Abstract
Probability models are instrumental in a wide range of applications by being able to accurately model real-world data. Over time, numerous probability models have been developed and applied in practical scenarios. This study introduces the AAP-X family of distributions—a novel, flexible framework for continuous data analysis named after authors Aadil Ajaz and Parvaiz. The proposed family effectively accommodates both symmetric and asymmetric characteristics through its shape-controlling parameter, an essential feature for capturing diverse data patterns. A specific subclass of this family, termed the “AAP Exponential” (AAPEx) model is designed to address the inflexibility of classical exponential distributions by accommodating versatile hazard rate patterns, including increasing, decreasing and bathtub-shaped patterns. Several fundamental mathematical characteristics of the introduced family are derived. The model parameters are estimated using six frequentist estimation approaches, including maximum likelihood, Cramer–von Mises, maximum product of spacing, ordinary least squares, weighted least squares and Anderson–Darling estimation. Monte Carlo simulations demonstrate the finite-sample performance of these estimators, revealing that maximum likelihood estimation and maximum product of spacing estimation exhibit superior accuracy, with bias and mean squared error decreasing systematically as the sample sizes increases. The practical utility and symmetric–asymmetric adaptability of the AAPEx model are validated through five real-world applications, with special emphasis on cancer survival times, COVID-19 mortality rates and reliability data. The findings indicate that the AAPEx model outperforms established competitors based on goodness-of-fit metrics such as the Akaike Information Criteria (AIC), Schwartz Information Criteria (SIC), Akaike Information Criteria Corrected (AICC), Hannan–Quinn Information Criteria (HQIC), Anderson–Darling (A*) test statistic, Cramer–von Mises (W*) test statistic and the Kolmogorov–Smirnov (KS) test statistic and its associated p-value. These results highlight the relevance of symmetry in real-life data modeling and establish the AAPEx family as a powerful tool for analyzing complex data structures in public health, engineering and epidemiology.
1. Introduction
Probability models play a crucial role in statistical analysis, data modeling and real-world applications. However, no single probabilistic model can consistently outperform all others across diverse datasets and scenarios. This inherent limitation fuels ongoing research, inspiring statisticians to craft new probability distributions tailored to specific needs. Modifying or extending existing models often yields distributions that fit real-world data more closely than their predecessors. This drive for better fit, greater flexibility and wider applicability spans multiple disciplines and motivates continuous innovation.
One of the earliest and most fundamental methods for generating new distributions was introduced by Mudholkar and Srivastava [1], who crafted the Exponentiated Family of Distributions. Their elegant approach enhances a baseline distribution by adding a shape parameter—a simple twist that unlocks a broader spectrum of data patterns. It is an undeniable truth that no model reigns supreme; hence, statisticians tirelessly explore fresh models to capture reality’s complexities. The cumulative distribution function (CDF) of the exponentiated family is given by
Here, adds a new dimension of shape and is the CDF of the base distribution, itself defined by parameters. This elegant formulation breathes flexibility into classical distributions, molding them to suit intricate datasets.
Following this, Reference [2] introduced the Marshall–Olkin family of distributions. Their transformation reshapes the baseline CDF with an parameter, yielding the following:
where represents the survival function. This structure grants finer control over tail behavior, making it invaluable for reliability and risk analysis.
Building on this foundation, Reference [3] proposed the Alpha Power Transformation (APT), introducing a parameter () that redefines the baseline CDF () as follows:
This transformation injects fresh flexibility, with the exponential distribution appearing as a special, illuminating case.
To further expand the toolkit, Reference [4] introduced the MIT transformation—named after its creators, Murtaza, Ishfaq and Tariq—enhancing a baseline CDF by weaving in a shape parameter ():
This simple yet powerful tweak opens doors to greater modeling flexibility.
Expanding horizons, Reference [5] applied the T-X family methodology by Reference [6] to create the New Exponent Power-X (NGEP-X) family, whose CDF reads as follows:
This formulation melds exponential power with a new perspective, enriching distribution shapes.
Meanwhile, Reference [7] unveiled the alpha-log-power transformed family (ALPTG), embedding a novel shape parameter independent of the parent model, offering fresh avenues for continuous distribution modeling:
Continuing the innovation, Reference [8] proposed the New Logarithmic Tangent-U (NLT-U) family, introducing the NLT-Wei model, a flexible Weibull-based sub-model. It showcases hazard functions that curve and twist—rising, falling, unimodal and bathtub-shaped functions—capturing real-world failure behaviors.
Riding the wave of novel approaches, Reference [9] embraced trigonometric transformations, crafting a family where the baseline distribution undergoes a sine- and cosine-inspired metamorphosis:
This infusion of trigonometry injects rhythm into the shape of distributions, enhancing their expressive power.
Further enriching this path, Reference [10] introduced the ASP family, a sine-based flexible distribution that masterfully handles skewed and heavy-tailed data:
This approach offers a robust alternative for engineering, medicine and beyond.
Finally, Reference [11] proposed the Alpha Beta Power-E (ABPE) Transformation, whose CDF is expressed as follows:
bringing yet another harmonious blend of parameters to the family of flexible distributions.
In light of the extensive developments in probability distribution generation techniques discussed above, our study aims to contribute to this ongoing evolution by introducing a novel and more adaptable probability model. Many existing methods enhance baseline distributions by incorporating additional parameters, but these modifications can sometimes lead to re-parameterization issues, increased model complexity and challenges in practical implementation. To address these concerns, we propose an innovative AAP family of distributions, a flexible framework that refines existing distributions while minimizing the complexities associated with parameter inflation. The AAP-X family is designed to enhance the fitting ability of probability distributions, providing a more efficient and adaptable modeling approach. Unlike conventional transformation techniques that merely add parameters, the AAP-X method strategically modifies the structural properties of the base distribution to achieve greater flexibility without excessive parameterization. This innovative framework can effectively capture both symmetric and asymmetric data patterns through its shape-controlling mechanism, making it applicable across diverse domains.
As a specific application of our framework, we introduce an improved version of the exponential distribution using the AAP-X transformation. This enhanced model demonstrates superior flexibility, particularly in terms of its probability density function (PDF) and hazard rate function (HRF), allowing for a more accurate representation of real-life data. To validate its practical relevance and establish its superiority over classical models, we conducted a comparative analysis against several well-known models, including the Alpha Power Exponential and Marshall–Olkin Exponential models. These comparisons, presented in the application section of the manuscript, reveal that the AAP-X family consistently outperforms these traditional models based on standard goodness-of-fit criteria such as the AIC, BIC, AICC, HQIC and KS statistics.
Overall, this study not only introduces a novel statistical transformation but also preserves the theoretical elegance while addressing practical challenges. Its ability to capture diverse data behaviors, particularly symmetry and asymmetry, makes it highly relevant for applications in reliability analysis, survival studies, epidemiology and beyond.
The remainder of this paper is structured as follows: Section 2 presents the research gap and highlights the need for developing the proposed AAP-X family of distributions. Section 3 introduces the proposed AAP-X family of distributions, along with its mathematical formulation and key properties. Section 4 presents a special case of the family based on the exponential distribution, which serves as a tractable and insightful example. It also discusses various structural characteristics of the model, including the hazard rate function, moments and the moment-generating function. In Section 5, estimation of parameters is addressed using six methods of estimation. Section 6 includes a detailed simulation study to compare the performance of estimators. In Section 7, the practical utility of the model is demonstrated through real-world data applications. Finally, Section 8 concludes the paper with a summary of findings and directions for future research.
2. Research Gap
The existing literature on extended distributions, particularly those enhancing baseline models such as the exponential distribution, reveals several limitations and gaps that warrant further investigation. This paper aims to address the following specific research gaps:
- Existing transformation techniques often increase model complexity through additional parameters, leading to parameter inflation without substantial gains in flexibility.
- Several extended models fail to offer adequate flexibility in capturing both symmetric and asymmetric data patterns observed in real-world datasets.
- There has been limited exploration of transformation frameworks that preserve theoretical tractability while enhancing distributional shapes.
- Many proposed models do not comprehensively evaluate estimation techniques, resulting in limited insights into their practical implementation.
- Prior works do not offer extensive empirical comparisons across diverse real-life datasets, especially from both medical and engineering domains.
- Limited research exists to assess the performance of new distributions using a wide range of goodness-of-fit criteria and graphical diagnostics.
- The integration of domain-specific datasets for validation of new models remains underexplored, especially in the context of survival and reliability data.
This study responds to the above research gaps by proposing the AAP-X family of distributions, which enhances model flexibility without excessive parameterization and is validated using extensive real-world applications and simulation-based performance comparisons.
3. The Model and Properties
In this section, we introduce a novel family of probability distributions, termed the Aadil Ajaz–Parvaiz-X (AAP-X) Family. The primary motivation behind the development of this family is to enhance the flexibility of existing probability models by incorporating an additional parameter that adjusts the shape and scale properties of the baseline distribution. This transformation aims to provide better modeling capabilities for complex real-world data structures across various disciplines, including engineering, finance and survival analysis. The AAP-X family is defined by modifying the cumulative distribution function (CDF) of a baseline distribution (, where represents the parameter vector of the base distribution). The transformation introduces a new shape control parameter () that governs the behavior of the tail and the adaptability of the resulting distribution. To understand the behavior and practical utility of the AAP-X family, we explore key statistical properties.
Definition 1.
Let represent the CDF of the reference model. Then, the CDF () of the AAP-X family, incorporating the additional shape parameter (γ), is obtained as follows:
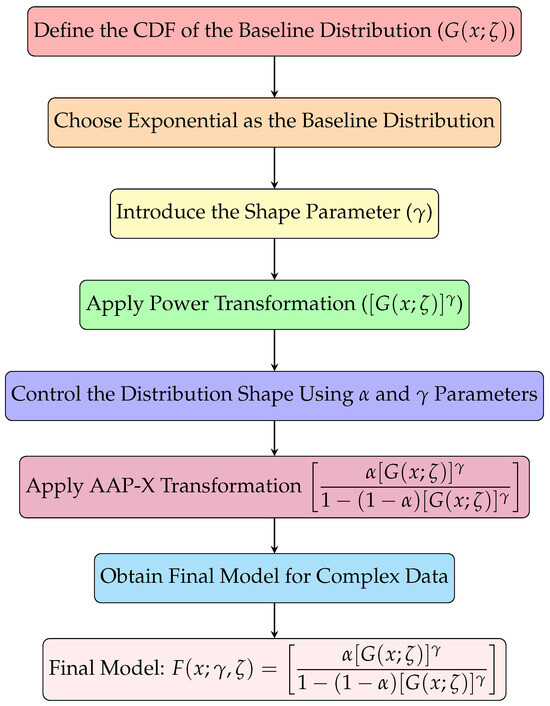
To aid in the understanding of the transformation process from the baseline distribution to the proposed AAP-X family, a schematic flow diagram is presented in Figure 1. This flowchart visually summarizes the key steps involved in deriving the cumulative distribution function (CDF) of the AAP-X family.

Figure 1.
Flowchart of the study.
It is straightforward to verify that the cumulative distribution function (CDF) defined in Equation (10) satisfies the essential properties of a valid distribution function. Specifically, the following holds:
and the function is right-continuous and differentiable. Hence, the expression in Equation (10) is a valid CDF. The probability density function (PDF) corresponding to the CDF in Equation (10) is given by Equation (11):
Here, is the PDF of the baseline distribution, and is its CDF.
Mathematically, the AAP-X family generalizes several well-established transformation techniques. Its flexibility is illustrated through the following special cases:
- When and , Equation (10) collapses to , the CDF of the baseline distribution.
These reductions confirm that the proposed AAP technique not only generalizes existing transformation approaches but also unifies them under a common framework. Such unification enables a comprehensive exploration of the effects of transformation parameters on the distributional shape, skewness and tail behavior.
3.1. Aging Properties
The term “aging properties” in statistical modeling and survival analysis refers to features of a survival distribution that characterize the evolution of the probability of failure or death. These properties are important in understanding the lifetime behavior and dependability of systems or individuals. For the proposed AAP-X family, the survival function (SF), hazard rate (HR) and cumulative hazard function (CHF) measures are calculated as follows:
3.2. Quantile Function
The quantile function is an important tool in many statistical methods, including Monte Carlo techniques. By inverting the CDF of a distribution, it is possible to extract random numbers from that distribution. Given a random variable (X) that belongs to the AAP-X family, the quantile function for X can be found in the manner described below:
Solving for , we obtain
In order to determine the quantile function () for the AAP-X family, we need the inversion function of . Thus,
Therefore, by applying Equation (17) for a specific baseline CDF-G, random variates from the AAP family can be generated.
3.3. Moments and Moment-Generating Function
For the AAP-X family introduced in this study, the ordinary rth moment is calculated as
using the following series representation:
where
Moreover, the MGF, denoted as for an AAP-X distributed random variable (X), is obtained as follows:
3.4. Order Statistics
In distribution theory, order statistics (OS) are essential because they characterize the operational time of systems, and components and are crucial to reliability analysis, estimate theory and life testing. Consider a random sample of observations () drawn from the AAP-X family with a CDF and PDF provided by Equations (10) and (11). The PDF of the rth order statistics—say, —is then calculated as follows:
The expansion of in binomial form is provided as follows:
By substituting Equation (21) in Equation (20), we derive
Replacing Equations (10) and (11) in Equation (22) yields the PDF of the statistics.
3.5. Residual and Reverse Residual Lifetime
The notions of residual lifetime (RL) and reverse residual lifetime (RRL) are widely applicable in several fields, including survival modeling, actuarial science, biometrics and risk assessment. Within the framework of the AAP-X family, the RL of a random variable (X), represented as , is described as follows:
In addition, we also derive the expression for the RRL of a random variable (X) following the AAP-X family. The calculated expression denoted as is defined as
4. Special Case
In this part, we present a specific case within the AAP-X family by extending the exponential distribution.
4.1. AAP Exponential (AAPEx) Distribution
In this subsection of the article, a particular subclass of the AAP-X family, characterized by three parameters is introduced. We refer to this particular model as the AAP Exponential (AAPEx) model. The CDF () and PDF () of the exponential distribution are defined as follows:
and
By inserting Equations (23) and (24) into Equations (10) and (11), we get the updated version of the exponential distribution (i.e., AAPEx) model as expressed by
and the PDF takes the form as
In order to enhance reader understanding, we provide the following clear explanations for the roles of each parameter in the proposed AAPEx model:
- The shape and tail behaviors are largely controlled by the parameter. It improves the flexibility of the model by modifying the weight of the modified function.
- The steepness of the cumulative distribution function is influenced by the parameter. It influences the skewness and kurtosis, making the model capable of capturing asymmetric patterns in data.
- The parameter is the scale parameter from the baseline exponential distribution. It affects the spread of distribution by controlling the rate of decay.
- For the transformation to stay valid and the resultant function to remain a suitable CDF, the formula expressed as is utilized.
The statistical properties, including SF (), HF () and CHF () for the special sub-model, are derived as follows:
and
In addition, for the QF of X, the AAPEx model is expressed as follows:
Specifically, are used in Equation (30) to determine the quartiles, Bowley’s skewness and Moor’skurtosis of the AAPEx model, respectively (see Table 1).

Table 1.
Quantiles, Bowley skewness and Moor’s kurtosis for the AAPEx model under varying parameters.
Figure 2 illustrates the PDF curves of the AAPEx model for different settings of the , and parameters. The plots demonstrate a wide variety of shapes, such as unimodal right-skewed (for , and ), J-shaped or strictly increasing (for , and ), and L-shaped or right-skewed (for , and ). Additionally, a bimodal and asymmetrical shape is observed when , and , while a left-skewed unimodal form arises for , and . A monotonically decreasing or exponential-type decay is evident for , and . These variations highlight the flexibility of the AAPEx model in representing both symmetric and asymmetric distributions, making it a suitable choice for modeling real-world lifetime data.
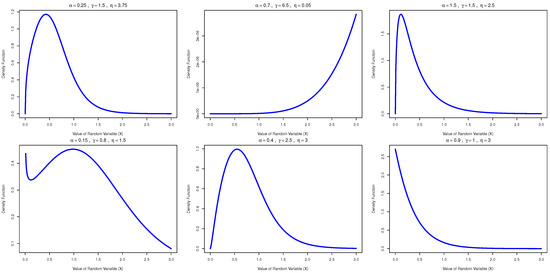
Figure 2.
PDF curves of the AAPEx model for different parameter settings of , and .
Figure 3 illustrates the hazard rate function (hrf) of the AAPEx model for various combinations of the , and parameters and demonstrates its ability to exhibit a wide range of asymmetrical forms. The plots reveal a rich variety of shapes. An increasing hazard rate is observed for , while a bathtub-shaped curve appears for . An upside-down bathtub shape is evident in the case of , and a decreasing hazard rate is shown for . Additionally, a reversed J-shaped hrfis observed for . These distinct forms highlight the flexibility of the AAPED model in capturing various lifetime behaviors and support its applicability to a broad spectrum of real-world reliability data.
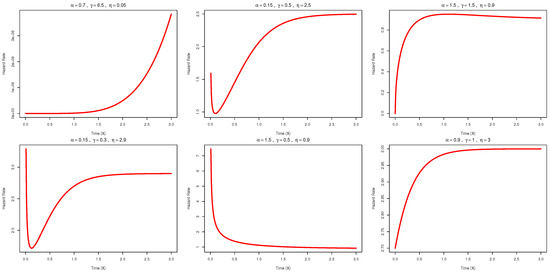
Figure 3.
Diverse hazard-rate shapes of the AAPEx model for different settings of , and .
4.2. Moments and Moment-Generating Function
The moment can be calculated by substituting Equations (25) and (26) into Equation (18), which yields
Using series expansion, we obtain the following:
Thus, the rth moment for AAPEx model is given as
Using the series representation of , we have
Substituting the value of Equation (34) into Equation (35), we get the moment-generating function of the AAPEx model as follows:
Table 1 provides a detailed computation of three key statistical measures—quantiles, Bowley skewness and Moor’s kurtosis—across a range of parameter combinations.
5. Parameter Estimation
In this section, we explore the estimation of the parameters of the AAPEx model through various frequentist estimation methods. Parameter estimation serves as a fundamental step for applied statisticians and reliability engineers, offering a structured approach to identify the most appropriate estimation technique for the model. Six estimation techniques are considered for the AAPEx model parameters, namely maximum likelihood estimation (), Cramér–von Mises estimation (), maximum product of spacings estimation (), ordinary least squares estimation (), weighted least squares estimation () and Anderson–Darling estimation ().
5.1. Maximum Likelihood Estimation () for a Complete Sample
Let random variables be a random sample whose observed values () are from the AAPEx model with the PDF () given in Equation (26). The likelihood function corresponding to —say, —is given as
Taking the natural logarithm of the likelihood function, the log-likelihood function () is given by
Substituting the PDF of the AAPEx model, i.e.,
we get
The derivatives of with respect to , and are delineated as
and
By evaluating the system of non-linear equations (, and ), the maximum likelihood estimates for the parameters of the AAPEx model can be obtained.
5.2. Cramer-von Mises Estimation (CVME)()
The CVME parameter estimation technique is also a powerful tool for parameter estimation. By minimizing the CVM statistic, the method provides parameter estimates that optimize the fit between the observed data and the expected distribution.
By evaluating the system of non-linear equations (, and ), we procure the Cramer-von Mises estimates for the parameters of the AAPEx model.
5.3. Maximum Product of Spacing Estimation (MPSE) ()
The MPS method is a parameter estimation technique used to fit probability distributions to data. It maximizes the product of the spacings (gaps) between ordered data points in the CDF. It provides robust parameter estimates by focusing on the spacing of data points rather than their exact values.
where and . The maximum product of spacing estimators is obtained by maximizing the following function:
By solving the non-linear equations (, and ), we procure the maximum product of spacing estimates for the parameters of the AAPEx distribution.
5.4. Ordinary Least Squares Estimation (LSE) () and Weighted Least Squares Estimation (WLSE) ()
LSE is a widely used method for estimating the parameters of a model by minimizing the sum of the squared differences (errors) between observed data and the values predicted by the model.
Similarly, WLS generalizes OLS by assigning weights to each data point, giving more importance to observations with lower variance or higher reliability.
5.5. Anderson–Darling Estimation (ADE) ()
The ADE parameter estimation technique is a powerful tool for fitting models to data, particularly when sensitivity to the tails of the distribution is important. By minimizing the AD statistic, the method provides parameter estimates that optimize the fit between the observed data and the hypothesized distribution.
By evaluating the non-linear equations (, and ), we obtain the Anderson–Darling estimates of the parameters of the proposed distribution.
6. Simulation Illustration
Here, we perform an extensive Monte Carlo simulation study to systematically evaluate the performance of several estimation strategies used for the suggested probability distribution. The main motivation of this simulation is to compare and investigate the performance of each estimator based on its accuracy, efficiency and robustness under different sample sizes and situations. Each sample is analyzed with six estimation procedures: MLE, CVME, MPSE, OLSE, WLSE and ADE. For comparison of these estimation methods, we evaluate the absolute bias (AB), mean relative estimate (MRE) and mean squared error (MSE) for each estimate. The following equations are employed to generate the statistics:
and
We perform exhaustive simulation analysis with many iterations in order to accomplish this. Random samples of the sizes and 400 are simulated from our proposed model based on three different sets of parameter values, as shown below.
- .
The results of the simulation analysis are reported in Table 2, Table 3 and Table 4. By reviewing these tables together, we can compare the relative strengths and efficiency of the various estimation approaches studied in this research. The obtained findings play a crucial role in determining the most appropriate estimation method for subsequent applications and research studies.

Table 2.
Simulation-based performance of estimators with , and .

Table 3.
Simulation-based performance of estimators with , and .

Table 4.
Simulation-based performance of estimators with , and .
A number of inferences can be made from the systematic examination of the simulation findings, such as the following:
- The bias of every estimator, regardless of the estimation technique, decreases as the sample size increases. This suggests that larger samples produce estimates that are more accurate and have less systematic error.
- The mean squared error (MSE) for any method of estimation decreases with a larger sample size. This means increased precision in the estimates resulting from a decrease in variance, as well as bias with larger samples.
- Similarly, for all estimators, the mean relative error (MRE) decreases with increasing sample size, demonstrating that larger samples minimize relative errors and yield more accurate and exact estimates.
- Regardless of sample size, the MLE and MPS approaches consistently exhibit the lowest bias, MSE and MRE, demonstrating their superior dependability for parameter estimation. These techniques have low errors across sample sizes and are very effective.
- In comparison to MLE and MPS, the CVM approach consistently has higher bias, MSE and MRE, despite showing some improvement with bigger sample numbers. Therefore, CVM is not as accurate or efficient as MLE or MPS, even if it might improve with larger samples.
- Particularly for smaller sample sizes, the OLS and WLS approaches typically have the highest bias, MSE and MRE. This indicates that these techniques may not be as appropriate for high-quality estimation in any situation because of their lower accuracy and precision when compared to MLE and MPS.
7. Applications in Medical Research and Engineering
In this section, the flexibility and efficiency of the AAPEx distribution are assessed by comparing its performance across real-world datasets from both medical and engineering domains, with particular emphasis on cancer survival, COVID-19 mortality and reliability data. Distributions considered in this study are the Exponential (Ex) distribution of Reference [12], the Exponentiated Exponential (EEx) distribution of Reference [13], the Weibull (W) distribution, the Sine Exponential (SEx) distribution of Reference [14], the Gamma (G) distribution, the Alpha Power Exponential (APEx) distribution of Reference [15], the Kumaraswamy Exponential (KwEx) distribution by Reference [16] and the Marshal–Olkin Exponential (MoEx) distribution of Reference [2]. For the purposes of comparison, multiple goodness-of-fit (GoF) measures are evaluated, including the Akaike Information Criteria (AIC), Schwartz Information Criteria (SIC), Akaike Information Criteria Corrected (AICC), Hannan–Quinn Information Criteria (HQIC), Anderson–Darling (A*) test statistic, Cramer-von Mises (W*) test statistic, and the Kolmogorov–Smirnov (KS) test statistic and its associated p-value. The optimal distribution has lower values for all GoF metrics, except for the p-value, which must be higher.
For clarity and completeness, the analytical expressions of the probability density functions (PDFs) of the competing models are presented below:
- The exponential (Ex) distribution with a PDF is expressed as
- The exponentiated exponential (EEx) distribution with a PDF is expressed as
- The Weibull (W) distribution with a PDF is expressed as
- The sine exponential (SEx) distribution with a PDF is expressed as
- The Gamma (G) distribution with PDF as:
- The alpha power exponential (APEx) distribution with a PDF is expressed as
- The Kumaraswamy exponential (KwEx) distribution with a PDF is expressed as
- The Marshal–Olkin exponential (MoEx) distribution with a PDF is expressed as
7.1. Application 1: Survival Time of 44 Head and Neck Cancer Patients
The first application analyzes the survival times of 45 patients diagnosed with head and neck cancer. The dataset was retrieved from Reference [17]. This real-world medical dataset, as detailed in [18], follows a progressive Type-I censoring scheme and involves patients treated with a combination of radiation and chemotherapy. The data is positively skewed, highlighting the presence of extreme survival times. The complete dataset is included in Appendix A.
Table 5 consists of the summary statistics for the survival times of 45 patients diagnosed with head and neck cancer disease. The data is extremely positively skewed, as the skewness value is greater than 1. The kurtosis value is greater than 3, indicating that the dataset is leptokurtic.

Table 5.
Summary statistics of 45 patients diagnosed with head and neck cancer disease.
The failure rate of application 1 is bathtub-shaped because the TTT-transform plot displayed in Figure 4 is initially convex below the line, then concave above the line, and the boxplot has few outliers, as in shown in Figure 4.
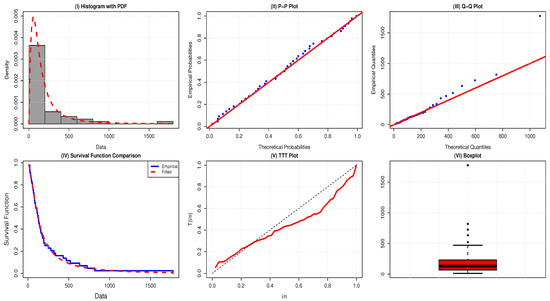
Figure 4.
Comparative plots evaluating the adequacy of the AAPEx model on survival times for the data of 45 head and neck cancer patients.
The maximum likelihood estimates (MLEs) and the GoF statistics, along with the corresponding p-values of the application described in Section 7.1 for the fitted models, including the AAPEx distribution and other competing distributions, are presented in Table 6. The negative logarithm and the information criteria are reported in Table 7. The results show that the AAPEx distribution has the smallest values of A*, W* and K-S, along with the largest p-value. This confirms that it provides a superior fit to the survival data compared to other models evaluated in the study.

Table 6.
MLEs With GoF statistics for 45 patients with head and neck cancer disease.

Table 7.
The negative logarithm and the information criteria results for 45 patients with head and neck cancer disease.
7.2. Application 2: Remission Times of 36 Bladder Cancer Patients
The second application represents the remission times (in months) for 36 patients diagnosed with bladder cancer, as already analyzed by Reference [19]. The dataset is fully observed, meaning that all remission times are complete, with no censored or truncated observations. No pre-processing was required beyond basic sorting and validation to ensure consistency with the original source. The observations of the dataset are reported in Appendix A.
The summary statistics of the remission times of 36 bladder cancer patients are provided in Table 8. The dataset is negatively skewed, as the skewness value is less than 0. The kurtosis value is less than 3, indicating that the dataset is platykurtic.

Table 8.
Summary statistics of 36 patients diagnosed with bladder cancer disease.
The maximum likelihood estimates (MLEs) and the GoF statistics, along with the corresponding p-values of the application described in Section 7.2 for the fitted models, including the AAPEx distribution and other competing distributions, are presented in Table 9. The negative logarithm and the information criteria are reported in Table 10. The results show that the AAPEx distribution has the smallest values of A*, W* and K-S, along with the largest p-value. This confirms that it provides a superior fit to the bladder cancer data compared to other models evaluated in this study.

Table 9.
MLEs With GoF statistics for 36 patients with bladder cancer disease.

Table 10.
The negative logarithm and the information criteria results for 36 patients with bladder cancer.
The TTT-transform plot of the data shows a concave pattern above the 45° line, providing insight into the shape of the hazard function, as shown in Figure 3, and no outlier is revealed in the boxplot illustrated in Figure 5.
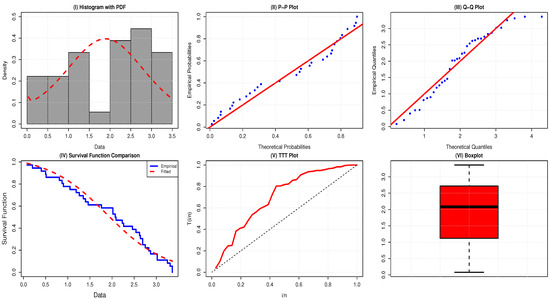
Figure 5.
Comparative plots for evaluation of the adequacy of the AAPEx model on remission times for the data of 36 bladder cancer patients.
7.3. Application 3: Remission Times of 128 Bladder Cancer Patients
The third application, comprising uncensored remission times (in months) for 128 patients diagnosed with bladder cancer, exhibits a pronounced right-skewed distribution, as discussed by Reference [20]. The remission times are presented in Appendix A.
The summary statistics of the remission times of 128 bladder cancer patients are provided in Table 11. The dataset is positively skewed and leptokurtic (skewness = 3.32 and kurtosis = 16.15).

Table 11.
Summary statistics of 128 patients diagnosed with bladder cancer.
The maximum likelihood estimates (MLEs) and the GoF statistics with respective p-values of the application described Section 7.3 are exhibited in Table 12. The information criteria and negative logarithm are provided in Table 13. All results indicate that the proposed model possesses the smallest A*, W* and K-S values, in addition to having the largest p-value. This attests to the fact that it offers the best fit of the bladder cancer data among models assessed in this study.

Table 12.
MLEs With GoF statistics for 128 patients with bladder cancer.

Table 13.
The negative logarithm and the information criteria results for 128 patients with bladder cancer.
The TTT plot of Dataset 3 is S-shaped—first convex below the line, then concave above it. This suggests a bathtub-shaped hazard rate, with decreasing, constant, then increasing failure rates over time, as shown in Figure 6.
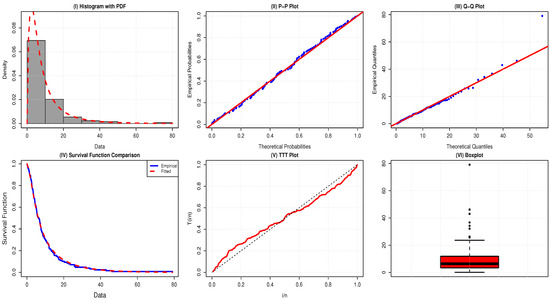
Figure 6.
Comparative plots for evaluation of the adequacy of the AAPEx model on remission times for the data of 128 bladder cancer patients.
7.4. Application 4: United Kingdom COVID-19 Mortality Rate Data
The fourth real-life application involves a dataset comprising COVID-19 mortality rates in the United Kingdom. This dataset was previously utilized by References [21,22]. The data were originally retrieved from the World Health Organization (WHO) COVID-19 [https://covid19.who.int/], which provides official global statistics on COVID-19 cases and deaths. The dataset is fully observed, with no censored or truncated values, and each observation represents a recorded mortality rate at a specific time point. Prior to analysis, basic preprocessing steps such as sorting and validation against the WHO database were performed to ensure consistency and accuracy.
Table 14 reports the summary statistics of the UK COVID-19 death rate data. The positive skewness means that the dataset is skewed to the right, and the kurtosis of less than 3 implies that it is platykurtic. The failure rate is of a bathtub shape, since the TTT-transform plot is first convex (below the 45° line), then concave (above the 45° line). Moreover, the boxplot indicates the existence of a few outliers, as shown in Figure 7.

Table 14.
Summary statistics of United Kingdom COVID-19 mortality rate data.
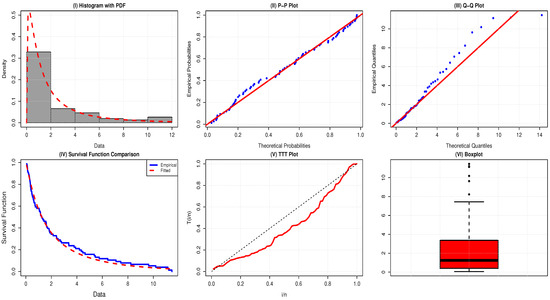
Figure 7.
Visual evaluation of the AAPEx model fitted to mortality rates of 76 UK COVID-19 patients.
The MLEs and the GoF statistics, along with corresponding p-values of the application described in Section 7.4, are reported in Table 15. The negative logarithm and information criteria are given in Table 16. The results reveal that the proposed model possesses the minimum values of A*, W* and K-S, along with the maximum p-value. This verifies that it yields a better fit to the COVID-19 mortality data than other models that were tested in this study.

Table 15.
MLEs with GoF statistics for United Kingdom COVID-19 mortality rate data.

Table 16.
The negative logarithm and the information criteria results for United Kingdom COVID-19 mortality rate data.
7.5. Application 5: Failure Time of Censored Lifetime Data
The fifth application, presented in the Appendix A, involves a dataset of 30 items tested under a Type-II censoring scheme, where the test is terminated after the failure. This dataset was previously utilized by [21,22].
Table 17 reports the summary statistics of the failure time of censored lifetime data. The dataset is almost symmetric, with skewness = 0.0025 and a kurtosis value of less than 3, which implies that it is platykurtic. The failure rate is of a bathtub shape, since the TTT-transform plot is first convex (below the 45° line), then concave (above the 45° line). Moreover, the boxplot indicates the existence of a few outliers, as shown in Figure 8.

Table 17.
Summary statistics of failure time of censored lifetime data.
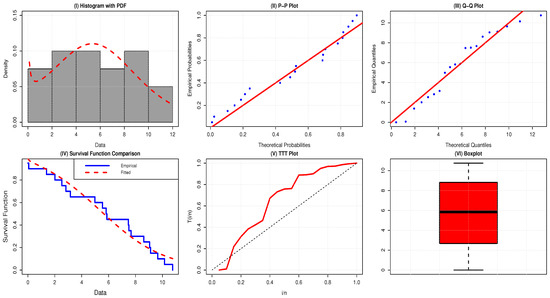
Figure 8.
Visual evaluation of the AAPEx model fitted to mortality rates of 76 UK COVID-19 patients.
The maximum likelihood estimates (MLEs) and goodness-of-fit (GoF) statistics, along with their corresponding p-values for the application described in Section 7.5, are presented in Table 18. The negative log-likelihood and associated information criteria are summarized in Table 19. The results demonstrate that the proposed model achieves the lowest values for the A*, W* and K–S test statistics, accompanied by the highest p-value among all the competing models. These findings confirm that the proposed model provides a superior fit to the failure time of censored lifetime data compared to the alternative models considered in this study.

Table 18.
MLEs with GoF statistics for failure time of censored lifetime data.

Table 19.
The negative logarithm and the information criteria results for failure time of censored lifetime data.
8. Concluding Remarks
This work advances statistical modeling through the development of the AAP-X family, a novel class of continuous distributions designed to address the inflexibility of classical models. Focusing on its exponential subclass, termed the AAPEx model, we resolve critical limitations of the traditional exponential distribution—most notably, its constant hazard rate—by introducing a shape parameter that enables more flexible hazard function modeling. The AAPEx framework supports increasing, decreasing and bathtub-shaped hazard rates, making it well-suited for complex real-world phenomena.
To lay the theoretical groundwork for the AAPEx model, we derived its structural properties, including moments, quantile functions and hazard-rate behavior. Through extensive Monte Carlo simulations, six frequentist estimation approaches (MLE, CVME, MPSE, OLSE, WLSE and ADE) were systematically implemented and compared. Practical applicability was demonstrated through four real-world datasets, including survival times of cancer patients and COVID-19 mortality rates. In all cases, the AAPEx model provided a superior fit compared to several established competing distributions, as measured by standard goodness-of-fit metrics.
This study is based on the following key assumptions: (i) the data are assumed to be independent and identically distributed (i.i.d.); (ii) the model employs a parametric form for the baseline distribution, such as the exponential; and (iii) the shape parameter () is well-defined and governs the distribution’s flexibility. Despite its strengths, the model may face convergence issues with small samples and computational challenges with large datasets, particularly in parameter estimation.
Future research can focus on several promising directions. These include extending the AAP-X model to incorporate survival regression structures for enhanced time-to-event analysis, developing multivariate and time-dependent versions and exploring Bayesian estimation methods to improve flexibility and quantify uncertainty. Furthermore, integrating machine learning techniques could significantly enhance the efficiency and scalability of the model, especially in high-dimensional data settings.
Author Contributions
Conceptualization, A.A.M.; Methodology, A.A.M.; Software, A.A.B. and Y.S.R.; Formal analysis, A.A.B. and B.S.A.; Investigation, S.P.A. and Y.S.R.; Resources, B.S.A. and A.A.; Data curation, A.A.B.; Writing—original draft, A.A.M. and A.A.B.; Writing—review & editing, A.A.B., S.P.A. and Y.S.R.; Visualization, B.S.A. and A.A.; Supervision, S.P.A.; Project administration, B.S.A.; Funding acquisition, B.S.A. and A.A. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by the Deanship of Graduate Studies and Scientific Research at Qassim University for financial support (QU-APC-2025).
Data Availability Statement
The data supporting the findings of this study are available within the article.
Acknowledgments
The researchers would like to thank the Deanship of Graduate Studies and Scientific Research at Qassim University for financial support (QU-APC-2025).
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A
The datasets listed below were analyzed in the section on applications in medical research.
- Application I: Survival Time of 44 Head and Neck Cancer Patients:
1.220, 2.356, 2.374, 2.587, 3.198, 3.700, 4.135, 4.300, 4.738, 5.546, 5.836, 6.347, 6.846, 7.447, 7.826, 8.100, 8.400, 9.200, 9.400, 11.00, 11.20, 11.90, 12.70, 13.00, 13.30, 14.00, 14.60, 15.50, 15.90, 17.30, 17.90, 19.40, 19.50, 20.90, 24.90, 28.10, 31.90, 33.90, 43.20, 46.90, 51.90, 63.30, 72.50, 81.70, 177.60
- Application II: Remission Times of 36 Bladder Cancer Patients:
0.08, 0.20, 0.40, 0.50, 0.51, 0.81, 0.87, 0.90, 1.05, 1.19, 1.26, 1.35, 1.40, 1.46, 1.76, 2.02, 2.02, 2.07, 2.09, 2.23, 2.26, 2.46, 2.54, 2.62, 2.64, 2.69, 2.69, 2.75, 2.83, 2.87, 3.02, 3.02, 3.25, 3.31, 3.36, 3.36
- Application III: Remission Times of 128 Bladder Cancer Patients:
0.08, 2.09, 3.48, 4.87, 6.94, 8.66, 13.11, 23.63, 0.20, 2.23, 3.52, 4.98, 6.97, 9.02, 13.29, 0.40, 2.26, 3.57, 5.06, 7.09, 9.22, 13.80, 25.74, 0.50, 2.46, 3.64, 5.09, 7.26, 9.47, 14.24, 25.82, 0.51, 2.54, 3.70, 5.17, 7.28, 9.74, 14.76, 26.31, 0.81, 2.62, 3.82, 5.32, 7.32, 10.06, 14.77, 32.15, 2.64, 3.88, 5.32, 7.39, 10.34, 14.83, 34.26, 0.90, 2.69, 4.18, 5.34, 7.59, 10.66, 15.96, 36.66, 1.05, 2.69, 4.23, 5.41, 7.62, 10.75, 16.62, 43.01, 1.19, 2.75, 4.26, 5.41, 7.63, 17.12, 46.12, 1.26, 2.83, 4.33, 5.49, 7.66, 11.25, 17.14, 79.05, 1.35, 2.87, 5.62, 7.87, 11.64, 17.36, 1.40, 3.02, 4.34, 5.71, 7.93, 11.79, 18.10, 1.46, 4.40, 5.85, 8.26, 11.98, 19.13, 1.76, 3.25, 4.50, 6.25, 8.37, 12.02, 2.02, 3.31, 4.51, 6.54, 8.53, 12.03, 20.28, 2.02, 3.36, 6.76, 12.07, 21.73, 2.07, 3.36, 6.93, 8.65, 12.63, 22.69
- Application IV: United Kingdom COVID-19 Mortality Rate Data:
0.0587, 0.0863, 0.1165, 0.1247, 0.1277, 0.1303, 0.1652, 0.2079, 0.2395, 0.2845, 0.2992, 0.3188, 0.3317, 0.3446, 0.3553, 0.3622, 0.3926, 0.3926, 0.4633, 0.4690, 0.4954, 0.5139, 0.5696, 0.5837, 0.6197, 0.6365, 0.7096, 0.7444, 0.8590, 1.0438, 1.0602, 1.1305, 1.1468, 1.1533, 1.2260, 1.2707, 1.4149, 1.5709, 1.6017, 1.6083, 1.6324, 1.6998, 1.8164, 1.8392, 1.8721, 2.1360, 2.3987, 2.4153, 2.5225, 2.7087, 2.7946, 3.3609, 3.3715, 3.7840, 4.1969, 4.3451, 4.4627, 4.6477, 5.3664, 5.4500, 5.7522, 6.4241, 7.0657, 8.2307, 9.6315, 10.1870, 11.1429, 11.2019, 11.4584, 0.2751, 0.4110, 0.7193, 1.3423, 1.9844, 3.9042, 7.4456.
- Application V: Failure Time of Censored Lifetime Data:
0.0014, 0.0623, 1.3826, 2.0130, 2.5274, 2.8221, 3.1544, 4.9835, 5.5462, 5.8196, 5.8714, 7.4710, 7.5080, 7.6667, 8.6122, 9.0442, 9.1153, 9.6477, 10.1547, 10.7582.
References
- Mudholkar, G.S.; Srivastava, D.K. Exponentiated Weibull family for analyzing bathtub failure-rate data. IEEE Trans. Reliab. 1993, 42, 299–302. [Google Scholar] [CrossRef]
- Marshall, A.W.; Olkin, I. A new method for adding a parameter to a family of distributions with application to the exponential and Weibull families. Biometrika 1997, 84, 641–652. [Google Scholar] [CrossRef]
- Mahdavi, A.; Kundu, D. A new method for generating distributions with an application to exponential distribution. Commun. Stat.-Theory Methods 2017, 46, 6543–6557. [Google Scholar] [CrossRef]
- Lone, M.A.; Dar, I.H.; Jan, T. A new family of generalized distributions with an application to weibull distribution. Thail. Stat. 2024, 22, 1–16. [Google Scholar]
- Shah, Z.; Khan, D.M.; Khan, I.; Ahmad, B.; Jeridi, M.; Al-Marzouki, S. A novel flexible exponent power-X family of distributions with applications to COVID-19 mortality rate in Mexico and Canada. Sci. Rep. 2024, 14, 8992. [Google Scholar] [CrossRef] [PubMed]
- Alzaatreh, A.; Lee, C.; Famoye, F. A new method for generating families of continuous distributions. Metron 2013, 71, 63–79. [Google Scholar] [CrossRef]
- Musekwa, R.R.; Gabaitiri, L.; Makubate, B. A New Technique to Generate Families of Continuous Distributions. Colomb. J. Stat. Colomb. Estad. 2024, 47, 329–354. [Google Scholar] [CrossRef]
- Alsolmi, M.M. A New Logarithmic Tangent-U Family of Distributions with Reliability Analysis in Engineering Data. Comput. J. Math. Stat. Sci. 2025, 4, 258–282. [Google Scholar] [CrossRef]
- Odhah, O.H.; Alshanbari, H.M.; Ahmad, Z.; Khan, F.; El-Bagoury, A.a.A.H. A New Family of Distributions Using a Trigonometric Function: Properties and Applications in the Healthcare Sector. Heliyon 2024, 10, e29861. [Google Scholar] [CrossRef] [PubMed]
- Mir, A.A.; Rasool, S.U.; Ahmad, S.; Bhat, A.; Jawa, T.M.; Sayed-Ahmed, N.; Tolba, A.H. A Robust Framework for Probability Distribution Generation: Analyzing Structural Properties and Applications in Engineering and Medicine. Axioms 2025, 14, 281. [Google Scholar] [CrossRef]
- Semary, H.; Hussain, Z.; Hamdi, W.A.; Aldahlan, M.A.; Elbatal, I.; Nagarjuna, V.B. Alpha–beta-power family of distributions with applications to exponential distribution. Alex. Eng. J. 2024, 100, 15–31. [Google Scholar] [CrossRef]
- Balakrishnan, K. Exponential Distribution: Theory, Methods and Applications; CRC Press: Boca Raton, FL, USA, 1996. [Google Scholar]
- Gupta, R.D.; Kundu, D. Exponentiated exponential family: An alternative to gamma and Weibull distributions. Biom. J. J. Math. Methods Biosci. 2001, 43, 117–130. [Google Scholar] [CrossRef]
- Isa, A.M.; Bashiru, S.O.; Ali, B.A.; Adepoju, A.A.; Itopa, I.I. Sine-exponential distribution: Its mathematical properties and application to real dataset. UMYU Sci. 2022, 1, 127–131. [Google Scholar]
- Nassar, M.; Afify, A.Z.; Shakhatreh, M. Estimation methods of alpha power exponential distribution with applications to engineering and medical data. Pak. J. Stat. Oper. Res. 2020, 16, 149–166. [Google Scholar] [CrossRef]
- Adepoju, K.; Chukwu, O. Maximum likelihood estimation of the Kumaraswamy exponential distribution with applications. J. Mod. Appl. Stat. Methods 2015, 14, 18. [Google Scholar] [CrossRef]
- Sule, I.; Doguwa, S.I.; Isah, A.; Jibril, H.M. The topp leone kumaraswamy-g family of distributions with applications to cancer disease data. J. Biostat. Epidemiol. 2020, 6, 40–51. [Google Scholar] [CrossRef]
- Abo-Kasem, O.; Ibrahim, O.; Aljohani, H.M.; Hussam, E.; Kilai, M.; Aldallal, R. Statistical Analysis Based on Progressive Type-I Censored Scheme From Alpha Power Exponential Distribution With Engineering and Medical Applications. J. Math. 2022, 2022, 3175820. [Google Scholar] [CrossRef]
- Hibatullah, R.; Widyaningsih, Y.; Abdullah, S. Marshall-Olkin extended power Lindley distribution with application. J. Ris. Apl. Mat. 2018, 2, 84–92. [Google Scholar] [CrossRef]
- Lee, E.T.; Wang, J. Statistical Methods for Survival Data Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2003; Volume 476. [Google Scholar]
- Kilai, M.; Waititu, G.A.; Kibira, W.A.; Alshanbari, H.M.; El-Morshedy, M. A new generalization of Gull Alpha Power Family of distributions with application to modeling COVID-19 mortality rates. Results Phys. 2022, 36, 105339. [Google Scholar] [CrossRef] [PubMed]
- Kargbo, M.; Gichuhi, A.W.; Wanjoya, A.K. A novel extended inverse-exponential distribution and its application to COVID-19 data. Eng. Rep. 2024, 6, e12828. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).