1. Introduction
Medical image segmentation is an important task at the intersection of computer vision and medicine, involving the identification and precise delineation of structures or tissues within medical images. Its purpose is to assist in clinical diagnosis and lesion studies, provide supportive evidence for treatment planning, and assist physicians in disease diagnosis. In addition, it can contribute to medical research by offering more data and evidence [
1,
2,
3].
With recent developments in deep learning, convolutional neural networks (CNNs) have been widely applied in the field of medical image processing. The U-Net architecture [
4] and its variants [
5,
6,
7] can generate high-resolution image segmentation maps. The success of these methods can be attributed to their ability to leverage upsampling and downsampling to address the varying sizes of medical lesions. They employ an encoder–decoder architecture with skip connections, enabling the model to perceive features generated at multiple scales simultaneously. However, CNNs struggle to establish long-range dependencies between pixels due to their relatively small receptive fields. In contrast, transformers that leverage their attention mechanism excel at capturing long-range dependencies between pixels [
8,
9]. Some methods have attempted to integrate the transformer architecture into U-shaped networks for medical image segmentation [
6]. However, effectively leveraging the global context captured by transformers while preserving the fine-grained local details crucial for precise medical image segmentation remains an ongoing challenge, and suboptimal integration can lead to blurred boundaries. While recent foundation models such as MedSAM [
10], developed by fine-tuning large pre-trained models, show promise for general-purpose segmentation, they typically require prompt guidance. To address these problems, we draw inspiration from both CNN and transformer architectures and propose a novel multiscale recombined channel attention (MRCA) module. Specifically, the MRCA module adopts a multiscale aggregation (MSA) module for the extraction of local and global features from images. Subsequently, a recombined channel attention (RCA) module is employed to reorganize and group features from multiple scales and executes attention operations to integrate features from different receptive fields. The U-Net architecture is then remodeled using the MRCA module (MRCA-UNet).
The main contributions of this study are outlined as follows.
- -
A novel MSA module is proposed that can extract features from targets through multiple branches, allowing for simultaneous perception of local and global features. Additionally, while extracting features, this module exhibits feature selection functionality, enabling the model to focus on more useful information.
- -
A new RCA mechanism is proposed that performs feature grouping and reorganization along the channel dimension. It can capture image features under different receptive fields using convolutional kernels of various sizes. Subsequently, multi-head attention is used to compensate for the deficiency in convolution in perceiving global information.
- -
Based on the MSA and RCA modules, an MRCA module is proposed. This module can extract and fuse multiscale image features by combining convolution operations and attention mechanisms. By applying this module to the U-Net architecture, we developed the MRCA-UNet network, which can achieve both local and global feature extraction and fusion while fully leveraging the capabilities of the channel features.
We conducted experiments on the Synapse and ISIC-2018 datasets, the results of which demonstrate that the MRCA-UNet method can achieve promising segmentation results.
2. Related Works
2.1. Convolutional Neural Networks
AlexNet uses CNNs for image recognition and classification tasks. ResNet [
11] introduced residual connections, making CNNs the main framework for computer vision tasks. Subsequently, the introduction of the Inception [
12], Res2Net [
13], and MixNet [
14] models continuously pushed the development of CNN design principles, including the proposal of multibranch parallel convolution ideas. SE-Net [
15] and nonlocal networks [
16] can enhance relevant features and suppress irrelevant features using attention mechanisms. CBAM [
17] can enhance the modeling ability of spatial and channel attention. EfficientNet [
18] uses compound coefficients to scale depth, width, and resolution to develop more efficient network architectures. Inspired by the structure of the Swin Transformer [
19], ConvNeXt [
20] adopts a hierarchical design that can improve CNN performance. Subsequently, FocalNet [
21] replaced the attention mechanisms with focal modulation, encoding both short- and long-term dependencies and aggregating token information through the modulation module. Moreover, In Conv2Former [
22], the attention blocks are modified into 11 × 11 large-kernel convolution blocks to achieve better recognition performance.
2.2. Vision Transformer
Transformers were initially applied in the field of natural language processing and later introduced to computer vision tasks through vision transformers [
23]. They compute the similarities between pixels using self-attention. However, the quadratic complexity of transformers for high-resolution images poses a considerable challenge in vision tasks. Consequently, various methods have been proposed to address the problem, including multiscale network frameworks [
24], local attention mechanisms [
25], and lightweight convolutional modules [
26].
2.3. Transformers for Medical Image Segmentation
Medical image segmentation involves identifying and precisely segmenting structures or tissues in medical images. Ronneberger et al. proposed the U-Net architecture [
4], which adopt an encoder–decoder architecture and uses skip connections to aggregate features from different stages. UNet++ [
7] employs dense skip connections to optimize multiscale feature extraction at different levels. UNet 3+ [
27] improved skip connections by using full-scale connections to extract features, and U-Net uses attention mechanisms [
6] to suppress irrelevant regions and highlight important features of local areas. Moreover, DC-UNet [
28] incorporates residual convolution and residual channel attention in the skip connections.
Large pre-trained models have garnered significant attention for their applications in medical image segmentation. Meta AI’s Segment Anything Model (SAM) [
29] has demonstrated remarkable zero-shot and few-shot segmentation capabilities across various domains. The DC-SAM [
30] method, based on prompt tuning, enhances the features of SAM’s prompt encoder, designs a cycle-consistent cross-attention mechanism, and adopts a dual-branch structure to achieve contextual segmentation of images and videos. Recently, the application of large pre-trained models to the field of medical image segmentation has garnered significant attention. For example, Ma et al. developed MedSAM [
10] by fine-tuning SAM on a large-scale dataset of over a million medical images. The model demonstrates strong zero-shot and few-shot segmentation capabilities across various modalities and tasks when provided with user prompts (such as points or boxes). However, a key limitation of MedSAM is its dependence on user prompts, making it difficult to apply directly in fully automatic segmentation scenarios.
Transformers have demonstrated superior performance in the field of image processing and are widely used in medical image segmentation. Chen et al. proposed TransUNet [
31], which combines a CNN and transformer to capture both local and global feature relationships using this hybrid architecture. Swin-UNet [
32] introduced a Swin transformer into the U-shaped network, and MISSFormer [
33] redesigned the transformer blocks in a U-shaped network to enhance long-range dependencies and local context. These architectures have shown superior performance in medical segmentation tasks.
3. Methods
This section provides a detailed introduction to the specific structure of the MRCA-UNet framework. We then describe the core component of MRCA-UNet, that is, the MRCA module, which includes MSA and RCA mechanisms.
3.1. MRCA-UNet Framework
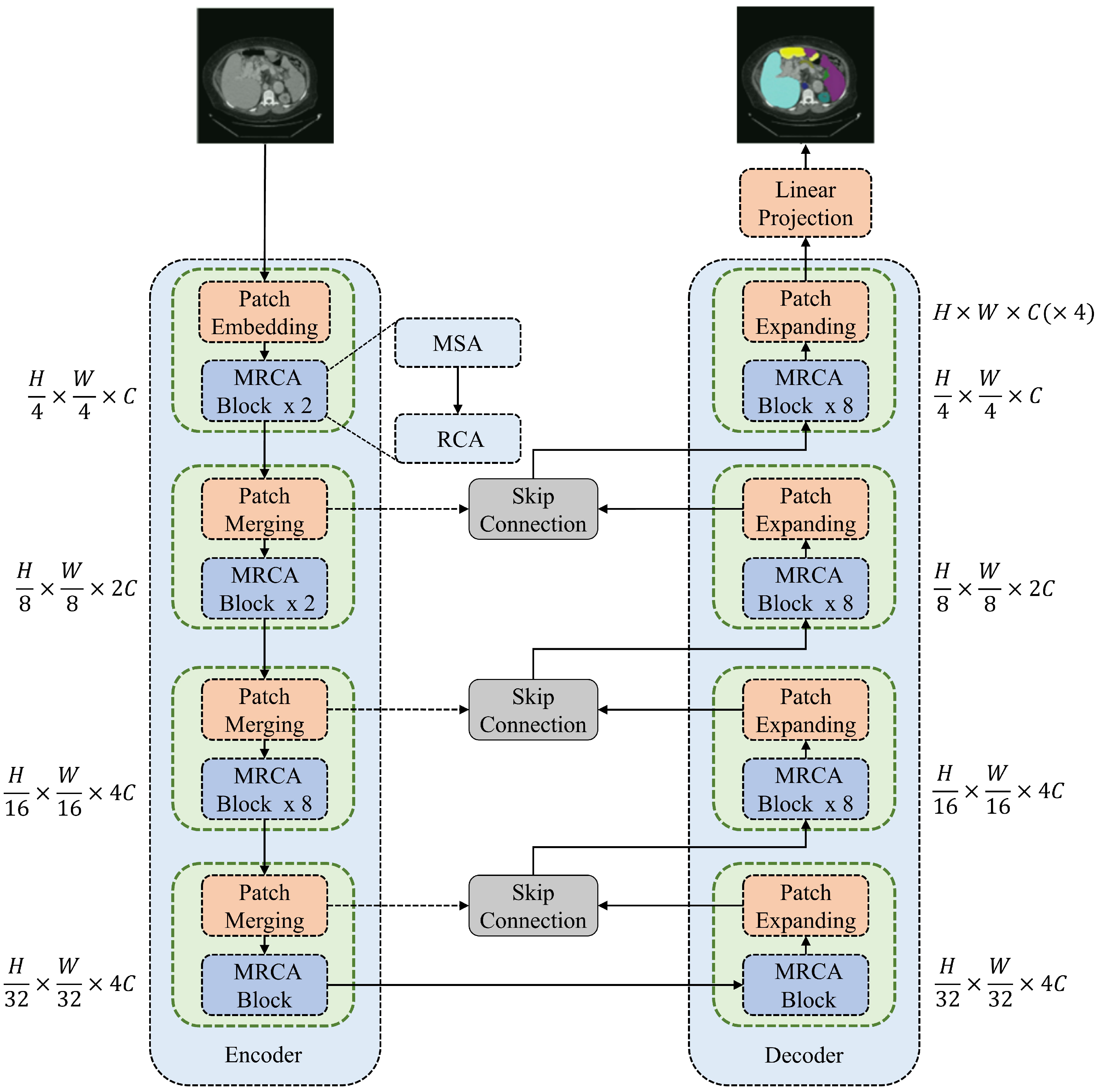
Figure 1 shows the framework of the proposed MRCA-UNet, which consists of three parts—namely, an encoder, a decoder, and the skip connection modules added between them.
An image of size is sent into the encoder. First, the encoder divides it into non-overlapping blocks of size and projects its feature dimensions to size C for processing through the MRCA module. The encoder processes the image features through the MRCA module after each downsampling operation, repeating this process three times. As the image resolution decreases, the number of channels increases, allowing the MRCA module to learn feature representations at different scales. The decoder adopts a structure completely symmetrical to that of the encoder, consisting of patch-expanding and MRCA modules. Information fusion is performed through skip connections between the encoder and the decoder to compensate for information loss during the downsampling process. When the feature map is restored to , it enters the linear projection layer for segmentation prediction. This U-shaped architecture trades off small local spatial losses for multiscale global dimension interactions, providing better support for pixel-level segmentation tasks.
3.2. Multiscale Recombined Channel Attention (MRCA)
The core idea of MRCA is to use multiple branches to extract global and local information from image features. In each branch, feature selection [
34] is employed to highlight more useful tokens and channels, thereby reducing the number of parameters and computational complexity of the model. Channel reorganization is then used to make full use of channel features; extract and aggregate the features of each group using convolution; and, finally, perceive the overall features with multi-head attention. Specifically, it is divided into two stages, that is, feature extraction and fusion. In this study, the MSA and RCA mechanisms are used to achieve these two stages. The MSA mechanism extracts multibranch features, whereas RCA completes feature fusion, with the two modules being used in a series structure. The MSA and RCA modules are discussed in detail in
Section 3.3 and
Section 3.4.
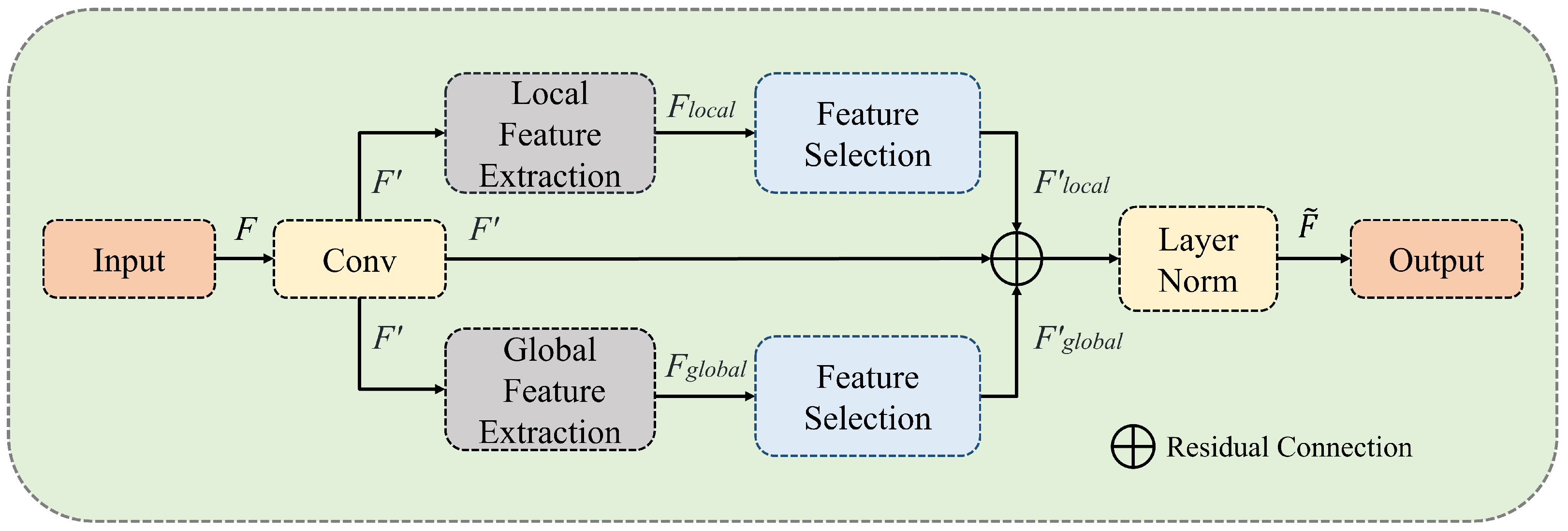
3.3. Multiscale Aggregation (MSA)
In medical image segmentation tasks, images of smaller organs and tissues can easily lose target information after multiple downsampling operations. As shown in
Figure 2, the MSA module effectively solves this problem by using multiscale branches for feature extraction, replacing traditional convolution operations. The MSA module is divided into three parallel branches: global feature extraction, local feature extraction, and residual connection branches. After feature extraction, feature selection is performed. Given an input feature tensor (
), a point-wise convolution is first performed to adjust the dimensions, resulting in
. The local and global feature extraction branches then compute
and
, respectively. Next, through feature selection, the results of the local branch (
), the global branch (
), and the initial convolution branch (
) are summed to obtain
.
The local and global branches can be achieved by dividing and aggregating the non-overlapping patches in the spatial dimension and controlling the number of divided patches. Finally, feature extraction is accomplished by calculating the attention matrix between the different patches.
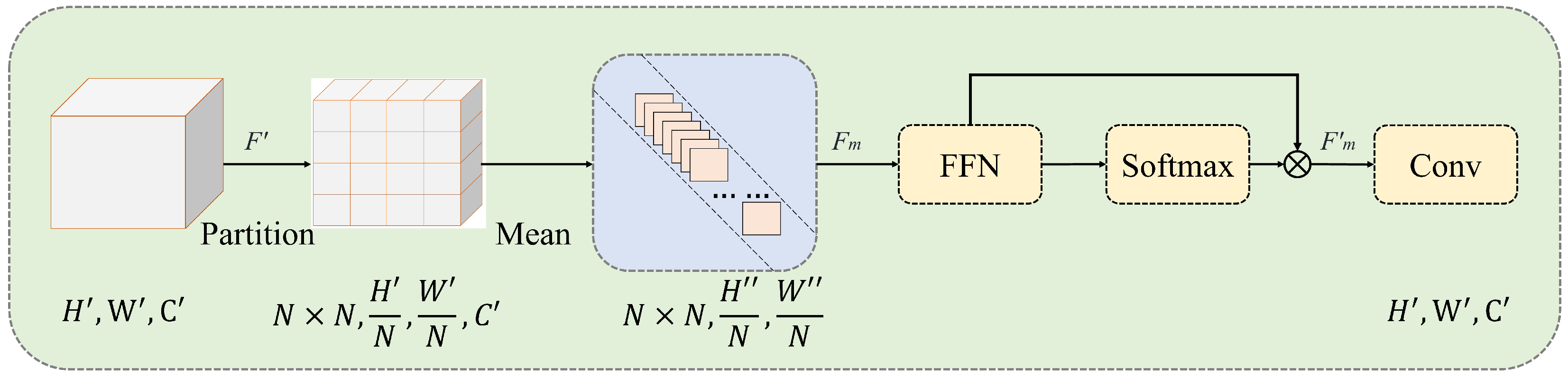
The feature extraction module is shown in
Figure 3. First, the feature map (
) of the input feature extraction module is divided into
patches of equal size (
). Average pooling is performed on the channels to obtain
. This is then sent to the feed-forward network (FFN) for linear computation. A softmax operation is applied to the extracted features to obtain their probability distribution in the spatial dimension. The features are then multiplied by the weights to obtain a weighted representation of the feature. Finally, a convolution is performed to restore the shape of the feature and to output the results of feature extraction.
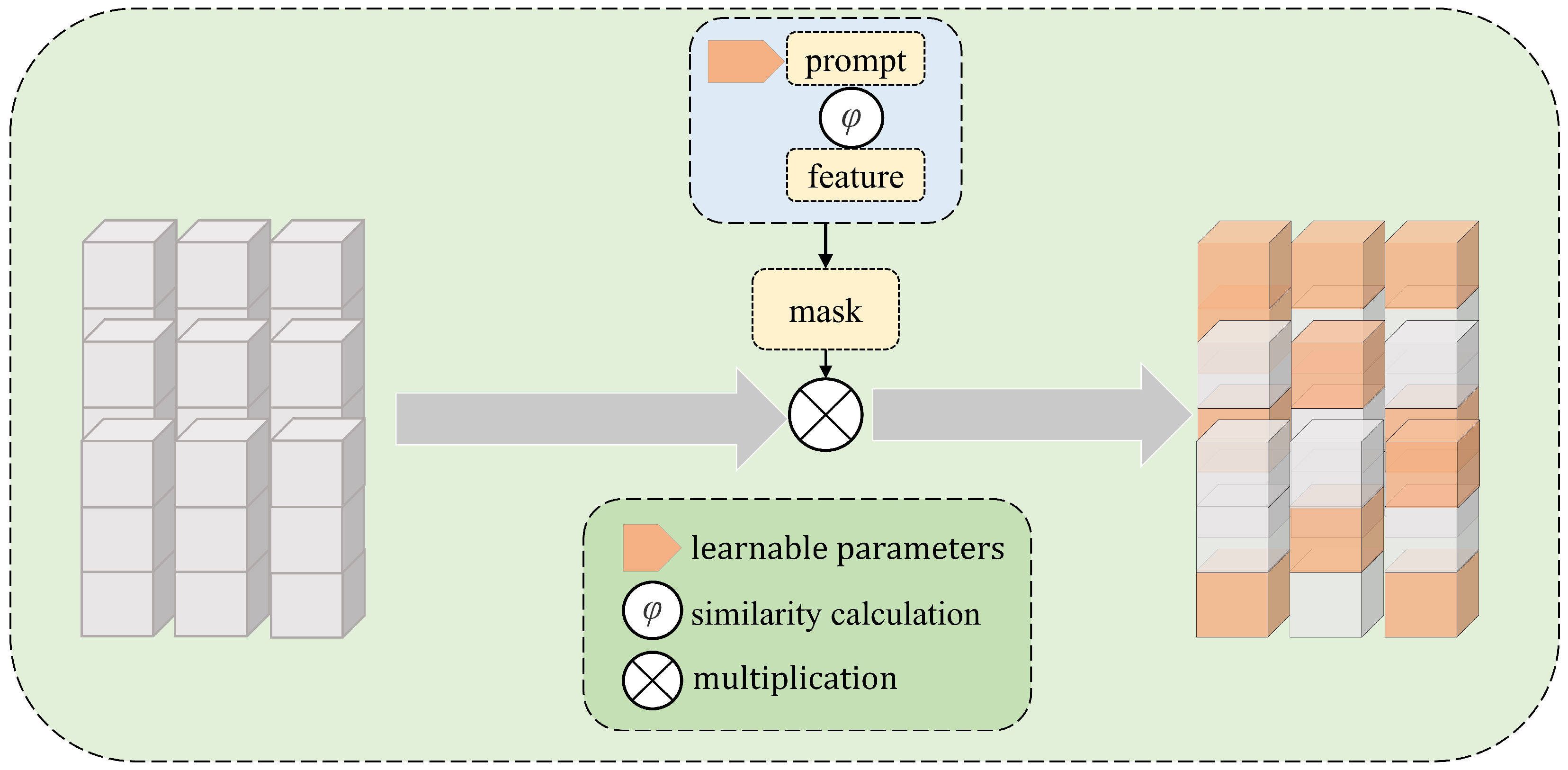
After feature extraction, the feature selection module is used to obtain effective features from the tokens. As shown in
Figure 4, the feature selection module generates a feature selection mask to weigh the image feature channels, highlighting important features and suppressing unimportant features. Specifically, a prompt parameter is introduced to perform a similarity comparison of the features to generate the mask. The mask then performs channel selection on the features, allowing the model to focus more effectively on key information in the input image. The prompt evolves from a randomly initialized tensor into parameters that can effectively select important features. This method leverages a combination of attention mechanisms and parameter optimization to enhance the feature representation capabilities of the model.
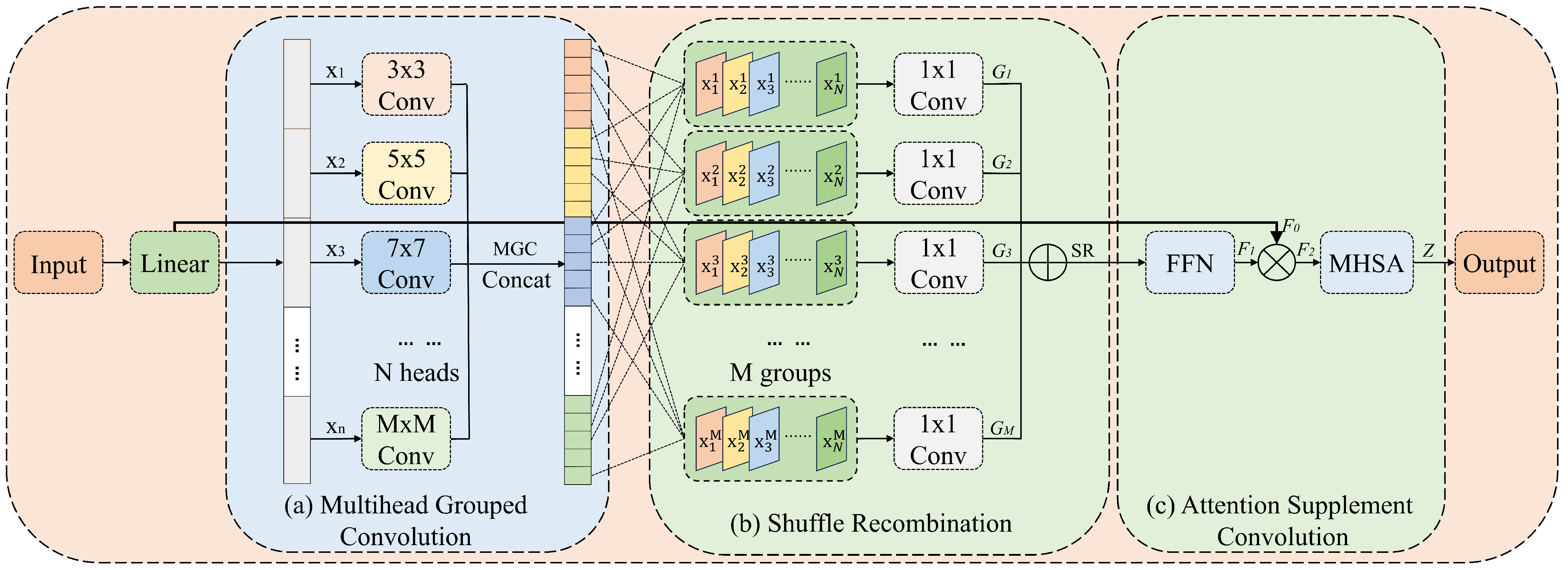
3.4. Recombined Channel Attention (RCA)
To fully integrate features from multiple branches, an RCA mechanism is proposed. This mechanism groups the features and uses convolution kernels of different sizes to capture various spatial features on multiple scales. In addition, channel reorganization can be applied to each group to enhance the diversity of multiscale features. Moreover, using large-kernel convolutions can expand the receptive field and improve the ability of the model to capture long-range dependencies. As shown in
Figure 5, the RCA module consists of three stages—namely, multi-head grouped convolution (MGC), shuffle recombination (SR), and attention supplement convolution (ASC).
3.4.1. Multi-Head Grouped Convolution (MGC)
As shown in
Figure 5a, MGC divides the input channels into
N heads and applies depth-wise separable convolutions with different kernel sizes to each head. A
convolution is used for the first head, and the kernel size is increased by two for each subsequent head. This approach allows the model to control the range of the receptive field and multiscale information by adjusting the number of heads. The proposed MGC can be expressed as follows:
where
represents the input image features divided into multiple heads along the channel dimension and
denotes the kernel size for each head.
3.4.2. Shuffle Recombination (SR)
To enhance information fusion between different groups, a grouping reorganization module called shuffle recombination (SR) is introduced. As shown in
Figure 5b, SR shuffles and recombines different fine-grained features. Specifically, a channel is selected from each head, and these channels form a new group that uses depth-wise convolution for feature fusion. Finally, the groups are concatenated along the channel. With a relatively small computational load, SR facilitates sufficient interaction between image features at different scales. The SR process can be expressed as follows:
where
is the depth-wise convolution operation and
denotes the
i-th channel of the
j-th head.
is the weight matrix of the point-wise convolution;
,
, where
N denotes the number of heads and
denotes the number of groups.
SR consists of M groups, where each group () is generated by the depth-wise convolution operation. Specifically, for the i-th channel of the j-th head, feature undergoes the depth-wise convolution operation, resulting in an output feature (). The features of the i-th channel from all heads comprise the weight matrix (), forming the i-th group (). Finally, all groups are concatenated to complete the SR operation.
3.4.3. Attention Supplement Convolution (ASC)
As shown in
Figure 5c, ASC feeds concatenated features into the FFN. Specifically, this FFN includes a convolutional layer, a normalization layer to accelerate network training and improve model stability, an activation function, and another convolutional layer. The feature maps processed by the FFN are multiplied by the features before grouping, helping the model better capture the nonlinear relationships and complex patterns in the data. Subsequently, a multi-head self-attention (MHSA) block is used for final feature processing. Its purpose is to complement the shortcomings of the convolution operation in global perception by effectively capturing long-range dependencies. Given an input feature of
, the features before grouping (
), and the intermediate states (
,
), we can compute the feature output (
Z) as follows:
where
denotes the output of the FFN, BN
denotes batch normalization operation,
denotes the result of element-wise multiplication between the feature maps and the original features, ⊗ denotes element-wise multiplication, and MHSA denotes the multi-head self-attention operation [
23].
3.5. Loss Function
During training, a combination of Dice and cross-entropy loss was used for the Synapse dataset. This hybrid loss function effectively addresses the problem related to class imbalance. For the ISIC-2018 dataset, only Dice loss was used for optimization. Dice loss
, cross-entropy loss
, and hybrid loss (
L) are defined as follows:
where
K denotes the number of classes;
and
denote the ground-truth label and predicted probability for class
k, respectively;
;
N denotes the number of pixels; and
denotes the weighting factor balancing the Dice loss and the cross-entropy loss. According to [
33],
and
are set to
and 0.6, respectively.
4. Experiments
The proposed MRCA-UNet model was trained using PyTorch 2.0.0 on an NVIDIA GeForce RTX 4090 GPU platform. The input image size was set to 224 × 224, and the initial learning rate was set to 0.05, using a multi-learning strategy. We trained and tested the proposed MRCA-UNet model on two publicly available medical image datasets—namely, the Synapse dataset for multi-organ segmentation and the ISIC-2018 challenge dataset. Both datasets are related to clinical diagnoses, and their segmentation results are beneficial for assisting in treatment. They comprise images and their corresponding ground-truth masks. These two datasets were used to effectively evaluate the accuracy and robustness of the proposed segmentation algorithm.
Table 1 shows detailed information for the medical segmentation datasets used in the experiments.
4.1. Synapse Multi-Organ Segmentation Comparison
The Synapse dataset contains 30 abdominal clinical CT images, totaling 3779 axial images. The training and testing datasets include 18 and 12 samples, respectively [
24,
31]. This dataset focuses on eight abdominal organs, that is, the aorta, gallbladder, spleen, left kidney, right kidney, liver, pancreas, and stomach. The evaluation metrics used in the experiment were the average Dice similarity coefficient (DSC) and the average Hausdorff distance (HD).The predictions can be separated into True Positive (TP), False Positive (FP), and True Negative (TN). DSC and HD can be described as follows:
where
Y denotes the ground-truth mask,
represents the segmentation map generated by the model, d(y,
) calculates the Euclidean distance between a specific point (y) (from the ground truth) and point
(from the prediction).
A comparison of the segmentation results of the proposed MRCA-UNet model and other methods on the Synapse dataset is shown in
Table 2. Note that the method indicated with an asterisk (*) represents the result reproduced in this work, which differs from the results reported in the original paper. The results indicate that the proposed model achieved accuracies of 81.61% in DSC (↑) and 23.36% in HD (↓). Compared with the other methods, the MRCA-UNet model achieved the second-best segmentation result in terms of DSC value, which is very close to the best result of MISSFormer [
33], and the best segmentation results for the kidneys (both left and right) and liver. For the kidneys, the model excels in extracting fine-grained features, and its global information processing is well suited for the liver, as incorporating information from surrounding tissues helps to better identify the boundaries of the liver. In addition, our model achieved the second-best segmentation results for the gallbladder and pancreas. This can be attributed to multiscale processing, which allows the model to better capture local information, particularly when it comes to handling the finer details of the gallbladder and pancreas.
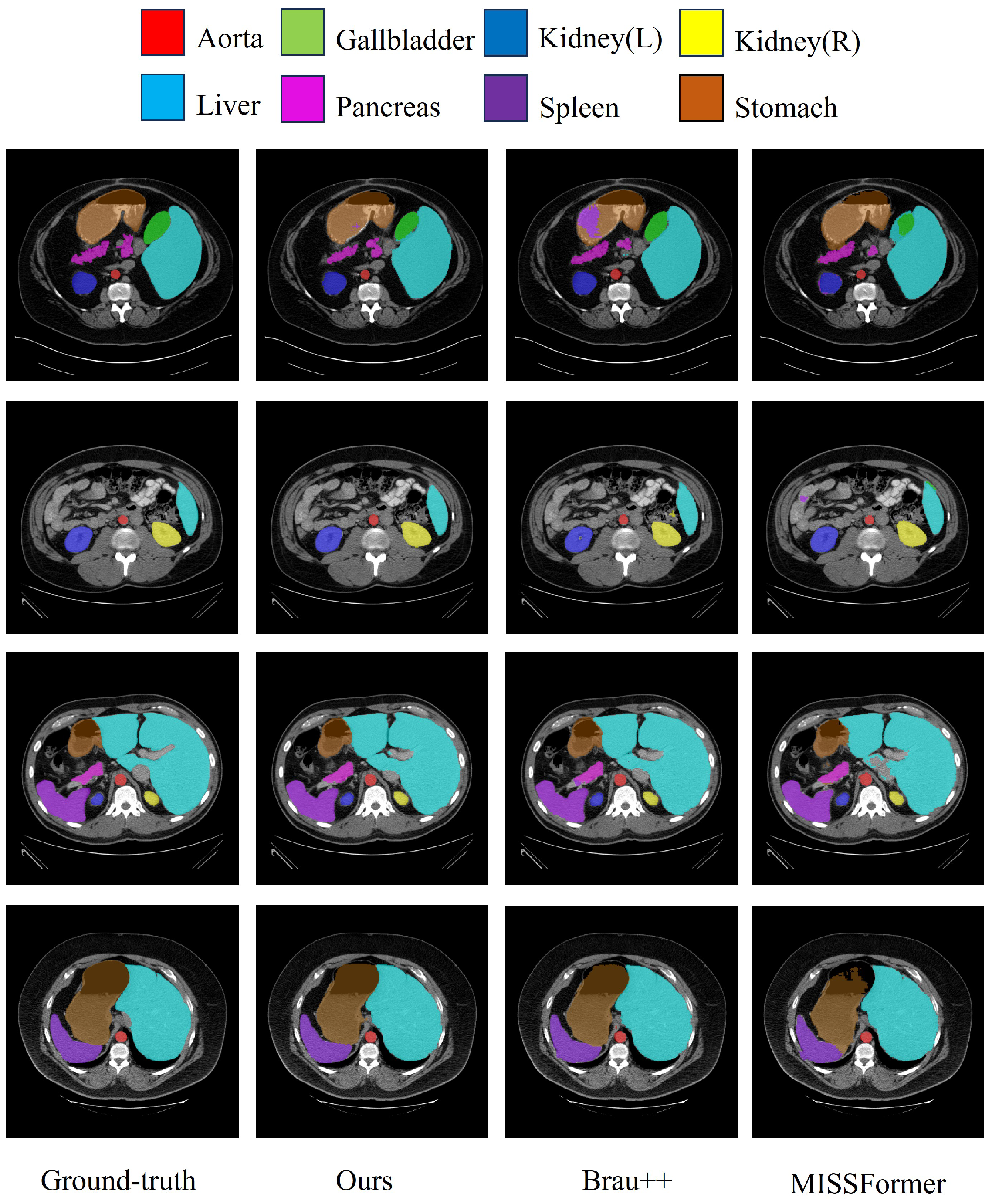
Figure 6 shows a visualization of the segmentation results for the Synapse multi-organ CT dataset.
4.2. ISIC-2018 Dataset Segmentation Results
The ISIC-2018 challenge lesion segmentation task tested the ability of the model to handle skin lesion analysis, covering a variety of skin lesion types. Professional dermatologists annotated the images to ensure data accuracy and reliability. The high image resolution helps the model to capture more detailed information. The ISIC-2018 dataset is significant in the field of automated skin lesion detection, providing researchers with a standardized benchmark dataset that allows for a fair comparison of different algorithms and models, thereby promoting the advancement of skin lesion analysis techniques. The IoU, accuracy, precision, and recall are calculated as follows:
This study explored the performance of skin lesion segmentation models using the ISIC-2018 dataset to provide new ideas and methods for the development of automated diagnostic tools. We reproduced all methods using publicly available code and present their results. The comparative results are shown in
Table 3. Compared with the other results, the MRCA-UNet model achieved an accuracy of 95.94% and an improvement of 0.33% over the BRAU-Net++ model, demonstrating that our model exhibited strong capabilities in general segmentation tasks and could effectively distinguish between lesion and non-lesion areas. Although MRCA-UNet does not outperform some advanced methods, such as BRAU-Net++, and Swin-Unet, in mIoU and DSC, it still delivers great results, surpassing most models. Thus, MRCA-UNet provides efficient and reliable segmentation results in medical segmentation tasks, offering strong support for applications requiring high classification accuracy.
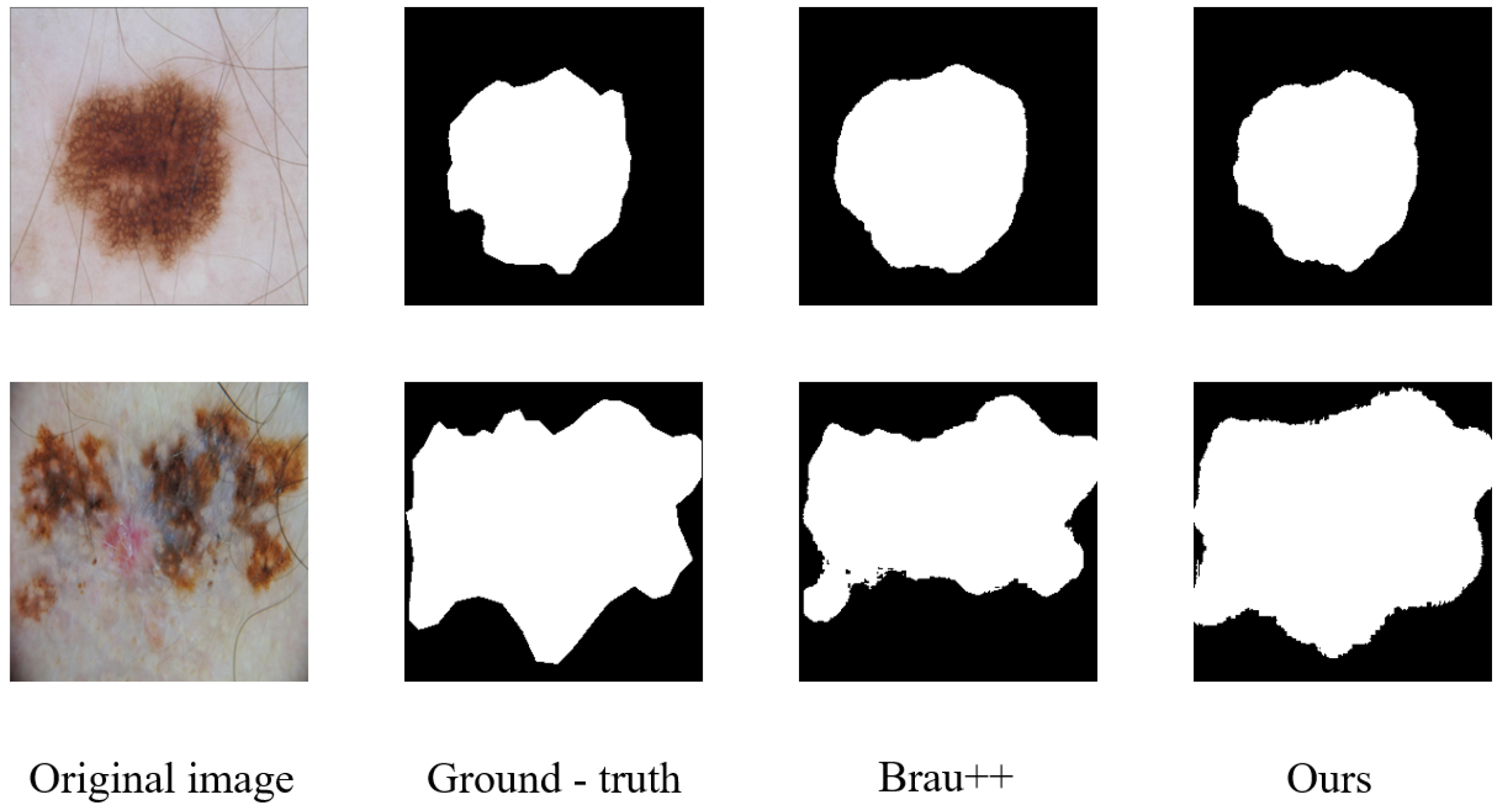
Figure 7 shows a visualization of the segmentation results for the ISIC-2018 dataset.
4.3. Ablation Study
To verify the effects of various factors on the MRCA-UNet model, we conducted ablation experiments on the Synapse dataset and the ISIC-2018 dataset to evaluate the effectiveness of each component. Specifically, we evaluated the effects of removing the multibranch feature extraction, channel reorganization, and ASC stages. We tested the model’s computational efficiency (including FLOPs, params, and FPS) on the Synapse dataset, using an input image size of
during testing.
Table 4 presents the detailed quantitative ablation results.
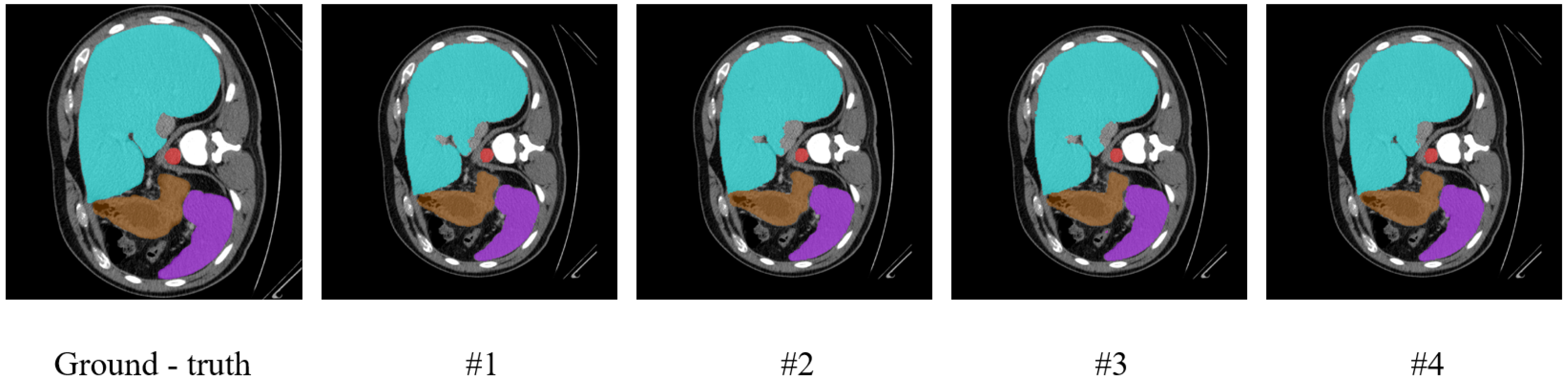
To further visually verify the effectiveness of the key components in our model, we conducted visualization experiments, the results of which are presented in
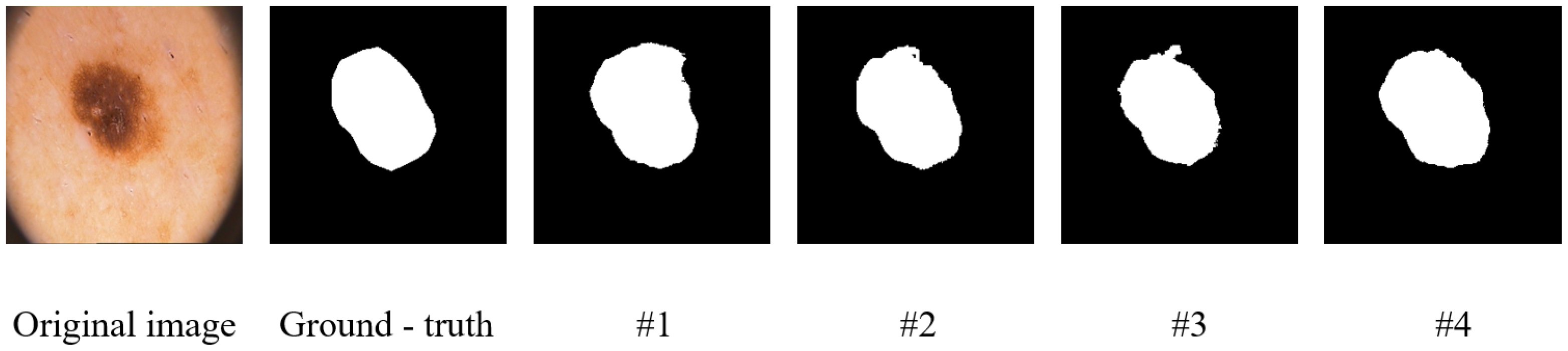
Figure 8 and
Figure 9. The figures show visualization maps of the segmentation results on the Synapse and ISIC-2018 datasets when different key components were removed (corresponding to settings #1, #2, and #3 in
Table 4), as well as for the complete MRCA-UNet model (#4). By comparing these visual results, the contribution of each module to the final segmentation effect can be more clearly understood.
4.3.1. The Effectiveness of the Multiscale Aggregation Module
Multibranch feature extraction is a crucial component of the MSA module that achieves multiscale information perception by extracting features locally and globally.
Table 4 shows the results for the Synapse dataset after removing the multibranch feature extraction stage and using the MRCA-UNet model with multibranch feature extraction. Specifically, we replaced multibranch feature extraction with a single-stream module. Compared to the single-stream module, multibranch feature extraction achieved better segmentation performance, with a DSC improvement of 1.68% and an HD decrease of 6.95%. The results indicate that multibranch feature extraction can more effectively capture information at different scales while focusing on both local details and global context. This enhances the model’s ability to perceive and process complex lesion structures, leading to more accurate boundary detection and better overall segmentation performance in segmentation tasks.
4.3.2. Effectiveness of Recombined Channel Attention
By grouping features and using convolutional kernels of different sizes, various spatial features could be captured at multiple scales, and channel reorganization could be performed to enhance the diversity of multiscale features. To study the effect of channel reorganization on segmentation, we removed the channel reorganization module. Specifically, regular attention computations were applied directly after linear projection. The results in
Table 4 show that removing the channel reorganization module from the Synapse dataset resulted in a DSC decrease of 0.94% and an HD increase of 0.86% compared to the MRCA-UNet model. Channel recombination plays a crucial role in optimizing the utilization of feature channels and enhancing the feature representation capability, thereby improving the model’s ability to handle features from different channels.
4.3.3. Effectiveness of Attention Supplement Convolution
To compensate for the shortcomings of the convolution operations in global perception, attention operations were used after the FFN. We trained and tested the MRCA-UNet model with and without attention operations on the Synapse dataset. The results in
Table 4 show that, with attention supplement convolution, the MRCA-UNet model improved the DSC value by 0.39% and decreased the HD value by 6.98%, indicating that the global perception of the attention operation can compensate for deficiencies in the model after convolution. These results indicate that the use of the attention module after the convolution operation significantly enhances the ability of the model to capture global features and effectively improves the accuracy of segmentation boundaries.
5. Discussion
The MRCA-UNet model proposed in this study, by integrating multiscale feature perception and channel attention mechanisms, achieves significant performance improvements in the field of medical image segmentation. It demonstrates particularly high segmentation accuracy when handling complex multi-organ and skin lesion segmentation tasks. However, during the research process, some issues requiring further investigation and refinement were also identified.
A notable concern regarding the MRCA-UNet model is its computational efficiency. The model comprises 70.32 million parameters, a scale that is significantly higher than that of other comparable models. The primary reason lies in the introduction of the multiscale aggregation (MSA) and recombined channel attention (RCA) modules within the model. While these modules enhance feature extraction and fusion capabilities, they inevitably increase the model’s complexity. For example, the MSA module extracts local and global features through multiple parallel branches, each requiring independent convolutional operations and parameter support, which undoubtedly increases the computational overhead. Meanwhile, the RCA module involves multiscale kernel operations and multi-head attention mechanisms, operations that also consume substantial computational resources. In practical applications, this characteristic of a high parameter count and computational load can lead to significantly longer training times and imposes stricter requirements on hardware resources. For instance, in resource-constrained medical environments, such as some primary healthcare institutions, access to high-performance GPU devices may be limited, which would greatly restrict the model’s applicability. Furthermore, in scenarios with high real-time requirements, such as image segmentation tasks in surgical navigation, the model’s high computational demands might prevent it from meeting real-time processing needs.
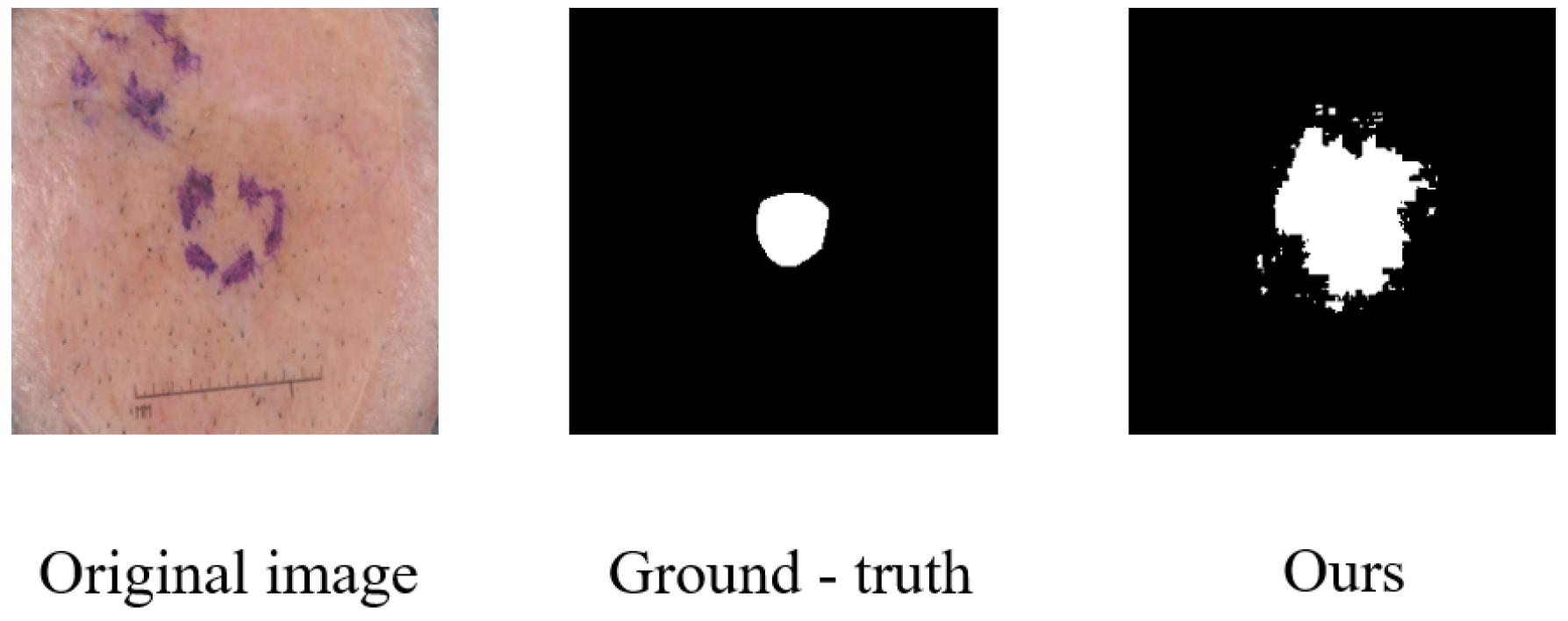
Beyond computational efficiency, another area identified for improvement relates to the model’s handling of fine boundary details in certain challenging cases. While MRCA-UNet demonstrates strong overall performance, we observed instances where precise boundary delineation could be further refined. A representative example is illustrated in
Figure 10. Although the model correctly identifies the general region of the skin lesion, the predicted segmentation mask exhibits noticeable over-segmentation and highly irregular, jagged edges compared to the smoother ground-truth boundary. This suggests that the model, despite its multiscale capabilities, may still struggle with lesions characterized by very subtle intensity transitions at the boundary or when fine textural details near the edge create ambiguity. Such inaccuracies in boundary precision, even if localized, can be critical in clinical applications requiring exact delineation. This highlights a need for future work focused specifically on enhancing boundary refinement mechanisms within the model architecture.
To address this issue, future research directions could focus on lightweight model design. On one hand, model compression techniques can be explored for the existing MRCA-UNet model, such as employing pruning techniques to remove redundant neurons or convolutional kernels, thereby reducing the parameter count and computational load. On the other hand, more efficient multibranch feature extraction and channel attention mechanisms could be designed, for instance, by incorporating techniques like deformable convolutions to reduce computational complexity. Additionally, model quantization techniques can be integrated to convert high-precision weight parameters into lower-precision representations, thereby further enhancing computational efficiency while potentially preserving model performance.
Finally, the application prospects of the model in three-dimensional (3D) medical image segmentation warrant exploration. Currently, most U-Net-based medical image segmentation models are primarily applied to two-dimensional (2D) tasks. However, in actual medical diagnosis, the application of 3D medical images (such as 3D CT and MRI scans) is becoming increasingly widespread. Three-dimensional image segmentation can provide more comprehensive anatomical information, aiding physicians in making more accurate diagnoses and treatment plans. However, directly applying the MRCA-UNet model to 3D image segmentation may face several challenges, such as limitations in computational resources and the high level of computational complexity associated with 3D convolutional operations. Future research could explore how to effectively extend the MRCA-UNet model to the 3D medical image segmentation domain, for example, by designing lightweight 3D convolutional modules or employing patch-based processing and fusion strategies to reduce computational costs. In summary, the MRCA-UNet model demonstrates significant potential in the field of medical image segmentation, but there are still issues that need to be addressed. Through further research and improvement, we anticipate overcoming these limitations and contributing further to the advancement of medical image segmentation technology.
6. Conclusions
This study proposes an MRCA-UNet framework for medical image segmentation. A novel multiscale aggregation module is employed to extract local and global features through multiple branches. Features from different receptive fields are fused by a recombined channel attention module. The restructured MRCA-UNet model not only perceives multiscale information but also fully utilizes channel features, which is crucial for accurately handling medical image segmentation tasks. Experiments on the Synapse and ISIC-2018 datasets demonstrated that the MRCA-UNet model can achieve good segmentation results in complex medical image segmentation tasks.
Despite the promising performance of MRCA-UNet on segmentation tasks, this study still has some limitations. One area requiring attention is the model’s computational efficiency. The model contains 70.32M parameters, a significant increase compared to other models. This increase in complexity is primarily attributed to the introduced multibranch aggregation module and the recombined channel attention mechanism. While these components enhance feature representation capabilities, they also significantly increase the model’s parameter count and computational load. This can lead to longer training times and higher demands on hardware resources and may potentially affect its deployment potential in real-time applications or resource-constrained environments.
Future work will focus on addressing the problems of model redundancy and parameter complexity to design more efficient medical image segmentation models.
Author Contributions
Conceptualization, L.L.; methodology, X.L.; software, X.L.; validation, J.W.; writing—original draft preparation, X.L.; writing—review and editing, L.L. and S.N.M.; investigation and analysis, S.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the Natural Science Foundation of Hebei Province (No. F2022201013); the Scientific Research Program of Anhui Provincial Ministry of Education (No. 2024AH051686); the Science and Technology Program of Huaibei (No. 2023HK037); the Anhui Shenhua Meat Products Co., Ltd., Cooperation Project (No. 22100084); and the Entrusted Project by Huaibei Mining Group (2023).
Data Availability Statement
The data used in this study is available from public datasets.
Conflicts of Interest
The authors declare that this study received funding from Anhui Shenhua Meat Products Co., Ltd., and Huaibei Mining Group. The funders were not involved in the study design; collection, analysis, or interpretation of data; the writing of this article; or the decision to submit it for publication.
References
- Félix, I.; Raposo, C.; Antunes, M.; Rodrigues, P.; Barreto, J.P. Towards markerless computer-aided surgery combining deep segmentation and geometric pose estimation: Application in total knee arthroplasty. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2021, 9, 271–278. [Google Scholar] [CrossRef]
- Srivastava, A.; Jha, D.; Chanda, S.; Pal, U.; Johansen, H.D.; Johansen, D.; Riegler, M.A.; Ali, S.; Halvorsen, P. MSRF-Net: A multi-scale residual fusion network for biomedical image segmentation. IEEE J. Biomed. Health Inform. 2021, 26, 2252–2263. [Google Scholar] [CrossRef] [PubMed]
- Elbatel, M.; Martí, R.; Li, X. FoPro-KD: Fourier prompted effective knowledge distillation for long-tailed medical image recognition. IEEE Trans. Med. Imaging 2024, 43, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted res-unet for high-quality retina vessel segmentation. In Proceedings of the 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018; pp. 327–331. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A nested u-net architecture for medical image segmentation. In Proceedings of the International Worshops on Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support (DLMIA 2018 and ML-CDS 2018), held in conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; pp. 3–11. [Google Scholar]
- Hou, Q.; Zhang, L.; Cheng, M.M.; Feng, J. Strip pooling: Rethinking spatial pooling for scene parsing. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 4003–4012. [Google Scholar]
- Huang, Z.; Wang, X.; Huang, L.; Huang, C.; Wei, Y.; Liu, W. CCNet: Criss-cross attention for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 603–612. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Zeng, Y.X.; Hsieh, J.W.; Li, X.; Chang, M.C. MixNet: Toward accurate detection of challenging scene text in the wild. arXiv 2023, arXiv:2308.12817. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-local neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7794–7803. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning (ICML), Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Yang, J.; Li, C.; Dai, X.; Gao, J. Focal modulation networks. In Proceedings of the 36th International Conference on Neural Information Processing Systems (NIPS), New Orleans, LA, USA, 28 November–9 December 2022; pp. 4203–4217. [Google Scholar]
- Hou, Q.; Lu, C.Z.; Cheng, M.M.; Feng, J. Conv2Former: A simple transformer-style convnet for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 8274–8283. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Fu, S.; Lu, Y.; Wang, Y.; Zhou, Y.; Shen, W.; Fishman, E.; Yuille, A. Domain adaptive relational reasoning for 3d multi-organ segmentation. In Proceedings of the 23rd International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Lima, Peru, 4–8 October 2020; pp. 656–666. [Google Scholar]
- Zhou, J.; Wang, P.; Wang, F.; Liu, Q.; Li, H.; Jin, R. ELSA: Enhanced local self-attention for vision transformer. arXiv 2021, arXiv:2112.12786. [Google Scholar]
- Wang, A.; Chen, H.; Lin, Z.; Han, J.; Ding, G. RepVit: Revisiting mobile cnn from vit perspective. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 16–22 June 2024; pp. 15909–15920. [Google Scholar]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. UNet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Lou, A.; Guan, S.; Loew, M. DC-UNet: Rethinking the U-Net architecture with dual channel efficient CNN for medical image segmentation. In Proceedings of the SPIE 11596, Medical Imaging 2021: Image Processing, Online, 15–19 February 2021; pp. 758–768. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.-Y.; et al. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 2–6 October 2023; pp. 4015–4026. [Google Scholar]
- Qi, M.; Zhu, P.; Li, X.; Bi, X.; Qi, L.; Ma, H.; Yang, M.-H. DC-SAM: In-context segment anything in images and videos via dual consistency. arXiv 2025, arXiv:2504.12080. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Tel Aviv, Israel, 23–27 October 2022; pp. 205–218. [Google Scholar]
- Huang, X.; Deng, Z.; Li, D.; Yuan, X. MISSFormer: An effective medical image segmentation transformer. arXiv 2021, arXiv:2109.07162. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Gai, S.; Darrell, T.; Wang, X. Refocusing is key to transfer learning. arXiv 2023, arXiv:2305.15542. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 4th International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Valanarasu, J.M.J.; Oza, P.; Hacihaliloglu, I.; Patel, V.M. Medical transformer: Gated axial-attention for medical image segmentation. In Proceedings of the 24th International Conference on Medical image computing and computer assisted intervention (MICCAI), 27 September–1 October 2021; pp. 36–46. [Google Scholar]
- Cai, P.; Lu, J.; Li, Y.; Lan, L. Pubic symphysis-fetal head segmentation using pure transformer with bi-level routing attention. arXiv 2023, arXiv:2310.00289. [Google Scholar]
- Xu, Q.; Ma, Z.; Na, H.; Duan, W. DCSAU-Net: A deeper and more compact split-attention U-Net for medical image segmentation. Comput. Biol. Med. 2023, 154, 106626. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Framework of the proposed MRCA-UNet. MRCA-UNet comprises three parts: the encoder, decoder, and skip connections. Both the encoder and decoder are built based on the MRCA module.
Figure 1.
Framework of the proposed MRCA-UNet. MRCA-UNet comprises three parts: the encoder, decoder, and skip connections. Both the encoder and decoder are built based on the MRCA module.
Figure 2.
The MSA module processes image features through multiple branches, performing feature selection after feature extraction to ensure the model focuses on more effective tokens.
Figure 2.
The MSA module processes image features through multiple branches, performing feature selection after feature extraction to ensure the model focuses on more effective tokens.
Figure 3.
Feature extraction module. The features are divided into patches, and based on the number of divided patches, either local or global feature extraction is performed.
Figure 3.
Feature extraction module. The features are divided into patches, and based on the number of divided patches, either local or global feature extraction is performed.
Figure 4.
Feature selection module. The model automatically adjusts the value of the prompt based on the training data. A similarity calculation between the learnable parameters and the image features generates a mask, which is used for weighted feature selection.
Figure 4.
Feature selection module. The model automatically adjusts the value of the prompt based on the training data. A similarity calculation between the learnable parameters and the image features generates a mask, which is used for weighted feature selection.
Figure 5.
The recombined channel attention consists of three stages: (a) multi-head grouped convolution, (b) shuffle recombination, and (c) attention supplement convolution.
Figure 5.
The recombined channel attention consists of three stages: (a) multi-head grouped convolution, (b) shuffle recombination, and (c) attention supplement convolution.
Figure 6.
Segmentation results on the Synapse multi-organ CT dataset.
Figure 6.
Segmentation results on the Synapse multi-organ CT dataset.
Figure 7.
Segmentation results on the ISIC-2018 dataset.
Figure 7.
Segmentation results on the ISIC-2018 dataset.
Figure 8.
Visualization results of the ablation study on the Synapse dataset.
Figure 8.
Visualization results of the ablation study on the Synapse dataset.
Figure 9.
Visualization results of the ablation study on the ISIC-2018 dataset.
Figure 9.
Visualization results of the ablation study on the ISIC-2018 dataset.
Figure 10.
Visual illustration of the boundary delineation limitations of MRCA-UNet.
Figure 10.
Visual illustration of the boundary delineation limitations of MRCA-UNet.
Table 1.
Details of the medical segmentation datasets used.
Table 1.
Details of the medical segmentation datasets used.
| Dataset | Input Size | Total | Train | Valid | Test |
|---|
| Synapse | | 3379 | 2212 | 1167 | - |
| ISIC-2018 | | 2594 | 1868 | 467 | 259 |
Table 2.
Comparison of various methods for segmentation on the Synapse dataset.
Table 2.
Comparison of various methods for segmentation on the Synapse dataset.
| Method | Params (M) | DSC (↑) | HD (↓) | Aorta | Gallbladder | Kidney (L) | Kidney (R) | Liver | Pancreas | Spleen | Stomach |
|---|
| V-Net [35] | - | 68.80 | - | 75.34 | 51.87 | 77.10 | 80.75 | 87.84 | 40.05 | 80.56 | 56.98 |
| DARR [24] | - | 69.77 | - | 74.74 | 53.77 | 72.31 | 73.24 | 94.08 | 54.18 | 89.90 | 45.96 |
| R50 U-Net [4] | - | 74.68 | 36.87 | 87.74 | 63.66 | 80.60 | 78.19 | 93.74 | 56.90 | 85.87 | 74.16 |
| U-Net [4] | 14.80 | 76.85 | 39.70 | 89.07 | 69.72 | 77.77 | 68.60 | 93.43 | 53.98 | 86.67 | 75.58 |
| Attn-UNet [6] | 34.88 | 77.77 | 36.02 | 89.55 | 68.88 | 77.98 | 71.11 | 93.57 | 58.04 | 87.30 | 75.75 |
| R50 ViT [4] | - | 71.29 | 32.87 | 73.73 | 55.13 | 75.80 | 72.20 | 91.51 | 45.99 | 81.99 | 73.98 |
| TransUNet [31] | 105.28 | 77.48 | 31.69 | 87.23 | 63.13 | 81.87 | 77.02 | 94.08 | 55.86 | 85.08 | 75.62 |
| Swin-UNet [32] | 27.17 | 79.13 | 21.55 | 85.47 | 66.53 | 83.28 | 79.61 | 94.29 | 56.58 | 90.66 | 79.60 |
| MISSFormer [33] | 42.46 | 81.96 | 18.20 | 86.99 | 68.65 | 85.21 | 82.00 | 94.41 | 65.67 | 91.92 | 80.81 |
| Brau-Net++ * [7] | 50.76 | 80.00 | 26.16 | 87.86 | 65.17 | 84.92 | 81.09 | 94.80 | 59.23 | 88.32 | 78.64 |
| MRCA-UNet (Ours) | 70.32 | 81.61 | 23.36 | 87.00 | 69.50 | 87.12 | 82.10 | 94.84 | 65.22 | 89.93 | 77.19 |
Table 3.
Segmentation comparison of various methods on the ISIC-2018 dataset.
Table 3.
Segmentation comparison of various methods on the ISIC-2018 dataset.
| Method | mIoU (↑) | DSC (↑) | Accuracy (↑) | Precision (↑) | Recall (↑) |
|---|
| U-Net [4] | 80.21 | 87.45 | 95.21 | 88.32 | 90.60 |
| Attn U-Net [6] | 80.80 | 86.31 | 95.44 | 91.52 | 89.01 |
| MedT [36] | 81.43 | 86.92 | 95.10 | 90.56 | 89.93 |
| TransUNet [31] | 77.05 | 84.97 | 94.56 | 84.77 | 89.85 |
| Swin-Unet [32] | 81.87 | 87.43 | 95.44 | 90.97 | 91.28 |
| BRAU-Net [37] | 82.81 | 89.32 | 95.10 | 90.27 | 92.25 |
| DCSAU-Net [38] | 82.17 | 88.74 | 94.75 | 90.93 | 90.98 |
| BRAU-Net++ [7] | 84.01 | 90.10 | 95.61 | 91.18 | 92.24 |
| MRCA-UNet (Ours) | 80.65 | 88.14 | 95.94 | 90.66 | 89.37 |
Table 4.
Comparison of segmentation performance between MRCA-UNet and models with individual modules removed.
Table 4.
Comparison of segmentation performance between MRCA-UNet and models with individual modules removed.
| | Method | Synapse | ISIC-2018 | Efficiency |
|---|
| DSC (↑) | HD (↓) | mIoU (↑) | Flops (G) | Params (M) | FPS (↑) |
|---|
| #1 | Without multibranch feature extraction | 79.93 | 30.31 | 77.78 | 25.51 | 59.98 | 20.33 |
| #2 | Without grouped recombination | 80.67 | 24.22 | 79.01 | 26.30 | 63.85 | 27.10 |
| #3 | Without attention | 81.22 | 30.34 | 79.90 | 23.68 | 57.18 | 21.21 |
| #4 | MRCA-UNet | 81.61 | 23.36 | 80.65 | 28.41 | 70.32 | 16.03 |
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).