Symmetry and Skewness in Weibull Modeling: Optimal Grouping for Parameter Estimation in Fertilizer Granule Strength
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Data
2.2. The Weibull Distribution for Ungrouped Data
2.3. Weibull Distribution for Grouped Data
2.3.1. Fisher Information Matrix and Covariance Matrix for Grouped Data
- is the probability that an observation falls into the n-th interval;
- θᵢ and θⱼ are model parameters;
- k is the number of intervals;
- N is the number of total observations.
2.3.2. Optimal Grouping
2.3.3. Construction of Optimal Intervals from Raw Data
2.3.4. Construction of Equal Intervals from Raw Data
2.4. Analysis of Estimation Accuracy
2.4.1. Asymptotic Relative Efficiency (ARE)
2.4.2. Confidence Intervals
2.5. Chi-Squared Test
3. Results and Discussion
3.1. Estimation of the Parameters of the Weibull Distribution
3.2. Chi-Squared Goodness-of-Fit Test for the Weibull Distribution
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Superfosfat, Fraction 1–2 mm, Data
Appendix A.2. Polifoska 8, Fraction 1–2 mm, Data
Appendix A.3. Salmag, Fraction 2.5–3.15 mm, Data
Appendix B
| import sympy as sp import numpy as np from scipy.optimize import minimize # Number of interval k = 5 z = sp.symbols(f’z1:{k + 1}’, real = True, positive = True) # Define expressions for A, B, P A = [-z [0] * sp.exp(-z [0])] for i in range(1, k − 1): A.append(z[i − 1] * sp.exp(-z[i − 1])–z[i] * sp.exp(-z[i])) A.append(z[k − 2] * sp.exp(-z[k − 2])) B = [z [0] * sp.exp(-z [0]) * sp.ln(z [0])] for i in range(1, k − 1): B.append(z[i] * sp.exp(-z[i]) * sp.ln(z[i])–z[i − 1] * sp.exp(-z[i − 1]) * sp.ln(z[i − 1])) B.append(-z[k − 2] * sp.exp(-z[k − 2]) * sp.ln(z[k − 2])) P = [1 − sp.exp(-z [0])] for i in range(1, k − 1): P.append(sp.exp(-z[i − 1]) − sp.exp(-z[i])) P.append(sp.exp(-z[k − 2])) # Fisher information matrix determinant phi = (sum(A[i]**2/P[i] for i in range(k)) * sum(B[i]**2/P[i] for i in range(k)) - sum(A[i] * B[i]/P[i] for i in range(k))**2) phi_func = sp.lambdify(z, phi, modules = “numpy”) def objective(z_vals): return -phi_func(*z_vals) z0 = np.linspace(0.1,10, k) bounds = [(0.0001, None)] * k constraints = {‘type’: ‘ineq’, ‘fun’: lambda x: np.diff(x)—0.0001} methods = [“SLSQP”, “trust-constr”] results = {} for method in methods: res = minimize(objective, z0, bounds = bounds, constraints = constraints, method = method) results[method] = res print(f”\nMethod: {method}”) print(“Status:”, res.success) for i, val in enumerate(res.x[:−1], 1): print(f”z_{i} = {val:.6f}”) print(“phi =“, -res.fun) best = max(results, key = lambda m: -results[m].fun if results[m].success else -np.inf) print(f”\nBest method: {best}”) for i, val in enumerate(results[best].x[:−1], 1): print(f”z_{i} = {val:.6f}”) print(“Maximum phi:”, -results[best].fun) |
Appendix C
| i | Z1 | Z2 | Z3 | Z4 | Z5 | Z6 | Z7 | Z8 | Z9 | Z10 | Z11 | Z12 | Z13 | Z14 | A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 4 5 6 7 8 9 10 11 12 13 14 15 | 0.2731 0.2109 0.1044 0.0774 0.0501 0.0377 0.0275 0.0214 0.0165 0.0132 0.0106 0.0087 0.0072 | 2.6067 1.3998 0.5122 0.3653 0.2321 0.1737 0.1269 0.0723 0.0769 0.0617 0.0500 0.0412 0.0344 | 3.4137 1.9592 1.2266 0.6759 0.4836 0.3432 0.1378 0.2042 0.1636 0.1326 0.1093 0.0912 | 3.8608 2.5735 1.7189 1.1904 0.7828 0.5770 0.4347 0.3431 0.2755 0.2259 0.1878 | 4.4106 2.9924 2.2039 1.6027 1.1806 0.8561 0.6512 0.5105 0.4122 0.3392 | 4.7952 3.4271 2.5710 1.9933 1.5307 1.1785 0.9023 0.7109 0.5736 | 5.2048 3.7672 2.9273 2.3149 1.8562 1.4808 1.1788 0.9393 | 5.5258 4.1025 3.2261 2.6155 2.1406 1.7598 1.4436 | 5.8478 4.3866 3.5095 2.8812 2.4009 2.0130 | 6.1215 4.6581 3.7612 3.1276 2.6402 | 6.3843 4.9016 3.9988 3.3568 | 6.6212 5.1306 4.2193 | 6.8437 5.3470 | 7.0565 | 0.4079 0.5572 0.6836 0.7571 0.8109 0.8480 0.8756 0.8963 0.9123 0.9248 0.9349 0.9431 0.9498 |
| i | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | P12 | P13 | P14 | P15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 4 5 6 7 8 9 10 11 12 13 14 15 | 0.2390 0.1901 0.0992 0.0745 0.0489 0.0370 0.0271 0.0211 0.0164 0.0131 0.0105 0.0087 0.0072 | 0.6872 0.5628 0.3016 0.2315 0.1582 0.1225 0.0921 0.0729 0.0577 0.0468 0.0383 0.0317 0.0265 | 0.0738 0.2142 0.4582 0.4012 0.2842 0.2240 0.1713 0.1378 0.1107 0.0910 0.0754 0.0632 0.0535 | 0.0329 0.1199 0.2164 0.3294 0.3125 0.2524 0.2066 0.1679 0.1395 0.1165 0.0986 0.0840 | 0.0211 0.0641 0.1291 0.1937 0.2558 0.2545 0.2215 0.1882 0.1592 0.1357 0.1165 | 0.0121 0.0419 0.0779 0.1250 0.1708 0.2095 0.2137 0.1947 0.1710 0.1488 | 0.0083 0.0270 0.0533 0.0827 0.1175 0.1515 0.1779 0.1835 0.1726 | 0.0055 0.0191 0.0370 0.0591 0.0831 0.1099 0.1356 0.1548 | 0.0040 0.0136 0.0273 0.0432 0.0615 0.0815 0.1025 | 0.0029 0.0102 0.0204 0.0329 0.0468 0.0622 | 0.0022 0.0078 0.0158 0.0255 0.0365 | 0.0017 0.0061 0.0124 0.0203 | 0.0013 0.0048 0.0099 | 0.0011 0.0039 | 0.0009 |
References
- Weibull, W. A statistical distribution function of wide applicability. J. Appl. Mech. 1951, 18, 293–297. [Google Scholar] [CrossRef]
- Rinne, H. The Weibull Distribution: A Handbook; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Mudholkar, G.S.; Srivastava, D.K.; Freimer, M. The exponentiated Weibull family: A reanalysis of the bus-motor-failure data. Technometrics 1995, 37, 436–445. [Google Scholar] [CrossRef]
- Ambrožič, M.; Vidovič, K. Reliability of the Weibull analysis of the strength of construction materials. J. Mater. Sci. 2007, 42, 9645–9653. [Google Scholar] [CrossRef]
- Cheng, C.; Chen, J.; Li, Z. A new algorithm for maximum likelihood estimation with progressive type-I interval censored data. Commun. Stat. Simul. Comput. 2010, 39, 750–766. [Google Scholar] [CrossRef]
- Ng, H.K.T.; Wang, Z. Statistical estimation for parameters of Weibull distribution based on progressively type-I interval censored sample. J. Stat. Comput. Simul. 2009, 79, 145–159. [Google Scholar] [CrossRef]
- Barraza-Contreras, J.M.; Piña-Monarrez, M.R.; Torres-Villaseñor, R.C. Reliability by Using Weibull Distribution Based on Vibration Fatigue Damage. Appl. Sci. 2023, 13, 10291. [Google Scholar] [CrossRef]
- Turygin, Y.; Bozek, P.; Abaramov, I.; Nikitin, Y. Reliability determination and diagnostics of a mechatronic system. Adv. Sci. Technol. Res. J. 2018, 12, 274–290. [Google Scholar] [CrossRef] [PubMed]
- Młynarski, S.; Pilch, R.; Smolnik, M.; Szybka, J. Analysis of the Modernised Railway Vehicle Component with Regard to Reliability and Operational Safety. Adv. Sci. Technol. Res. J. 2024, 18, 21–32. [Google Scholar] [CrossRef]
- Fedorov, A.; Gulayeva, Y.K. Strength statistics for porous alumina. Powder Technol. 2019, 343, 783–791. [Google Scholar] [CrossRef]
- Tumidajski, P.J.; Fiore, L.; Khodabocus, T.; Lachemi, M.; Pari, R. Comparison of Weibull and normal distributions for compressive strengths. Can. J. Civ. Eng. 2006, 33, 1287–1292. [Google Scholar] [CrossRef]
- Subero-Couroyer, C.; Ghadiri, M.; Brunard, N.; Kolenda, F. Weibull analysis of quasi-static crushing strength of catalyst particles. Chem. Eng. Res. Des. 2003, 81, 953–962. [Google Scholar] [CrossRef]
- Datsiou, K.D.; Overend, M. Weibull parameter estimation and goodness-of-fit for glass strength data. Struct. Saf. 2018, 73, 29–41. [Google Scholar] [CrossRef]
- Quinn, J.B.; Quinn, G.D. A practical and systematic review of Weibull statistics for reporting strengths of dental materials. Dent. Mater. 2010, 26, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.F.; Zhou, J.C.; Li, Y.D. Distribution of the mechanical strength of solid catalysts. Chem. Eng. Res. Des. 2006, 84, 1152–1157. [Google Scholar] [CrossRef]
- Rozenblat, Y.; Portnikov, D.; Levy, A.; Kalman, H.; Aman, S.; Tomas, J. Strength distribution of particles under compression. Powder Technol. 2011, 208, 215–224. [Google Scholar] [CrossRef]
- Gorjan, L.; Vidovič, K. Bend strength of alumina ceramics: A comparison of Weibull statistics with other statistics based on very large experimental data set. Ceram. Int. 2012, 32, 1221–1227. [Google Scholar] [CrossRef]
- Burgos-Peñaloza, J.A.; Lambert-Arista, A.A.; García-Cueto, O.R.; Santillán-Soto, N.; Valenzuela, E.; Flores-Jiménez, D.E. Comparative Analysis of Estimated Small Wind Energy Using Different Probability Distributions in a Desert City in Northwestern México. Energies 2024, 17, 3323. [Google Scholar] [CrossRef]
- Teimourian, H.; Abubakar, M.; Yildiz, M.; Teimourian, A. A Comparative Study on Wind Energy Assessment Distribution Models: A Case Study on Weibull Distribution. Energies 2022, 15, 5684. [Google Scholar] [CrossRef]
- Alsaqoor, S.; Marashli, A.; At-Tawarah, R.; Borowski, G.; Alahmer, A.; Aljabarin, N.; Beithou, N. Evaluation of Wind Energy Potential in View of the Wind Speed Parameters—A Case Study for the Southern Jordan. Adv. Sci. Technol. Res. J. 2022, 16, 275–285. [Google Scholar] [CrossRef]
- Zaindin, M. Parameter estimation of the modified Weibull model based on grouped and censored data. Int. J. Basic Appl. Sci. 2010, 10, 122–132. [Google Scholar]
- Chien, Z.; Mi, J. Statistical estimation for the scale parameter of the gamma distribution based on grouped data. Commun. Stat. Theory Methods 1998, 27, 3035–3045. [Google Scholar] [CrossRef]
- Cheng, K.F.; Chen, C.H. Estimation on the Weibull parameters with grouped data. Commun. Stat. Theory Methods 1988, 17, 325–341. [Google Scholar] [CrossRef]
- Basu, B.; Tiwari, D.; Kundu, D.; Prasad, R. Is Weibull distribution the most appropriate statistical strength distribution for brittle materials? Ceram. Int. 2009, 35, 237–246. [Google Scholar] [CrossRef]
- Cramér, H. Mathematical Methods of Statistics; Princeton University Press: Princeton, NJ, USA, 1951. [Google Scholar]
- Fisz, M. Probability Theory and Mathematical Statistics; Wiley and Sons: New York, NY, USA, 1967. [Google Scholar]
- Kulldorff, G. Contributions to the Theory of Estimation from Grouped and Partially Grouped Samples; Almqvist and Wiksell: Stockholm, Sweden, 1961. [Google Scholar]
- Archer, N.P. Maximum likelihood estimation with Weibull models when the data are grouped. Commun. Stat. Theory Methods 1982, 11, 199–207. [Google Scholar]
- Rosaiah, K.; Kantam, R.R.L.; Narasimham, V.L. Optimum class limits for ML estimation in 2-parameter gamma distribution from grouped data. Commun. Stat. Simul. Comput. 1991, 20, 1173–1189. [Google Scholar] [CrossRef]
- Rao, A.V.; Rao, A.V.D.; Narasimham, V.L. Asymptotically optimal grouping for maximum likelihood estimation of Weibull parameters. Commun. Stat. Simul. Comput. 1994, 23, 1077–1096. [Google Scholar]
- Kantam, R.R.L.; Rao, A.V.; Rao, G.S. Optimum group limits for estimation in scaled log-logistic distribution from a grouped data. Stat. Pap. 2005, 46, 359–377. [Google Scholar] [CrossRef]
- Marwa, A.A.; Zaher, H.; Elsherpieny, E.A. Optimum group limits for maximum likelihood estimation on the exponentiated Fréchet distribution based on grouped data. Br. J. Appl. Sci. Technol. 2013, 3, 1464–1480. [Google Scholar] [CrossRef]
- Mohan, C.R.; Rao, A.V.; Anjaneyulu, G.V.S.R. Comparison of least square estimators with rank regression estimators of Weibull distribution—A simulation study. J. Stat. 2013, 20, 1–10. [Google Scholar]
- Lin, C.-T.; Balakrishnan, N.; Wu, S.J.S. Planning life tests based on progressively type-I grouped censored data from the Weibull distribution. Commun. Stat. Simul. Comput. 2011, 40, 574–595. [Google Scholar] [CrossRef]
- Leszczyński, N.; Przystupa, W.; Nowak, J.; Rusek, P. Compression strength of superphosphate and urea granules. Przem. Chem. 2017, 96, 1963–1967. [Google Scholar]
- Przystupa, W.; Rusek, P.; Nowak, J. Evaluation of the strength of mineral fertilizer granules by using Weibull distribution. Przem. Chem. 2018, 97, 2124–2127. [Google Scholar]
- Watkins, A.J. On expectations associated with maximum likelihood estimation in the Weibull distribution. J. Ital. Stat. Soc. 1998, 7, 15–26. [Google Scholar] [CrossRef]
- Xiao, X.; Mukherjee, A.; Xie, M. Estimation procedures for grouped data—A comparative study. J. Appl. Stat. 2016, 43, 859–875. [Google Scholar] [CrossRef]
- Kendall, M.G.; Stuart, A. The Advanced Theory of Statistics; Charles Griffin and Company: London, UK, 1973; Volume 2. [Google Scholar]
- Chimitova, E.V.; Lemeshko, B.Y. Chi-Squared Goodness-of-Fit Tests: The Optimal Choice of Grouping Intervals. In Recent Advances in Systems, Control and Information Technology; Shakirov, R., Ed.; Springer: Cham, Switzerland, 2017; Volume 543, pp. 760–774. [Google Scholar]
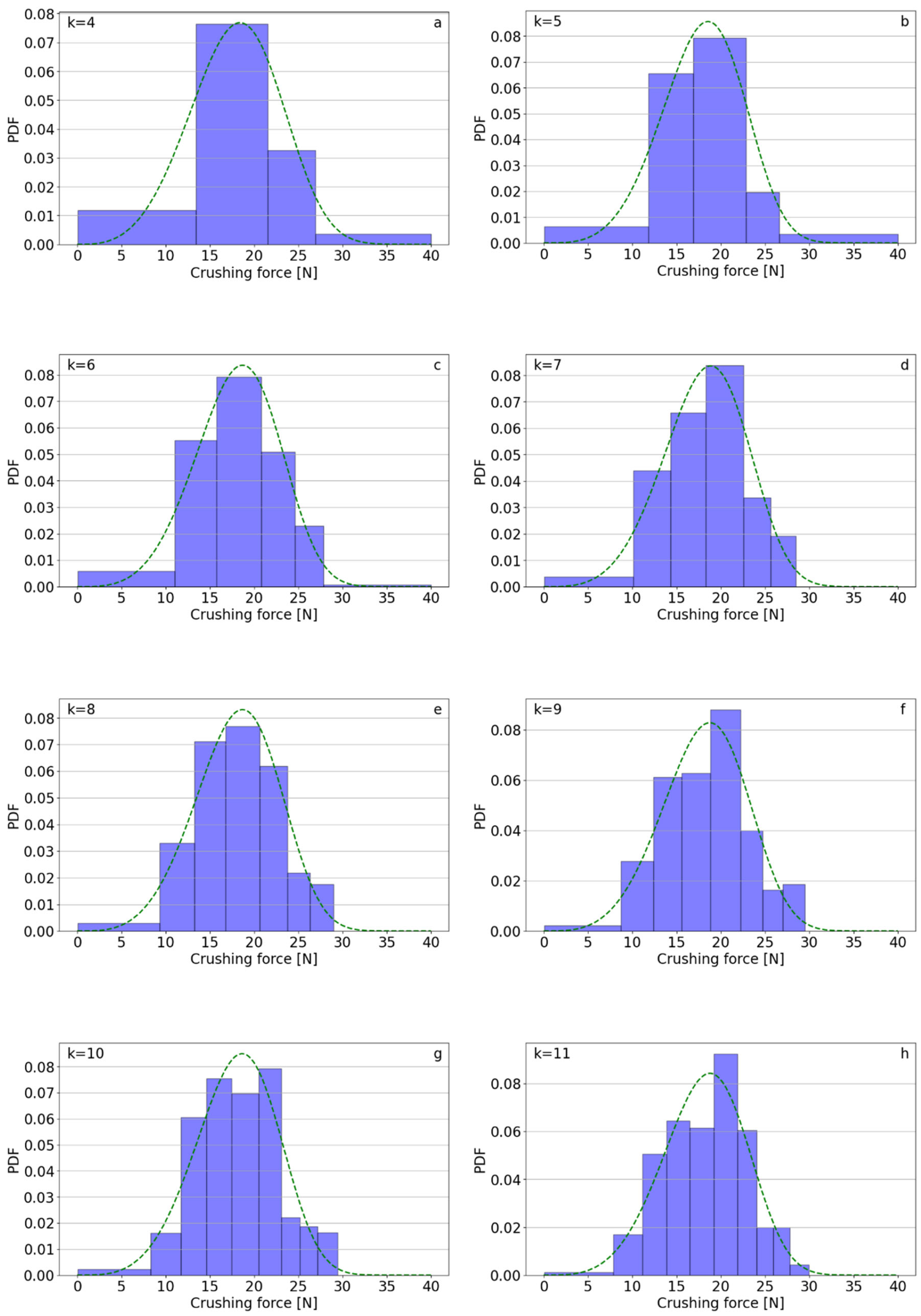
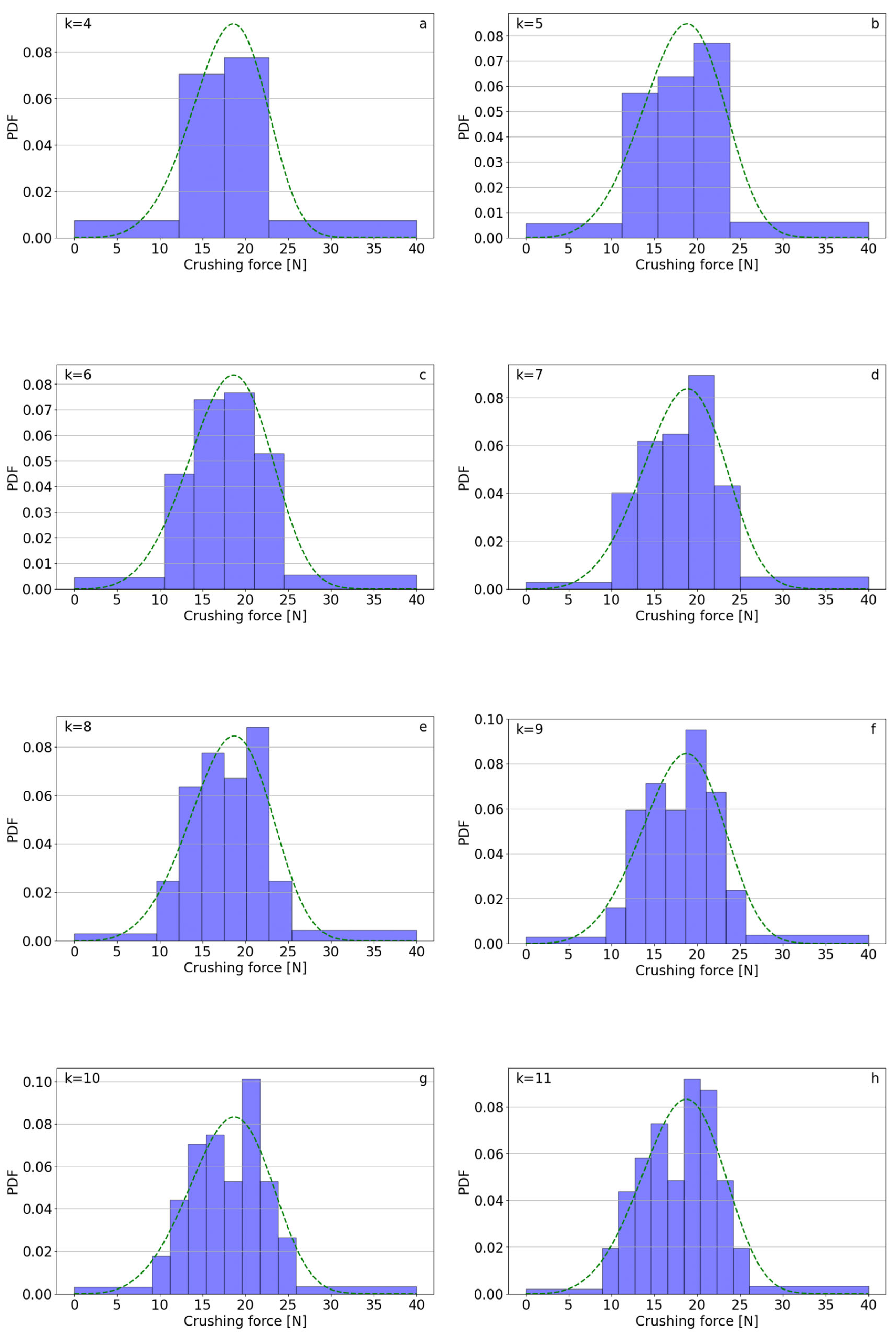

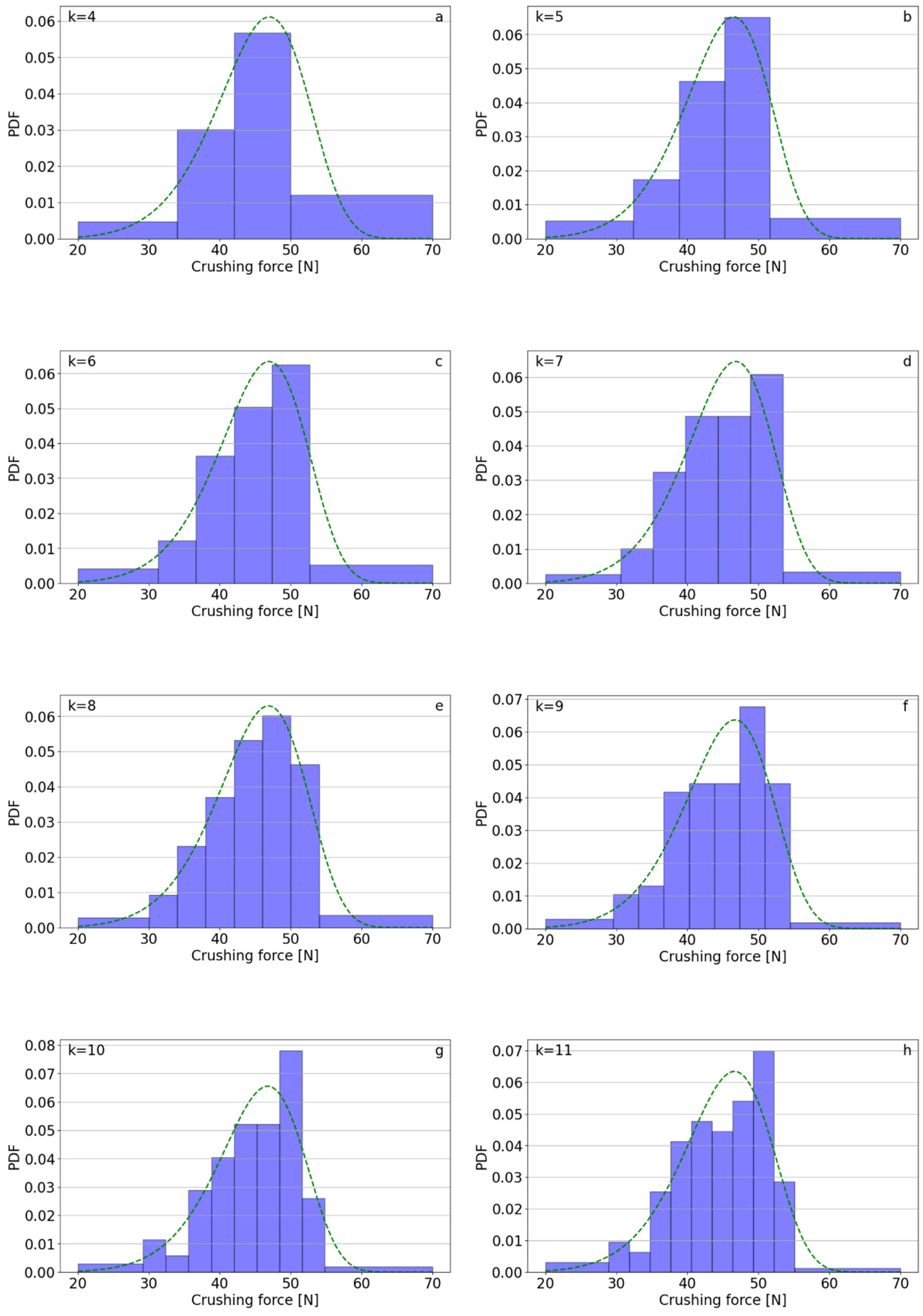

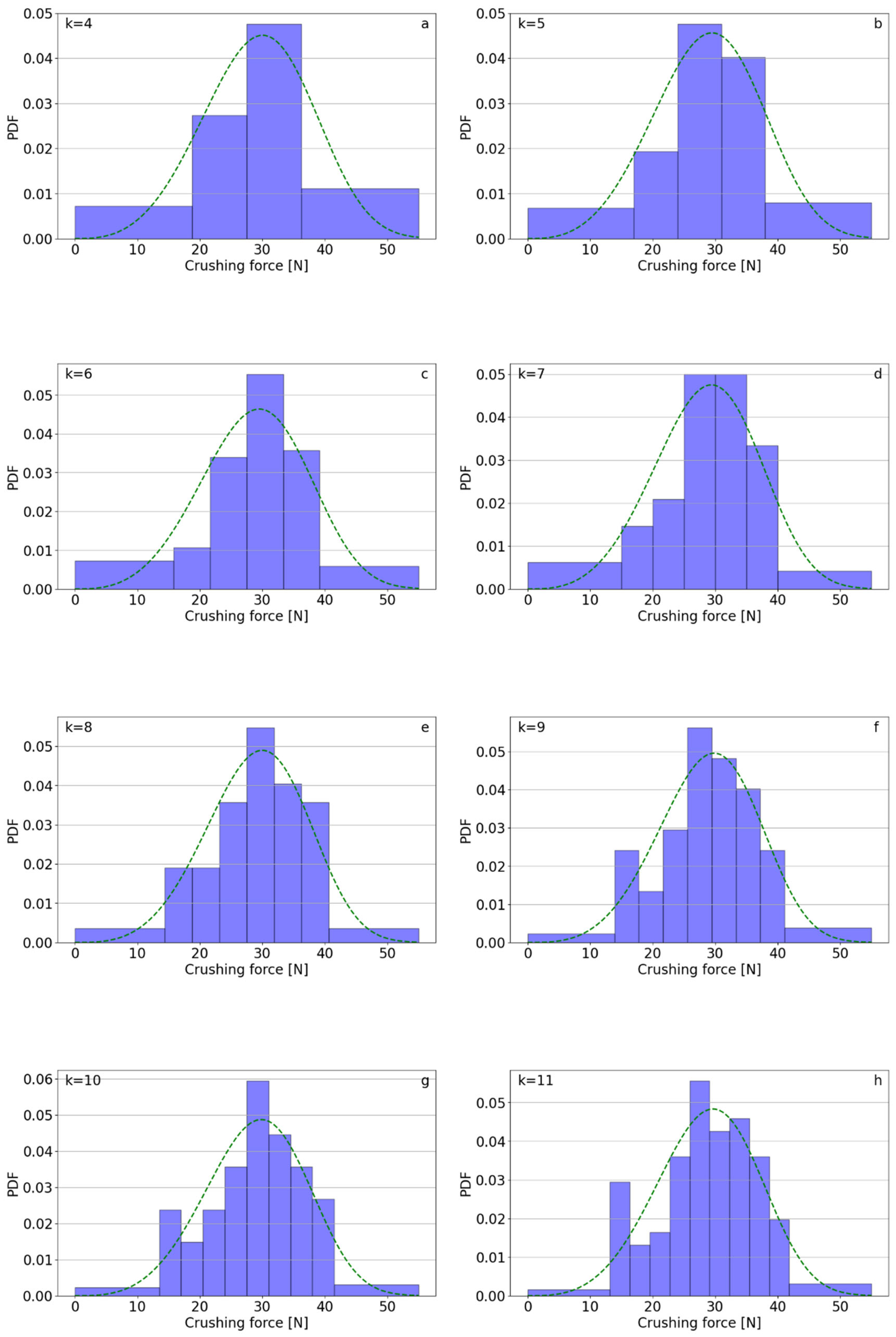
| k | Polifoska 8 | Salmag | Superfosfat | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Optimal Bin Width | ||||||||||||
| 95% CI for | 95% CI for |
95% CI for |
95% CI for |
95% CI for |
95% CI for | |||||||
| 4 | 19.779 | 3.990 | 19.779 ± 1.075 | 3.990 ± 0.723 | 47.470 | 8.318 | 47.470 ± 1.237 | 8.318 ± 1.506 | 31.941 | 4.341 | 31.941 ± 1.595 | 4.341 ± 0.786 |
| 5 | 19.642 | 4.446 | 19.642 ± 0.933 | 4.446 ± 0.739 | 47.039 | 8.299 | 47.039 ± 1.197 | 8.299 ± 1.378 | 31.572 | 3.782 | 31.572 ± 1.763 | 3.782 ± 0.628 |
| 6 | 19.827 | 4.382 | 19.827 ± 0.936 | 4.382 ± 0.709 | 47.649 | 8.342 | 47.649 ± 1.181 | 8.342 ± 1.35 | 31.792 | 4.181 | 31.792 ± 1.572 | 4.181 ± 0.677 |
| 7 | 19.976 | 4.414 | 19.976 ± 0.927 | 4.414 ± 0.696 | 47.413 | 8.068 | 47.413 ± 1.204 | 8.068 ± 1.272 | 31.829 | 4.133 | 31.829 ± 1.578 | 4.133 ± 0.652 |
| 8 | 19.830 | 4.354 | 19.830 ± 0.926 | 4.354 ± 0.677 | 47.333 | 8.141 | 47.333 ± 1.183 | 8.141 ± 1.266 | 31.808 | 4.213 | 31.808 ± 1.536 | 4.213 ± 0.655 |
| 9 | 19.913 | 4.353 | 19.913 ± 0.926 | 4.353 ± 0.67 | 47.400 | 8.330 | 47.400 ±1.152 | 8.330 ± 1.281 | 31.979 | 4.194 | 31.979 ± 1.544 | 4.1942± 0.645 |
| 10 | 19.745 | 4.434 | 19.745 ± 0.898 | 4.434 ± 0.677 | 47.354 | 8.275 | 47.354 ± 1.155 | 8.275 ± 1.263 | 31.719 | 4.079 | 31.719 ± 1.569 | 4.079 ± 0.623 |
| 11 | 19.891 | 4.430 | 19.891 ± 0.903 | 4.430 ± 0.672 | 47.441 | 8.287 | 47.441 ± 1.152 | 8.287 ± 1.257 | 31.860 | 4.230 | 31.860 ± 1.516 | 4.230 ± 0.642 |
| Equal Bin Width | ||||||||||||
| 4 | 19.513 | 4.777 | 19.513 ± 0.877 | 4.777 ± 0.891 | 47.793 | 7.870 | 47.793 ± 1.352 | 7.870 ± 1.611 | 32.503 | 3.837 | 32.503 ± 1.961 | 3.837 ± 0.834 |
| 5 | 19.926 | 4.466 | 19.926 ± 0.933 | 4.466 ± 0.795 | 47.208 | 8.297 | 47.208 ± 1.205 | 8.297 ± 1.48 | 31.946 | 3.813 | 31.946 ± 1.865 | 3.813 ± 0.747 |
| 6 | 19.752 | 4.359 | 19.752 ± 0.932 | 4.359 ± 0.741 | 47.708 | 8.156 | 47.708 ± 1.219 | 8.156 ± 1.410 | 31.880 | 3.871 | 31.880 ± 1.804 | 3.871 ± 0.719 |
| 7 | 20.012 | 4.432 | 20.012 ± 0.922 | 4.432 ± 0.737 | 47.545 | 8.276 | 47.545 ± 1.181 | 8.276 ± 1.375 | 31.673 | 3.946 | 31.673 ± 1.739 | 3.9456 ± 0.705 |
| 8 | 19.815 | 4.425 | 19.815 ± 0.908 | 4.425 ± 0.717 | 47.645 | 8.087 | 47.645 ± 1.202 | 8.087 ± 1.323 | 32.073 | 4.133 | 32.073 ± 1.673 | 4.133 ± 0.724 |
| 9 | 19.859 | 4.441 | 19.859 ± 0.903 | 4.441 ± 0.711 | 47.448 | 8.144 | 47.448 ± 1.181 | 8.144 ± 1.181 | 31.878 | 4.157 | 31.878 ± 1.645 | 4.1574 ± 0.714 |
| 10 | 19.838 | 4.361 | 19.838 ± 0.916 | 4.361 ± 0.693 | 47.466 | 8.391 | 47.466 ± 1.143 | 8.391 ± 1.324 | 31.952 | 4.098 | 31.952 ± 1.668 | 4.098 ± 0.700 |
| 11 | 19.889 | 4.373 | 19.889 ± 0.914 | 4.373 ± 0.691 | 47.391 | 8.106 | 47.391 ± 1.178 | 8.106 ± 1.272 | 31.814 | 4.039 | 31.814 ± 1.681 | 4.039 ± 0.684 |
| Raw Data | 19.849 | 4.438 | 19.849 ± 0.888 | 4.438 ± 0.653 | 47.427 | 8.297 | 47.427 ± 1.135 | 8.297 ± 1.220 | 31.808 | 4.196 | 31.808 ± 1.506 | 4.196 ± 0.617 |
| k | Polifoska 8 | Salmag | Superfosfat | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 0.683 | 0.816 | 1.026 | 0.537 | 0.842 | 0.656 | 0.704 | 0.573 | 0.891 | 0.616 | 0.663 | 0.616 |
| 5 | 0.906 | 0.781 | 0.907 | 0.673 | 0.899 | 0.784 | 0.887 | 0.679 | 0.729 | 0.965 | 0.733 | 0.767 |
| 6 | 0.901 | 0.847 | 0.908 | 0.776 | 0.924 | 0.817 | 0.868 | 0.748 | 0.917 | 0.832 | 0.784 | 0.828 |
| 7 | 0.918 | 0.879 | 0.928 | 0.785 | 0.889 | 0.920 | 0.924 | 0.787 | 0.91 | 0.896 | 0.843 | 0.861 |
| 8 | 0.919 | 0.929 | 0.957 | 0.828 | 0.921 | 0.928 | 0.892 | 0.850 | 0.961 | 0.886 | 0.911 | 0.818 |
| 9 | 0.920 | 0.950 | 0.967 | 0.843 | 0.971 | 0.907 | 0.923 | 0.875 | 0.951 | 0.915 | 0.942 | 0.840 |
| 10 | 0.978 | 0.930 | 0.940 | 0.886 | 0.967 | 0.933 | 0.986 | 0.850 | 0.921 | 0.982 | 0.916 | 0.875 |
| 11 | 0.967 | 0.943 | 0.944 | 0.893 | 0.971 | 0.942 | 0.929 | 0.920 | 0.987 | 0.924 | 0.902 | 0.914 |
| k | Polifoska 8 | Salmag | Superfosfat | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Optimal Grouping | Equi-Class | Optimal Grouping | Equi-Class | Optimal Grouping | Equi-Class | |||||||
| p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | |||||||
| 4 | - | - | 0.415 | 0.519 | - | - | 0.011 | 0.917 | - | - | 1.957 | 0.168 |
| 5 | 1.416 | 0.234 | 3.124 | 0.210 | 0.439 | 0.508 | 0.900 | 0.343 | 4.315 | 0.038 | 3.241 | 0.198 |
| 6 | 0.809 | 0.667 | 0.950 | 0.622 | 4.086 | 0.130 | 1.720 | 0.423 | 6.972 | 0.030 | 9.573 | 0.02 |
| 7 | 2.056 | 0.561 | 1.976 | 0.577 | 2.817 | 0.421 | 4.720 | 0.193 | 1.019 | 0.313 | 4.202 | 0.2404 |
| 8 | 0.493 | 0.781 | 5.890 | 0.207 | 5.061 | 0.080 | 0.583 | 0.965 | 2.026 | 0.363 | 4.577 | 0.333 |
| 9 | 2.581 | 0.461 | 4.820 | 0.306 | 8.344 | 0.039 | 6.562 | 0.255 | 3.481 | 0.323 | 5.273 | 0.383 |
| 10 | 5.611 | 0.230 | 5.555 | 0.352 | 7.353 | 0.118 | 5.071 | 0.280 | 3.560 | 0.469 | 5.986 | 0.425 |
| 11 | 3.607 | 0.462 | 4.829 | 0.566 | 4.152 | 0.386 | 5.215 | 0.390 | 7.322 | 0.120 | 7.324 | 0.292 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Przystupa, W.; Kurasiński, P.; Leszczyński, N. Symmetry and Skewness in Weibull Modeling: Optimal Grouping for Parameter Estimation in Fertilizer Granule Strength. Symmetry 2025, 17, 1566. https://doi.org/10.3390/sym17091566
Przystupa W, Kurasiński P, Leszczyński N. Symmetry and Skewness in Weibull Modeling: Optimal Grouping for Parameter Estimation in Fertilizer Granule Strength. Symmetry. 2025; 17(9):1566. https://doi.org/10.3390/sym17091566
Chicago/Turabian StylePrzystupa, Wojciech, Paweł Kurasiński, and Norbert Leszczyński. 2025. "Symmetry and Skewness in Weibull Modeling: Optimal Grouping for Parameter Estimation in Fertilizer Granule Strength" Symmetry 17, no. 9: 1566. https://doi.org/10.3390/sym17091566
APA StylePrzystupa, W., Kurasiński, P., & Leszczyński, N. (2025). Symmetry and Skewness in Weibull Modeling: Optimal Grouping for Parameter Estimation in Fertilizer Granule Strength. Symmetry, 17(9), 1566. https://doi.org/10.3390/sym17091566






