1. Introduction
Missing values frequently occur in data analysis due to various factors such as incomplete manual data entry, measurement errors, or sensor malfunctions [
1]. These unobserved values cannot be directly used in analyzing the characteristics of the data, potentially degrading model performance or even preventing the application of subsequent procedures [
2,
3]. As a result, most studies considered missing value imputation (MVI) before conducting their analysis, indicating that an accurate imputation is essential for reliable analysis and modeling [
4]. Although a variety of naïve MVI methods, such as mean imputation [
5], have been widely applied, these methods refer to simple approaches that fill in missing values based solely on the statistical properties of observed data and often fail to capture complex feature interactions [
6]. In contrast, neural network (NN)-based approaches have recently attracted significant attention due to their ability to effectively infer missing values from incompletely observed inputs, resulting in high imputation accuracy [
7].
The strategy to impute the missing values can be divided into chain and non-chain approaches. Among them, the chain approach can be regarded as an asymmetric process because the chain approach iteratively imputes feature values and progressively adds the newly imputed features into the training dataset. As a result, the input feature space grows step by step—similar to a natural number sequence (1, 2, 3, 4, 5, …), which introduces an asymmetric structure. In contrast, non-chain approaches can be regarded as symmetric approaches due to maintaining a fixed number of features throughout the process, resulting in a geometrically symmetric training dataset. For example, if we plot the number of features used in each iteration, the chain-based approach forms a right triangle as the feature set grows progressively over time. In this regard, non-chain imputers maintain a constant number of features across iterations, resulting in a rectangular shape. The advantage of the asymmetric chain approach is that it is more focused and detailed since each iteration only tries to impute one feature while using other already imputed features. This can lead to more accurate results at each step. In contrast, the symmetric non-chain approach looks at the whole dataset at once, so it might struggle to predict accurate values because it includes noisy or irrelevant features in the input.
In conventional NN-based MVI methods, the missing values are replaced with the outputs of the NN [
8]. To train the NN for missing value prediction, all the features except the target feature are used as a training dataset. Because the training dataset might also contain missing values, a naïve MVI method, such as mean imputation, is typically applied before the training process since conventional NN-based methods cannot handle missing values directly. However, such an approach can lead to inaccurate predictions, as the model is trained on a dataset containing roughly estimated values. This issue becomes more severe when the original dataset contains a large number of missing values, as the NN fits into a significantly distorted dataset.
Ideally, all the distorted features should be excluded from the training dataset to avoid an inaccurate NN model, which can be achieved by considering only features without missing values. However, in practice, there is no guarantee that there will always be a sufficient number of features without missing values, especially when the original dataset contains a large amount of missing data. As an alternative, feature imputation can be performed as an iterative process, where features carefully imputed by the neural network in previous iterations are included in the training dataset to impute the values of the next feature. This approach helps to minimize distortion while ensuring a sufficient quality of features. Despite the widespread use of advanced data generation mechanisms in NN-based MVI methods, such as autoencoders [
9,
10,
11,
12], generative adversarial networks (GANs) [
13,
14,
15,
16], and attention mechanisms [
17,
18,
19], the aforementioned chain approach, although widely used in multilabel learning (e.g., classifier chains), has been rarely explored in NN-based MVI studies.
In this study, we focus on an effective MVI method following asymmetric strategy. Specifically, the proposed method first identifies the features without missing values from the original dataset. Then, the employed NN is trained on the selected features, considering the next feature with missing values as a target. After the target feature is imputed, it is added to the input of the subsequent imputation steps. The experimental results and statistical tests on 25 publicly available datasets showed that the proposed method significantly outperforms the conventional methods regarding missing value prediction accuracy. The implementation is accessible via GitHub at
https://github.com/KhrTim/ChainImputer, accessed on 18 May 2025, supporting reproducibility and further research.
The remainder of this paper is organized as follows.
Section 2 reviews existing approaches to missing value imputation, including classical, neural, attention-based, and diffusion-based methods. Furthermore, we describe our proposed solution and provide the mathematical foundation and time complexity analysis in
Section 3.
Section 4 contains an experimental evaluation of our method in comparison with counterparts. Finally, in
Section 5, we provide additional experimental results comparing variations of the proposed method.
2. Related Work
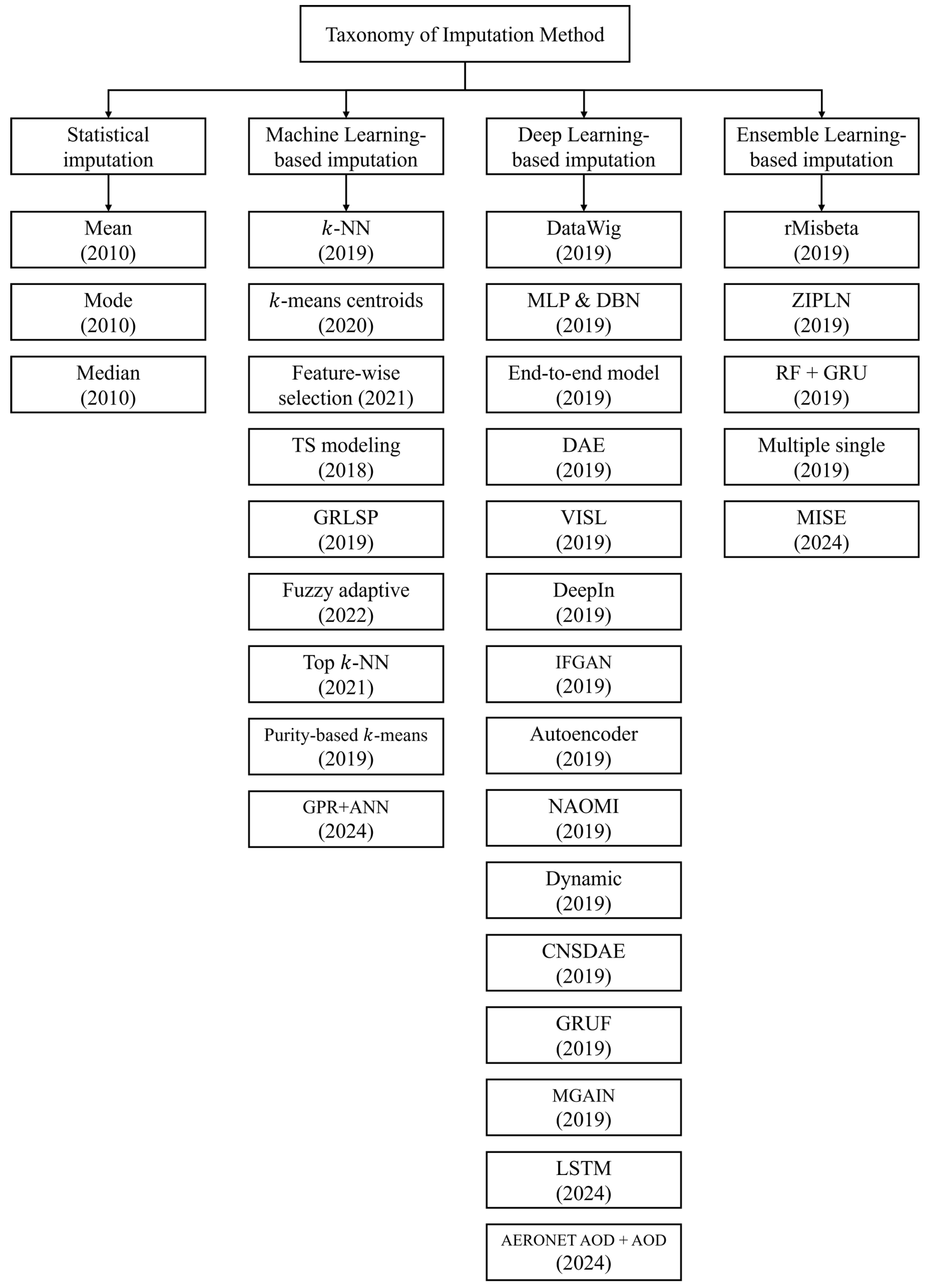
As MVI is a steady topic in machine learning, statistical imputation methods are widely used for handling missing data.
Figure 1 shows a taxonomy of the imputation methods. For example, in the work of [
20], statistical methods are chosen for their simplicity. Mean, mode, and median imputations are simple techniques that fill in missing values by computing the corresponding statistic from the observed values within each feature. Statistical imputation methods offer the advantage of being fast and straightforward in imputing missing values. However, the drawback is that they tend to underestimate variance and ignore relationships with other variables, making it challenging to impute accurate values [
5].
Machine learning-based imputation involves constructing predictive models to estimate missing values in a dataset [
21]. One of the advantages of machine learning-based imputation is its significantly greater flexibility compared to statistical methods, enabling better predictions by capturing higher-order interactions within the data [
5]. The
k-nearest neighbor (
k-NN) imputation is one type of machine learning-based imputation methods [
22].
k-NN imputation replaces missing values using information from the
k-nearest neighboring data points. This enhances the completeness of the dataset while maintaining the valuable information for further analysis. However, inaccuracies may occur if the missing feature patterns are complex or unique. Additionally, locating neighbors for imputation can be challenging if the data distribution is highly imbalanced.
There are many ongoing studies related to machine learning-based imputation [
23]. The experimental studies demonstrate the validity of various machine learning-based MVI methods [
21]. While the chain approach has been widely applied in various domains, such as classifier chains in multilabel learning [
24], the first imputation method based on this approach was proposed by Van Buuren and Groothuis-Oudshoorn [
25], introducing the widely used Multiple Imputation by Chained Equations (MICE). Conventionally, Tsai C. et al. [
26] conducted a comparative performance experiment among five widely used supervised learning methods:
k-nearest neighbors (
k-NN), classification and regression trees (CART), multi-layer perceptron neural networks (MLP), Naive Bayes, and support vector machines (SVMs). In their study, Yadav et al. [
27] compared the experimental results of missing value imputation using visualization and imputation of missing values (VIM), MICE, nonparametric missing value imputation using random forest (MissForest), and Harrell miscellaneous (HMISC) methods. Wang et al. [
28] performed a comparative analysis using two renowned imputation methods—autoregressive integrated moving average (ARIMA) and linear interpolation (LI) models—as well as three machine learning approaches:
k-NN, multi-layer perceptron (MLP), and support vector regression (SVR) (a Support Vector Machine (SVM) regression used for missing value imputation). Palanivinayagam et al. [
29] analysed the performance of five machine learning models: Naive Bayes, SVM,
k-nearest neighbours, random forest, and linear regression. The experimental results showed that the SVM classifier achieves the highest accuracy, highlighting its importance in diabetes research and addressing missing data. Li et al. [
30] tested six machine learning algorithms to predict hospital readmissions within 30 days for elderly patients (aged 65 and above). Experiments were conducted using RandomForest, LogisticRegression, XGBoost, LGBM, MLP, and Random + XGBoost and the results indicated that the RF + XGBoost algorithm exhibited superior overall performance in terms of Area Under the Curve (AUC) for prediction.
Raja et al. [
31] introduced a novel imputation method based on rough
k-means centroids that utilizes unsupervised machine learning methods to handle missing values. Another clustering-based method performs the imputation using a similarity-based spectral clustering approach, top k-nearest neighbor approach, and clustering [
32]. Alternatively, the data-driven missing value imputation technique incorporates feature-wise selection to perform accurate imputations [
33]. The Takagi–Sugeno (TS) modeling method is a promising approach for incomplete datasets, utilizing Bayesian networks for missing value imputation [
34]. The GRLSR method precisely imputes missing values in incomplete data by combining sample self-representation and local data structure [
35]. Meanwhile, the fuzzy adaptive imputation approach (FAIA) is a fuzzy-based information decomposition method to address missing value problems in imbalanced data streams [
36]. Another novel imputation approach is a query selection method that considers the imputation uncertainty in active learning with missing values [
37]. In addition, the purity-based
k-NN imputation method considers data purity and improves results by estimating high-purity instances as candidates [
38]. Yet another K-NN method extension named Focalize K-NN leverages correlated features and temporal lags to improve the performance of the traditional K-NN imputer [
39].
Deep learning-based imputation involves feeding data into the input layer of a neural network and computing the predicted output. The difference between each predicted value and its corresponding ground truth is then calculated [
4]. In other words, training through multiple layers enables the model to discover complex relationships embedded within the data structure and improve imputation performance through fine-tuning [
40]. Missing values can be imputed using traditional deep learning techniques such as long-short-term memory (LSTM) networks and Convolutional Neural Networks (CNN) [
35]. A more recent type of deep learning-based imputation is DataWig [
8]. DataWig (DW) creates a model for each feature to perform imputation. It aims to estimate the probabilities of possible values for a target feature using information from other features, including those with missing values. Accurately capturing the full range of data characteristics can be challenging in datasets with many missing values. As a result, the model may struggle to understand and handle missing values fully. Lin et al. [
41] evaluated the performance of two supervised methods—Multi-layer perceptron (MLP) and deep belief networks (DBN). Morales-Alvarez et al. [
42] introduced VISL—a novel scalable structure learning approach that simultaneously infers the structure between variable groups and imputes missing values using deep learning in the context of missing data. Another approach, DeepIn, is a missing value imputation solution for continuous missing patterns found in Internet-of-Things devices in smart spaces [
43]. Alternatively, dynamic imputation is a method to enhance the training of neural networks in the presence of missing values [
44]. Yet another DNN model imputes missing values in the hourly aerosol optical depth product by combining AERONET with a numerical model [
45]. Furthermore, Bidirectional Recurrent Imputation for Time Series (BRITS) [
46] treats missing values as trainable variables within a bidirectional recurrent neural network, allowing end-to-end optimization over time-series data.
The autoencoder introduces a new way to model the characteristics of a given dataset [
47]. For example, an autoencoder can leverage information from neighboring data points to perform data imputation [
9]. Expanding the applications of autoencoders, a different method was developed by modifying a denoising autoencoder (DAE) into a cluster-based imputation framework [
10]. Gjorshoska et al. [
11] utilized the autoencoder imputation method for resolving missing values in the food composition database (FCDB). A completely modified denoising stacking autoencoder (CMSDAE) is used for missing value imputation, particularly to enhance the quality of the MTL version [
12]. Psychogyios et al. [
48] proposed a method based on DAE with kNN for the pre-imputation task for the missing value imputation in the electronic health records.
An alternative solution comes from using the GAN, as was done by Zhang et al. [
13], who applied the end-to-end GAN for multivariate time series, addressing the problem of imputed values being significantly different from actual values. Generative adversarial imputation networks (GAIN) [
49] introduced a framework in which a generator imputes missing values conditioned on observed data, while a discriminator learns to distinguish imputed from real values using a hint mechanism, achieving state-of-the-art results. Another GAN-based solution named IFGAN is a missing value imputation algorithm based on feature-specific GAN [
14]. Furthermore, multiple generative adversarial imputation networks (MGAIN) is an imputation method that simplifies the network structure of GAIN and reduces the demand for data [
15]. Non-autoregressive multi-resolution imputation (NAOMI) is a new deep generative model for imputing long-range sequences [
16].
More recent studies explore the attention mechanism [
50] capabilities in the MVI area tasks. One method of attention application was demonstrated by Koswar et al. [
17], who proposed a framework that leverages between-feature or between-sample attentions. Another approach, named SAITS, operates with a weighted combination of two diagonally masked self-attention blocks, which explicitly capture temporal dependencies and feature correlations between time steps [
18]. ImputeFormer combines strengths of low-rank and deep learning models, introducing a low rankness-induced transformer to balance strong inductive bias and high expressivity [
19].
Another recent branch of MVI studies employs diffusion models for missing values generation. For example, Chen et al. apply a schrödinger bridge problem to probabilistic time series imputation by generating missing values conditioned on observed data [
51]. Alternatively, in the work of Wang et al. [
52], the evidence lower bound (ELBO) was re-derived in the scenario of multivariate time series imputation to take into account the correlations between observed and missing values. In the proposed multivariate imputation diffusion model (MIDM), the newly derived ELBO was enhanced with noise sampling and denoising mechanisms for multivariate time series imputation. Biloš et al. propose yet another method that adapts the noise generation and denoising mechanisms for time-series oriented scenario with irregularly sampled observations [
53]. To overcome difficulties occurring in temporal electronic health records analysis, Dai et al. [
54] proposed a Similarity-Aware Diffusion Model-Based Imputation (SADI), an imputation method that utilizes information across dependent variables. Following the diffusion ideas, Yang et al. [
55] utilized a high-frequency filter to boost the residual term imputation, supplemented by a dominant-frequency filter for the trend and seasonal imputation in their proposed solution named multivariate time series imputation (FGTI). Lastly, to achieve an imputation consistency in terms of intra-consistency between observed and imputed values, and inter-consistency between adjacent windows, Zhou et al. [
56] employed a contrastive complementary mask in Multivariate Time Series Consistent Imputation (MTSCI) to generate dual views during the forward noising process.
Ensemble-based imputation uses a combination of machine learning-based imputation and deep learning-based imputation. One type of ensemble-based imputation is stacked ensemble (SE) [
57]. After filling the NaN values with the
k-NN imputer, stacked ensemble (SE) creates a stacked ensemble classifier using models such as Extreme Gradient Boosting (XGB), random forest (RF), and Extra Tree Classifier (ETC). It is crucial to be aware that the predictions from each alternative imputation method are combined to make the final prediction. However, this approach can lead to a degradation in the performance of the final prediction if each model is overfitting.
Currently, there are many ongoing studies related to ensemble-based imputation. rMisbeta utilizes robust estimators based on the minimum beta divergence method. Experimental results show that data matrices imputed by rMisbeta outperform other statistical tools such as Zero,
k-NN, SVD, EM, and RF. Furthermore, rMisbeta is an accurate, simple, and fast tool for missing value imputation [
58]. Meanwhile, zero-inflated Poisson log-normal (ZIPLN) model is a feasible imputation method that uses a mixture distribution [
59]. In particular, the missing data imputation method based on random forest proposes a multi-step prediction for imputation and an ensemble model combining attention-based GRU models [
60]. In addition, the multiple single imputation method based on ensemble learning applies missing value prediction using bootstrap sampling in the ensemble method, assigns weights to these predictions, aggregates them, and generates the final prediction [
61]. Rao et al. proposed a multimodal imputation-based stacked ensemble (MISE) model to classify and predict air quality [
62]. It has demonstrated superior performance through experimental validation. Jung et al. [
63] utilized a bagging ensemble of multi-layer perceptrons, known as a Softmax ensemble network, to determine the ensemble weights for each MLP. Samad et al. [
64] demonstrated that the linear regressor of MICE, replaced by ensemble learning and deep neural networks, improves both the imputation accuracy of MICE and the classification accuracy of imputed data.
As missing value imputation methods continue to evolve, increasing emphasis is being placed on their applicability to real-world scenarios, where data may be irregular, incomplete, and collected under complex conditions. For instance, GRUF captures the past states of time series data collected from devices and appropriately fills in missing values using both historical sensor readings and temporally aligned data from neighboring nodes via edge computing [
65]. A recent study by Jiang and Zhang [
66] proposed an interpretable diffusion-based imputation framework tailored for industrial soft sensing, incorporating resampling strategies and Fourier-based components to enhance both accuracy and model transparency.
3. Proposed Method
Table 1 summarizes the notations used to describe the proposed method.
F denotes a set of datasets, comprising an input vector
and a corresponding label
, represented as
. Here,
f denotes a feature, while each observation in the dataset consists of multiple such features. MVI aims to fill in missing values within the dataset (
). It selects input features based on the provided list of features (train_col) and trains an imputation model denoted as
M to predict missing values using the selected input features. After filling in missing values with the model
M, denoted as
, the dataset is updated.
The procedure of conventional NN-based MVI methods is as follows [
8]. Suppose we have an original feature set
F where its features may contain missing values. Then, the algorithm chooses a target feature
f containing missing values from
F. Next, all the other features
are chosen to form the training dataset for NN. After that, a naïve MVI, such as mean imputation, is performed on each feature in
if it contains missing values. Thus,
can be filled with many roughly guessed values, resulting in an inaccurate prediction model. Lastly,
, which is an imputed version of
f by trained NN, replaces
f from
F, and then these steps are repeated until there are no features with missing values in
F. We argue that the advantage of employing NN, which is expected to yield accurate predictions on the missing value based on its strong fitting capability, can be nullified because NN will learn the roughly guessed values for its prediction at all repetitions. Instead, we may consider a natural imputation strategy as follows. Starting from
F, the algorithm chooses
with missing values and tries to train an NN using features
. Because the training process cannot proceed if there are missing values in
, the algorithm chooses
, which is the second target feature of imputation, and then tries to train an NN using features
. At the end of this recursive procedure, a set of features will remain that contains no missing values. Hence, a new feature, a carefully imputed version of the original feature, can be obtained after the employed NN is trained and imputation is performed. Next, in the backward stage of the recursive procedure, these features are inherently involved in the training of NN for the current recursion. When the recursion process is completed, all the missing values in
F are imputed.
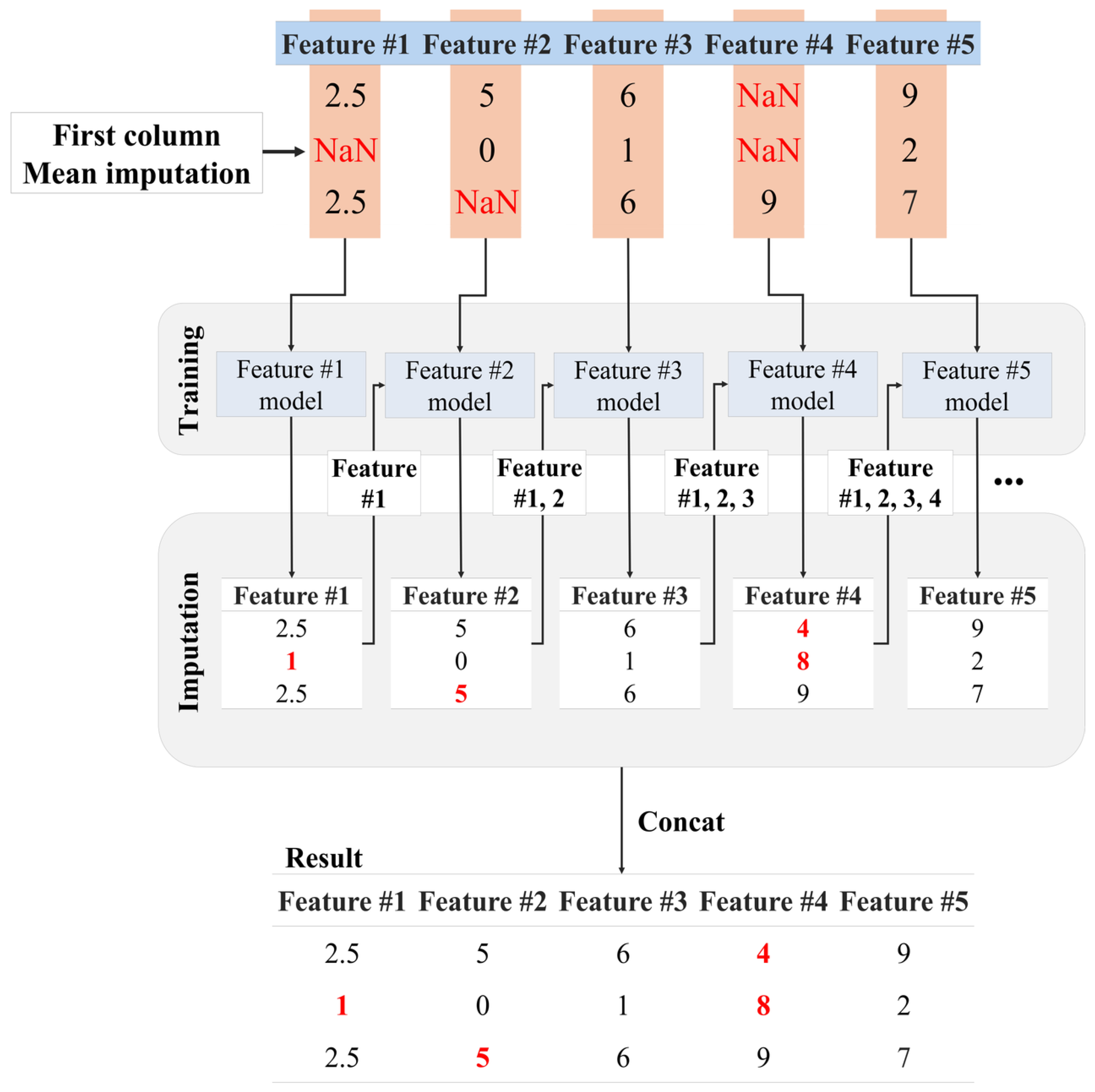
In
Figure 2, the proposed method models for each feature using the available data. When missing values are encountered in the first feature, a common approach consists of replacing them with the mean of the existing values within the feature because there are no trainable features without missing values. If there are features without missing values, an NN can be used to train on those features and then impute the missing value of the first feature. Next, the missing values in the second feature are imputed based on the NN trained on the first feature. This iterative process continues sequentially for each feature. It allows for the gradual refinement of the models and incorporates more complex relationships between features. We argue that this iterative approach helps reduce the impact of missing values and adjusts the imputation process according to the characteristics and dependencies present in each dataset feature.
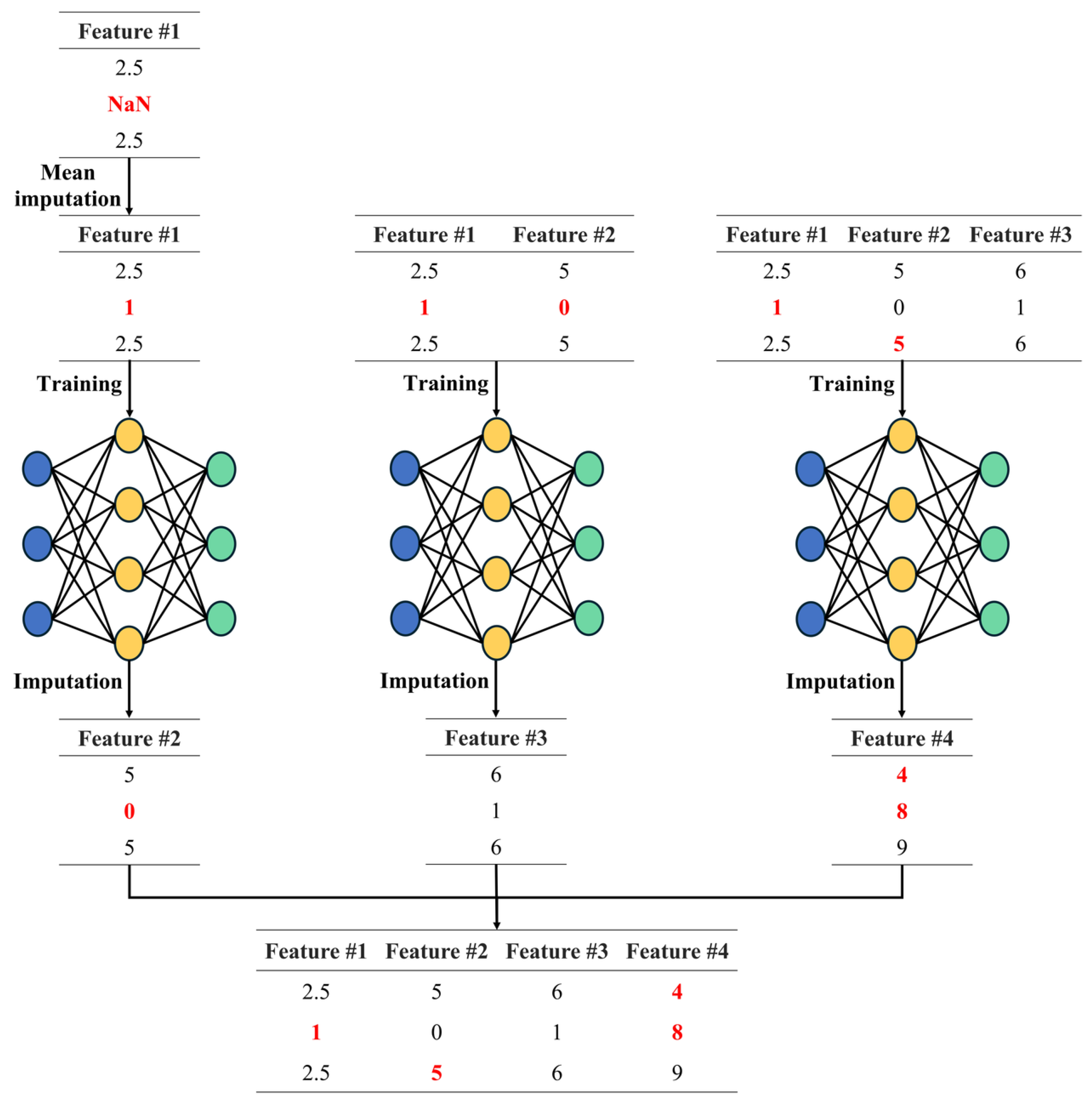
In
Figure 3, the proposed method iteratively trains models to handle missing values, utilizing previously imputed missing values to train each new model and replacing all missing values in the dataset. The proposed method trains a model using the current input features and leverages it to replace missing values, ensuring a robust and reliable data analysis process. The training process of the proposed method involves constructing independent models for each feature and using the imputed results from the preceding feature to predict missing values sequentially. This process can be expressed as follows. First, the training for the first feature is conducted as
where
represents the dataset after filling in the missing values for the first feature. Subsequently, the training for the second feature is carried out as
where
denotes the dataset after filling in missing values for both the first and second features, and
represents the dataset used to train the model for the second feature. This iterative process continues for each subsequent feature. Specifically, the model
for the
i-th feature is trained using the imputed results from features 1 through
i. This can be written as
where
represents the dataset after filling in missing values for features 1 through
i, and
represents the dataset used to train the model for the
i-th feature. This process ensures that each feature-wise model is trained using the imputed results from previous features, facilitating accurate imputation of missing values across the dataset.
It is worth noting that, unlike traditional chain-based methods such as MICE, the proposed method introduces two key differences. First, the feature ordering for imputation is not determined randomly, but is predefined based on the ascending order of missingness ratios. Second, the proposed method adopts an asymmetric structure by incrementally constructing a cumulative feature set. Each newly imputed feature is incorporated into the input of the subsequent imputation step, enabling more informed training and progressive refinement of the neural models. This progressive structure helps reduce the distortion caused by unreliable inputs in the early stages of the imputation process.
Algorithm 1 presents a method for imputing missing values in a dataset. The proposed method, called ChainImputer, employs a chain approach, leveraging initially complete features or mean imputation, followed by model-based imputation using MXNet. First, the original feature set
F is initialized (Line 2), and
S, which will contain features without missing values, is set to an empty set (Line 3). The algorithm iterates over each feature
f in the feature set
F (Line 4). For each feature, it checks if there are missing values (Line 5). If a feature
f has no missing values, it is added to the set
S (Line 6.) If, after processing all features, the set
S is empty, indicating that all features in
F have missing values, the algorithm selects a random feature
f from the feature set
F (Line 10). It then uses a mean imputation method to fill in the missing values of
f to create the initial feature (Line 11). The imputed feature
is then added to the set
S and removed from
F (Lines 12–13). For each remaining feature
f in the feature set
F, the algorithm uses an MXNet model trained on the features in
S to impute the missing values in
f (Line 16). Next, the original feature
f is removed from the feature set
F, and the imputed feature
is then added to the set
S (Lines 17–18). This process is repeated until all the features in
F are considered.
| Algorithm 1 Procedures of proposed imputation method |
- 1:
procedure ChainImputer(F) ▹ Proposed missing value imputer - 2:
Initialize F to the original feature set; - 3:
; - 4:
for each feature do - 5:
if f has no missing values then - 6:
; - 7:
end if - 8:
end for - 9:
if then ▹ When there is no features for training MXNet - 10:
Choose a random feature ; - 11:
; - 12:
; - 13:
; - 14:
end if - 15:
for each feature do - 16:
; ▹ complete f using MXNet trained based on S - 17:
; - 18:
; - 19:
end for - 20:
end procedure
|
An imputed entry is an estimate and may deviate from the true value. If that entry is repeatedly reused as an input during later stages of the chain, its estimation error can propagate and accumulate, potentially degrading subsequent predictions. Therefore, an imputation schedule that minimizes the number of times previously imputed entries are reused provides a principled way to limit cumulative noise. We now show that the ascending-missingness order achieves this minimum.
Let denote the missing-value counts sorted in ascending order. If the feature with missing entries is imputed at position i, its column will be reused in exactly subsequent models. The total number of imputed entries reused for an ordering is therefore formalized as follows:
Proposition 1 (Re-use cost)
. For any permutation π of , the cumulative count of imputed entries that are reused during the chain is Lemma 1. Consider two positions, with , placed in reverse order inside π. Swapping their positions decreases .
Proof. Let
and
so that
. Before the swap, the contribution of these two features to (
4) is
. After swapping, it becomes
. Their difference satisfies
so the swap strictly reduces
. □
Theorem 1 (Optimality of ascending order)
. The ascending-missingness schedule minimizes in (4) over all permutations π. Proof. Starting from any permutation, repeatedly apply the swap described in Lemma 1 to every inverted pair. Each swap lowers and strictly reduces the inversion count. The process terminates only when no inversions remain, i.e., when the list is in ascending order, which therefore attains the global minimum. □
By Theorem 1, the ascending-missingness order minimizes ; hence, it reuses the fewest imputed entries and introduces the least cumulative distortion in subsequent training steps.
Based on Algorithm 1, we analyze the time complexity of the proposed ChainImputer and compare it with the neural network-based baseline DW for a balanced perspective. Let
n denote the number of instances,
the number of features,
a the overall missing rate,
T the number of training epochs,
the cost of applying a simple filler to one column (for example, mean imputation so that
); and
the cost of one network epoch over the entire data set. DW first imputes the complete table once and then trains for
T epochs [
8], which leads to
ChainImputer processes the
d columns in sequence: for each feature it produces predicted values for that column and then trains the network for
T epochs, resulting in
Although this worst-case big-
bound is larger, ChainImputer begins with a single input feature, and the effective
increases gradually as the model incorporates more columns during the imputation process. Consequently, for data sets with substantial sparsity or moderate feature counts, its observed running time can be comparable to, or even less than, that of DW in practice, despite the higher asymptotic bound.
6. Conclusions
In this paper, the proposed chain imputer method not only demonstrates a systematic approach to handling missing values but also significantly enhances imputation accuracy. The results show the superiority of the proposed method in imputing missing values across diverse datasets. The proposed method proves to be both robust and effective, outperforming the comparison models by yielding lower values in 22 out of 25 datasets. Its high performance in datasets such as Abalone and Adult is particularly notable, suggesting that it can provide a reliable solution for imputing missing values across various fields. The Friedman and Bonferroni–Dunn post hoc tests further validate the superiority of the proposed method. The significant differences observed between the groups confirm that the proposed method consistently outperforms the comparison models. The Bonferroni–Dunn test highlights the clear superiority of the proposed method. Such statistical validation contributes to establishing the proposed method as a practical solution for MVI tasks and enhances confidence in the experimental results. In conclusion, the experimental results and in-depth analysis presented in this study suggest that by combining chain strategy and neural network modeling, a robust and adaptable methodology can ensure the reliability and performance of the data analysis process.
Despite its effectiveness, the proposed method has some limitations. First, the current implementation is tightly coupled with the MXNet framework, which may restrict portability and optimization opportunities in other environments. Future work will explore alternative architectures and platforms to improve scalability. Second, the imputation order, which is currently determined in a heuristic manner, can impact performance. A more principled approach based on entropy-driven feature informativeness will be investigated to derive an optimal imputation sequence. Third, the hyperparameters for each comparison method, including the proposed model, were not independently optimized but were adopted from previously published experimental settings. Although this ensures consistency and reproducibility, it may affect the comparative performance. As a future direction, systematic tuning procedures such as grid search or Bayesian optimization could be applied to each method to ensure a fair and balanced evaluation. These limitations point to important directions for future research that can further improve the robustness and generalizability of the proposed method.
Furthermore, valuable insights and outcomes are anticipated from applying the proposed method in various fields, potentially leading to advancements in research, industry, and decision-making practices. Enhancing imputation performance leads to more reliable results in various tasks such as predictive modeling, classification, and clustering. Imputation of missing values helps minimize the risk of distorted data and unreliable insights in data-driven analyses. Additionally, the advantage of utilizing the proposed method is its applicability to datasets with missing values across various fields. Extending this approach to larger and more diverse datasets, including real-world applications, is expected to produce reliable results.