C. elegans, do not degrade SAS-6 in the same manner; hence, template-based assembly of new SAS-6 cartwheels cannot account for strict centriole symmetry in these species.
Cryo-electron tomography of Trichonympha sp. [46,47] and C. reinhardtii [48] centrioles revealed that the spokes of successive SAS-6 rings merge as they radiate outwards, leading to stacking of SAS-6 rings with a periodicity of approximately 8.5 nm at the central hub and 17 nm at the end of spokes (Figure 6A). SAS-6 stacking periodicity aligns with the axial periodicity of microtubules at the outer centriole wall. C. elegans centrioles, in contrast, likely use a different mechanism to reach the same periodicity. In EM of fixed C. elegans embryos an electron lucent centriole centre was observed with no evidence of aligned, or merged, cartwheel spokes [49,50], which was interpreted as possible evidence for a centriole cartwheel formed by CeSAS-6 spirals rather than stacked rings. Reconstructions of such CeSAS-6 spirals from the crystallographic structures show successive spokes with 16 nm axial periodicity, thereby aligning with microtubules, but offset by a ~20° angle [30]. In this manner, CeSAS-6 spokes do not align to give rise to strong electron scattering (Figure 6B). Thus, we surmise that both spiral and ring SAS-6 arrangements may be present in centrioles, as they both provide contact points with the microtubule wall at equivalent spacing and hence can be accommodated with no additional alterations of the centriole architecture.
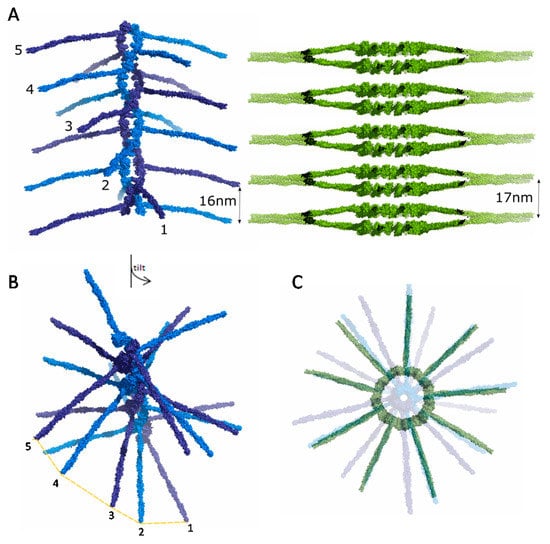
Figure 6.
SAS-6 oligomer structural comparison. (A) A CeSAS-6 spiral oligomer is shown in blue alongside a stack of LmSAS-6 cartwheels in green. The distance between equivalent spokes is indicated; (B) A tilted view of a CeSAS-6 spiral as in panel A with consecutive spoke ends indicated; the rotational offset between spokes is evident; (C) Top view of panel A with a CeSAS-6 spiral and LmSAS-6 cartwheel stack overlaid, showing the spoke distribution from the alternative SAS-6 oligomeric structures.
In summary, the conformational flexibility observed in SAS-6 oligomers suggests it is unlikely this protein alone dictates centriole symmetry. Rather, SAS-6 and other protein factors, including Bld10/Cep135 and microtubules, collaborate to strictly define the radial appearance of this organelle. In contrast, different SAS-6 oligomeric architectures, spirals and rings, can be accommodated equally well within a cylindrical microtubule wall, and thus may be both present in centrioles from different species.
Acknowledgments
This work was supported by the Medical Research Council project grant MR/N009274/1 to IV. Costs related to open access publishing were provided by the RCUK Open Access Block Grant to Oxford.
Author Contributions
Jodie E. Ford analysed literature and performed MD simulations, Phillip J. Stansfeld supervised the MD simulations. Jodie E. Ford and Ioannis Vakonakis wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
SAS-6 dimer loops were modelled using modeller v9.12 and the resulting models used to initiate 100-ns long MD simulations in simple point charge (SPC) water using the OPLS-AA/L forcefield at 310 K and Na and Cl ions added to neutalise charge. Pressure was maintained using the Parrinello-Rahman barostat [51] and temperature was maintained using the V-rescale thermostat [52]. Simulations were run and analysed in GROMACS v4.6 [53] with a 2-fs timestep and frames were recorded every 10 ps. Trajectories were visualised in PyMOL v1.7.x [54] and Chimera v1.10.2 [55]. Protein data bank (PDB) codes used in MD simulations: 4CKN, 4CKM [28], 4GFC, 4G79 [30].
References
- Conduit, P.T.; Wainman, A.; Raff, J.W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Bornens, M. The centrosome in cells and organisms. Science 2012, 335, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Arquint, C.; Gabryjonczyk, A.M.; Nigg, E.A. Centrosomes as signalling centres. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt-Dias, M.; Hildebrandt, F.; Pellman, D.; Woods, G.; Godinho, S.A. Centrosomes and cilia in human disease. Trends Genet. 2011, 27, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Gönczy, P. Centrosomes and cancer: Revisiting a long-standing relationship. Nat. Rev. Cancer 2015, 15, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A.; Raff, J.W. Centrioles, centrosomes, and cilia in health and disease. Cell 2009, 139, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Chavali, P.L.; Putz, M.; Gergely, F. Small organelle, big responsibility: The role of centrosomes in development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Thornton, G.K.; Woods, C.G. Primary microcephaly: Do all roads lead to Rome? Trends Genet. 2009, 25, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Venghateri, J.B.; Jindal, B.; Panda, D. The centrosome: A prospective entrant in cancer therapy. Expert Opin. Ther. Targets 2015, 19, 957–972. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.S.; Bakker, B.; Boeckx, B.; Moyett, J.; Lu, J.; Vitre, B.; Spierings, D.C.; Lansdorp, P.M.; Cleveland, D.W.; Lambrechts, D.; et al. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell 2017, 40, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Winey, M.; O’Toole, E. Centriole structure. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Jana, S.C.; Marteil, G.; Bettencourt-Dias, M. Mapping molecules to structure: Unveiling secrets of centriole and cilia assembly with near-atomic resolution. Curr. Opin. Cell Biol. 2014, 26, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Gönczy, P. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 2012, 13, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Hirono, M. Cartwheel assembly. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Leidel, S.; Delattre, M.; Cerutti, L.; Baumer, K.; Gönczy, P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005, 7, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, L.; Ozlu, N.; Hannak, E.; Cowan, C.; Habermann, B.; Ruer, M.; Muller-Reichert, T.; Hyman, A.A. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 2004, 14, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Kemp, C.A.; Kopish, K.R.; Zipperlen, P.; Ahringer, J.; O’Connell, K.F. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 2004, 6, 511–523. [Google Scholar] [CrossRef]
- Delattre, M.; Leidel, S.; Wani, K.; Baumer, K.; Bamat, J.; Schnabel, H.; Feichtinger, R.; Schnabel, R.; Gönczy, P. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 2004, 6, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Leidel, S.; Gönczy, P. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 2003, 4, 431–439. [Google Scholar] [CrossRef]
- Kirkham, M.; Muller-Reichert, T.; Oegema, K.; Grill, S.; Hyman, A.A. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 2003, 112, 575–587. [Google Scholar] [CrossRef]
- Dammermann, A.; Muller-Reichert, T.; Pelletier, L.; Habermann, B.; Desai, A.; Oegema, K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 2004, 7, 815–829. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, K.F.; Caron, C.; Kopish, K.R.; Hurd, D.D.; Kemphues, K.J.; Li, Y.; White, J.G. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 2001, 105, 547–558. [Google Scholar] [CrossRef]
- Firat-Karalar, E.N.; Stearns, T. The centriole duplication cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef]
- Azimzadeh, J. Exploring the evolutionary history of centrosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Mennella, V.; Keszthelyi, B.; McDonald, K.L.; Chhun, B.; Kan, F.; Rogers, G.C.; Huang, B.; Agard, D.A. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 2012, 14, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Hiraki, M.; Kamiya, R.; Hirono, M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 2007, 17, 2169–2174. [Google Scholar] [CrossRef] [PubMed]
- Keller, D.; Orpinell, M.; Olivier, N.; Wachsmuth, M.; Mahen, R.; Wyss, R.; Hachet, V.; Ellenberg, J.; Manley, S.; Gönczy, P.; et al. Mechanisms of hssas-6 assembly promoting centriole formation in human cells. J. Cell Biol. 2014, 204, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Van Breugel, M.; Wilcken, R.; McLaughlin, S.H.; Rutherford, T.J.; Johnson, C.M. Structure of the SAS-6 cartwheel hub from leishmania major. eLife 2014, 3, e01812. [Google Scholar] [CrossRef] [PubMed]
- Van Breugel, M.; Hirono, M.; Andreeva, A.; Yanagisawa, H.A.; Yamaguchi, S.; Nakazawa, Y.; Morgner, N.; Petrovich, M.; Ebong, I.O.; Robinson, C.V.; et al. Structures of SAS-6 suggest its organization in centrioles. Science 2011, 331, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Hilbert, M.; Erat, M.C.; Hachet, V.; Guichard, P.; Blank, I.D.; Flückiger, I.; Slater, L.; Lowe, E.D.; Hatzopoulos, G.N.; Steinmetz, M.O.; et al. Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry. Proc. Natl. Acad. Sci. USA 2013, 110, 11373–11378. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, D.; Vakonakis, I.; Olieric, N.; Hilbert, M.; Keller, D.; Olieric, V.; Bortfeld, M.; Erat, M.C.; Flückiger, I.; Gönczy, P.; et al. Structural basis of the 9-fold symmetry of centrioles. Cell 2011, 144, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ochi, T.; Matak-Vinkovic, D.; Robinson, C.V.; Chirgadze, D.Y.; Blundell, T.L. Non-homologous end-joining partners in a helical dance: Structural studies of XLF-XRCC4 interactions. Biochem. Soc. Trans. 2011, 39, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chirgadze, D.Y.; Bolanos-Garcia, V.M.; Sibanda, B.L.; Davies, O.R.; Ahnesorg, P.; Jackson, S.P.; Blundell, T.L. Crystal structure of human XLF/cernunnos reveals unexpected differences from XRCC4 with implications for nhej. EMBO J. 2008, 27, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Junop, M.S.; Modesti, M.; Guarne, A.; Ghirlando, R.; Gellert, M.; Yang, W. Crystal structure of the XRCC4 DNA repair protein and implications for end joining. EMBO J. 2000, 19, 5962–5970. [Google Scholar] [CrossRef] [PubMed]
- Hammel, M.; Rey, M.; Yu, Y.; Mani, R.S.; Classen, S.; Liu, M.; Pique, M.E.; Fang, S.; Mahaney, B.L.; Weinfeld, M.; et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J. Biol. Chem. 2011, 286, 32638–32650. [Google Scholar] [CrossRef] [PubMed]
- Andres, S.N.; Vergnes, A.; Ristic, D.; Wyman, C.; Modesti, M.; Junop, M. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 2012, 40, 1868–1878. [Google Scholar] [CrossRef] [PubMed]
- Hilbert, M.; Noga, A.; Frey, D.; Hamel, V.; Guichard, P.; Kraatz, S.H.; Pfreundschuh, M.; Hosner, S.; Flückiger, I.; Jaussi, R.; et al. SAS-6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Nat. Cell Biol. 2016, 18, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Pfreundschuh, M.; Alsteens, D.; Hilbert, M.; Steinmetz, M.O.; Muller, D.J. Localizing chemical groups while imaging single native proteins by high-resolution atomic force microscopy. Nano Lett. 2014, 14, 2957–2964. [Google Scholar] [CrossRef] [PubMed]
- Hodges, M.E.; Scheumann, N.; Wickstead, B.; Langdale, J.A.; Gull, K. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 2010, 123, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-Santos, Z.; Machado, P.; Branco, P.; Tavares-Cadete, F.; Rodrigues-Martins, A.; Pereira-Leal, J.B.; Bettencourt-Dias, M. Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 2010, 123, 1414–1426. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, M.; Nakazawa, Y.; Kamiya, R.; Hirono, M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr. Biol. 2007, 17, 1778–1783. [Google Scholar] [CrossRef] [PubMed]
- Riparbelli, M.G.; Dallai, R.; Mercati, D.; Bu, Y. Centriole symmetry: A big tale from small organisms. Cell Motil. Cytoskelet. 2009, 1105, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, M.; Callaini, G.; Riparbelli, M.G. The drosophila centriole—Conversion of doublets into triplets within the stem cell niche. J. Cell Sci. 2015, 128, 2437–2442. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.S.; Kim, M.; Yang, T.T.; Liao, J.C.; Tsou, M.F. SAS-6 assembly templated by the lumen of cartwheel-less centrioles precedes centriole duplication. Dev. Cell 2014, 30, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Acehan, D.; Kao, C.H.; Jane, W.N.; Uryu, K.; Tsou, M.F. De novo centriole formation in human cells is error-prone and does not require SAS-6 self-assembly. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Guichard, P.; Hachet, V.; Majubu, N.; Neves, A.; Demurtas, D.; Olieric, N.; Flückiger, I.; Yamada, A.; Kihara, K.; Nishida, Y.; et al. Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Curr. Biol. 2013, 23, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Guichard, P.; Desfosses, A.; Maheshwari, A.; Hachet, V.; Dietrich, C.; Brune, A.; Ishikawa, T.; Sachse, C.; Gönczy, P. Cartwheel architecture of trichonympha basal body. Science 2012, 337, 553. [Google Scholar] [CrossRef] [PubMed]
- Guichard, P.; Hamel, V.; Le Guennec, M.; Banterle, N.; Iacovache, I.; Nemcikova, V.; Fluckiger, I.; Goldie, K.N.; Stahlberg, H.; Levy, D.; et al. Cell-free reconstitution reveals centriole cartwheel assembly mechanisms. Nat. Commun. 2017, 8, 14813. [Google Scholar] [CrossRef] [PubMed]
- Sugioka, K.; Hamill, D.R.; Lowry, J.B.; McNeely, M.E.; Enrick, M.; Richter, A.C.; Kiebler, L.E.; Priess, J.R.; Bowerman, B. Centriolar SAS-7 acts upstream of SPD-2 to regulate centriole assembly and pericentriolar material formation. eLife 2017, 6, e20353. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, L.; O’Toole, E.; Schwager, A.; Hyman, A.A.; Muller-Reichert, T. Centriole assembly in Caenorhabditis elegans. Nature 2006, 444, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- The Pymol Molecular Graphics System, version 1.6; Schrödinger, LLC.; Available online: https://www.pymol.org/ (accessed on 15 May 2017).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. Ucsf chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).