1. Introduction
Body Mass Index (BMI) is a commonly used indicator to measure the degree of obesity, classifying people as underweight, normal weight, overweight, or obese [
1,
2]. In recent years, research focused on the prediction of BMI has attracted increasing attention. Traditionally, BMI is obtained by measuring an individual’s height and weight. However, this method may encounter multiple challenges due to the physical limitations of certain patients. To address this issue, vision-based BMI measurement methods have emerged as a viable solution [
3]. Previous studies have proposed predicting BMI based on facial images by analyzing the relationship between facial features and BMI [
4,
5,
6]. Xiang et al. [
7] proposed a three-dimensional deep model named STNet for visual BMI estimation. However, compared to 2D body images, clear facial images are generally more challenging to obtain due to privacy protection concerns. In addition, BMI prediction based on facial images also faces certain accuracy challenges. To overcome these challenges, some researchers have turned to using 3D human body data or RGB-D images to calculate BMI [
8,
9]. Although these methods can estimate BMI with reasonable accuracy under certain conditions, the acquisition of RGB-D images is highly sensitive to sunlight exposure and measurement distance, making it difficult to obtain high-quality 3D human body data in outdoor environments.
Symmetry refers to the invariance of an object or system under certain transformations, such as reflection, rotation, or translation. This concept has wide-ranging applications across multiple disciplines, from physics and chemistry to biology and engineering. By incorporating symmetry into our analysis, we can uncover deeper patterns and relationships within complex systems, leading to a more comprehensive understanding of the phenomena under investigation.
With the continuous development of computer vision and image-processing technology, some scholars have utilized 3D human reconstruction methods [
10] to extract 3D anthropometric features, such as waist circumference and hip circumference, from 2D images and have then used regression methods to estimate BMI [
11]. These 3D anthropometric features have been proven by Coetzee et al. [
12] to be closely related to BM, and can also be predicted using parametric models. However, learning from these 3D anthropometric features still cannot strongly represent body shape information. This is the problem with the incomplete consideration of anthropometric features.
Recently, numerous health science studies have shown that certain 2D anthropometric indicators are closely related to BMI [
13,
14]. However, most 2D anthropometric measurement features, such as waist-to-thigh width ratio and hip-to-head width ratio, only represent the lateral proportions of the body and lack longitudinal information.
With the development of computational power, big data and deep learning, particularly Convolutional Neural Networks (CNNs), have made significant progress in the field of image processing [
15]. A deep network model that extracts deep features has been applied to BMI estimation tasks; Sui et al. [
11] proposed a three-branch BMI estimation method that integrates hand-defined 3D anthropometric features with features extracted using a CNN model to improve the accuracy of BMI estimation. It is evident that deep features can effectively represent the semantic characteristics of images and enhance the accuracy of BMI estimation.
Therefore, to address these issues, such as the incomplete consideration of anthropometric features, the neglect of multiplex networks, and the low performance of BMI estimation, we propose a BMI estimation method based on multiplex networks. In the proposed method, 3D anthropometric features, 2D anthropometric features, and deep features are extracted by constructing a multichannel network, and BMI estimation is performed by Kernel Ridge Regression (KRR). The 3D anthropometric extraction network calculates 3D anthropometric characteristics, such as waist circumference (WC), hip circumference (HC), and the ratio of waist circumference to hip circumference (WCHCR), using human pose estimation [
16] and the reconstruction of 3D body shapes. The 2D anthropometric extraction network computes 2D anthropometric features, including the waist-to-thigh width ratio (WTR), the waist-to-hip width ratio (WHpR), the waist-to-head width ratio (WHdR), the hip-to-head width ratio (HpHdR), the number of pixels per unit area between the waist and hip (Area), the ratio of the distance from nose to knee to waist (HWR), the waist-to-width ratio (WSR), and the ratio of the distance from nose to knee to hip (H2H), through skeleton joint detection [
17] and body contour detection [
18]. These features comprehensively represent human body information. The deep learning extraction network extracts deep features from images using the attention-enhanced VGG model proposed in this paper. The experimental results indicate that the proposed method improves the accuracy of BMI prediction on public datasets while enhancing estimation accuracy without relying on additional information. The main contributions of this research are summarized as follows:
- (1)
To address the issue of insufficient consideration of anthropometric features in existing methods, three 3D anthropometric features, one 2D anthropometric feature, and a deep feature extraction method are proposed.
- (2)
A framework based on multiplex networks is proposed for estimating BMI from a single body image, addressing the neglect of multiplex networks and outperforming conventional single-network designs.
- (3)
A comprehensive comparison between the state-of-the-art methods and extensive ablation studies is conducted to verify the effectiveness of the proposed method.
3. Proposed BMI Estimation Framework
Figure 1 illustrates the framework of the proposed method. In this method, the 3D anthropometric feature, 2D anthropometric feature, and deep feature are extracted using multiplex networks, and BMI estimation is performed via KRR. Specifically, the input body image is processed by three major networks: the 3D anthropometric feature extraction network, which calculates 3D features through human pose estimation, 3D human shape reconstruction, virtual measurements, the 2D anthropometric feature extraction network, which computes 2D features, and the deep learning feature extraction network, which extracts deep features from the image. Finally, the three types of features are concatenated and mapped to BMI using a regression model.
3.1. Three-Dimensional Anthropometric Feature Extraction
In the process of extracting 3D anthropometric features, data preprocessing is first performed on the input RGB image using OpenPose to obtain human pose information. OpenPose was chosen for its real-time performance and robustness in detecting human poses across diverse datasets and scenarios. Its widespread adoption and community support facilitated seamless integration into our pipeline. Although alternative methods like AlphaPose [
38] offer similar capabilities, OpenPose’s extensive documentation and pre-trained models make it a more practical choice for implementation in our research.
Then, the SMPL-X [
39] model was employed for 3D human reconstruction, acquiring parameters such as body shape, body offsets, and pose. SMPL-X provides a high-fidelity parametric model that captures not only body shapes and poses but also facial expressions and hand gestures. This level of detail is crucial for our application, where generating realistic and detailed human models is essential. Compared to other models, like SMPL [
40], SMPL-X’s ability to model the full body, including hands and face, made it the preferred choice. SMPL-X is a 3D body shape reconstruction model that computes body poses, hand poses, and facial expressions from a single body image. It maps shape parameters
, pose parameters
, and expression parameters
to a 3D mesh M with N = 10,475 vertices and K = 54 joints. SMPL-X includes joints of the neck, jaw, eyeball, and fingers. The shape parameter
(
) has coefficients in a low-dimensional PCA space, which are calculated as follows:
where
is the pose parameter,
.
K is the number of keypoints,
is the shape parameter
, and
is the expression parameter
.
Finally, 3D anthropometric features are calculated using the Shapy method [
41], which leverages linguistic annotations to improve the accuracy of 3D shape reconstruction. In this study, after obtaining parameters such as body shape, body offset, and pose, WC and HC are calculated, followed by the HCHCR. The three 3D anthropometric features are calculated as follows:
(1) WC: The waist circumference is calculated as follows:
where
i and
j are two points in the convex hull and
is the SMPL-X shape parameter.
are the coordinates of the waist landmark points, and
is the body triangle index.
denotes the Euclidean norm, and
and
represent transformation matrices associated with the body triangle indices
and
.
(2) HC: The hip circumference is calculated given the coordinates of the hip landmarks
and the body triangle index
. The HC can be calculated according to Equation (
2), where the hip landmark points replace the waist landmark points in the calculation.
(3) WCHCR: The waist circumference-to-hip circumference ratio, which is based on the circumference of the waist and hips, can be calculated using the following formula:
3.2. Two-Dimensional Anthropometric Feature Extraction
Two-dimensional anthropometric feature extraction involves skeleton joint detection, human contour detection, and feature calculation. First, skeleton joint detection is performed using Mask R-CNN [
17]. Mask R-CNN was selected for its state-of-the-art instance segmentation capabilities, which are essential to isolate and analyze specific regions of interest in our study. Compared with a traditional two-stage method, such as R-FCN [
42], Mask R-CNN’s performance is better. To meet the study’s requirements, the detected skeleton joints were converted from the original mask to a binary mask, where non-zero pixels indicate the exact location of the skeleton joints. To ensure accurate body part positioning, 17 key skeletal joints were selected for detailed contour detection [
17], as shown in
Figure 1. Accurate skeleton joints improve the detection accuracy of human contours and perform well even in occluded body images.
Secondly, human contour detection was performed using Pose2Seg [
18], a human instance segmentation-based approach that separates instances based on human poses rather than proposal region detection, thereby reducing the influence of pose and occlusion. Pose2Seg leverages pose information to improve segmentation accuracy, which is particularly beneficial for human contour detection. However, during 2D anthropometric feature calculation, it was found that waist width measurements may include the width of the detected arm, particularly when the arm is naturally positioned next to the body. This can lead to the overestimation of waist width, negatively impacting BMI estimation accuracy. To address this issue, a self-correcting human parsing method was employed to subtract the arm regions from the contour map [
43], as shown in
Figure 1.
Finally, after skeleton joint detection and human contour detection, the next step was to calculate 2D anthropometric features. Jin et al. [
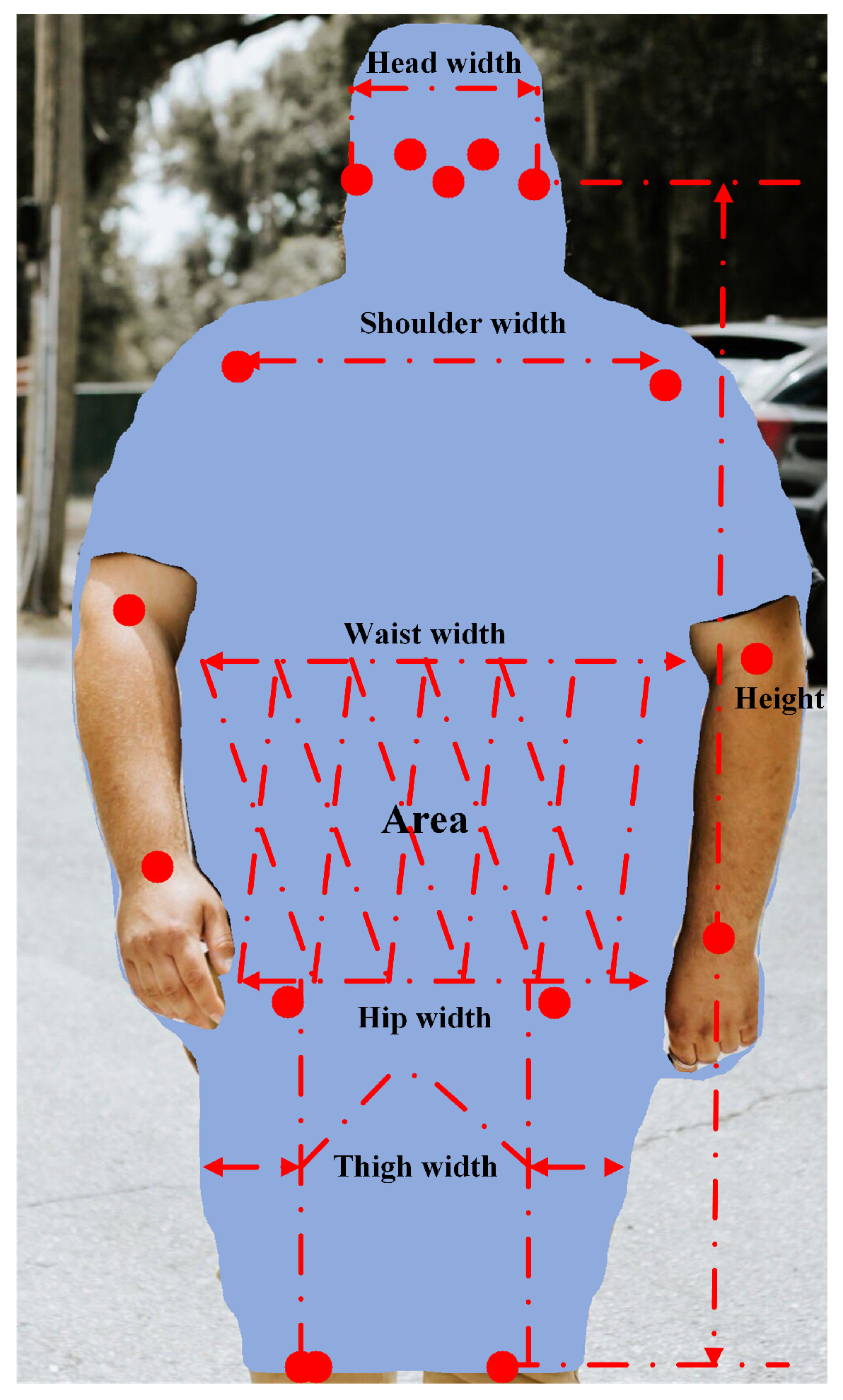
29] provided calculation methods for seven anthropometric features: WSR, WTR, WHpR, WHdR, HpHdR, Area, and HWR. The definitions of these features are as follows: WTR refers to the waist width-to-thigh width ratio; WHpR denotes the waist width-to-hip width ratio; WHdR is the waist width-to-head width ratio; HpHdR represents the hip width-to-head width ratio; Area indicates the number of pixels per unit area (noted by P) between the waist and hip; HWR corresponds to the ratio of the distance from nose to knee to the waist width; and WSR represents the waist width-to-shoulder width ratio. Please see
Figure 2 for more details.
The formula for H2H proposed in this study is as follows:
where
is the Euclidean distance from the nose to the knee,
is hip circumference, the nose coordinate point is
, the knee coordinate point is
, the left knee coordinate point is
, and the right knee coordinate point is
.
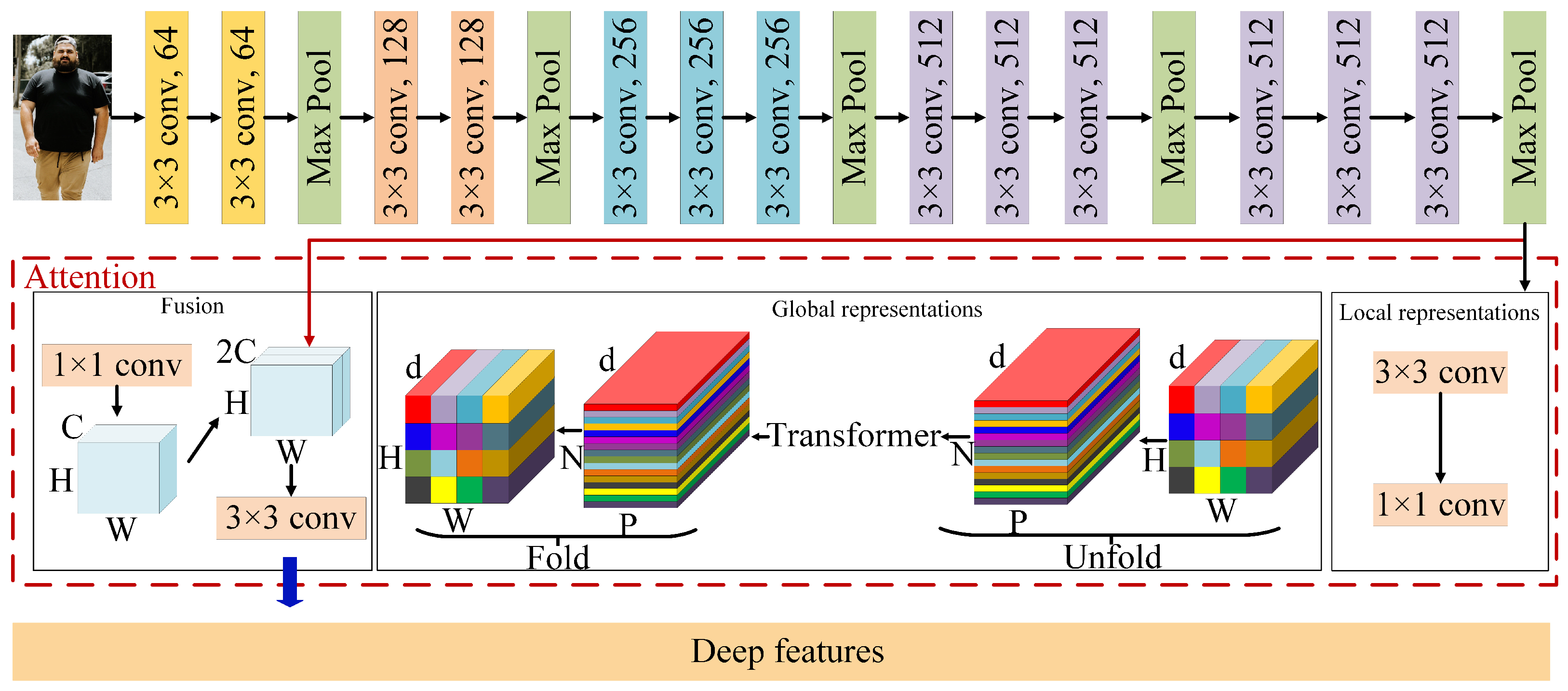
3.3. Deep Feature Extraction
Deep feature extraction employs VGG as the backbone network, which has been shown to significantly improve performance in image classification, object detection, and segmentation [
44]. In this study, an attention-enhanced VGG model is proposed to extract deep features, as shown in
Figure 3. The proposed model in this study is based on the VGG model and incorporates the MobileViT attention mechanism [
45], enhancing the network’s focus on important image regions and improving its feature extraction capability. The MobileViT attention mechanism works by selectively emphasizing informative features while suppressing less relevant ones. This selective focus allows the model to better understand the spatial relationships between different parts of the body, which is crucial for accurate BMI estimation from body images.
The attention-enhanced VGG network model proposed in this study utilizes VGG pre-trained weights on ImageNet [
46] to extract more image features and initialize network parameters effectively. Then, it is trained on the proposed dataset to accomplish a specific task. Additionally, the number of outputs in the last fully connected layer is modified from 1000 to a length of 10 for the deep feature DFs.
3.4. Feature Concatenation
BMI estimation can be defined as a regression problem that maps deep and anthropometric features to BMI. In this study, this paper uses regression models for BMI estimation. First of all, each image, given eight 2D anthropometric features, three 3D anthropometric features, and eleven anthropometric features, where F = [WSR, WTR, WHpR, WHdR, HpHdR, Area, HWR, H2H, WC, HC, WCHCR], undergoes normalized processing. Then, the deep feature vector DFs and the anthropometric feature vector F are concatenated into the KRR model to produce more accurate results.
3.5. Implementation Detail
The attention-enhanced VGG employs a VGG16 backbone with pre-trained weights. To align images of different sizes, the images are first scaled diagonally, resizing the longer side to 224. The shorter side is then padded with zeros (zero-padding) until the final image size is 224 × 224. Since the images are scaled diagonally, the short and long sides are adjusted proportionally while maintaining their original ratio. The input image tensor is normalized using mean values [0.485, 0.456, 0.406] and standard deviation values [0.229, 0.224, 0.225] before feeding into the network. The Adam optimizer [
47], with a learning rate of 0.0001, a batch size of 32, and 100 training epochs, is adopted to update the weights, and the Mean Square Error (MSE) is used as the loss function.
5. Conclusions
In this study, we propose three 3D anthropometric features, one 2D anthropometric feature, and a deep feature extraction method to address the incomplete consideration of anthropometric features. In addition, a BMI estimation method based on a multiplex network is proposed. In this method, three types of features are extracted by constructing a multiple network, and BMI estimation is performed using KRR. Compared with existing state-of-the-art methods, the proposed method significantly improves performance. These findings highlight the potential of our framework for practical applications in health informatics, such as remote health monitoring and personalized healthcare.
This study currently has the following limitations: First, the diversity of the dataset remains constrained by collection conditions, with insufficient coverage of extreme BMI values and special populations. Second, the simulation of complex environmental factors in real-world scenarios, such as lighting variations and occlusions, has not yet been systematically addressed. Third, although the model enhances resistance to image quality interference through a multi-scale feature fusion mechanism, its sensitivity to low-resolution images still requires further optimization. To address these issues, future work will focus on the following key improvements: First, a cross-regional, multimodal BMI dataset that incorporates medical-grade measurement device data should be constructed to strengthen the reliability of biometric characteristics. Second, an adaptive environmental perception module should be developed to improve stability in complex scenarios through domain generalization techniques. These enhancements will systematically elevate the clinical utility of the BMI estimation model in real-world medical applications.