Revisiting Chirality in Slime Mold: On the Emergence and Absence of Lateralized Movement in Physarum polycephalum Influenced by Various Stimuli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. Experiments
2.3. Data Analysis
3. Results
4. Discussion
4.1. Experiments 1 and 2: Lateralized Movement and the Influence of the Genome
4.2. Experiment 3: Phototaxis
4.3. Experiment 4: Magnetotaxis
4.4. Experiments 5 and 6: Inclination
4.5. Experiment 7: Vibration
4.6. General Discussion
4.7. Outlook
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rogers, L.J.; Vallortigara, G. When and Why Did Brains Break Symmetry? Symmetry 2015, 7, 2181–2194. [Google Scholar] [CrossRef]
- Frasnelli, E.; Vallortigara, G.; Rogers, L.J. Left–right asymmetries of behaviour and nervous system in invertebrates. Neurosci. Biobehav. Rev. 2012, 36, 1273–1291. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, L.N.; Levin, M. A unified model for left–right asymmetry? Comparison and synthesis of molecular models of embryonic laterality. Dev. Biol. 2013, 379, 1–15. [Google Scholar] [CrossRef]
- Lobikin, M.; Wang, G.; Xu, J.; Hsieh, Y.W.; Chuang, C.F.; Lemire, J.M.; Levin, M. Early, nonciliary role for microtubule proteins in left–right patterning is conserved across kingdoms. Proc. Natl. Acad. Sci. USA 2012, 109, 12586–12591. [Google Scholar] [CrossRef] [PubMed]
- Sauer, S.; Klar, A.J.S. Left-right symmetry breaking in mice by left-right dynein may occur via a biased chromatid segregation mechanism, without directly involving the Nodal gene. Front. Oncol. 2012, 2, 166. [Google Scholar] [CrossRef]
- Lyon, P.; Keijzer, F.; Arendt, D.; Levin, M. Reframing cognition: Getting down to biological basics. Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20190750. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Lemire, J.M.; Levin, M. It’s never too early to get it Right: A conserved role for the cytoskeleton in left-right asymmetry. Commun. Integr. Biol. 2013, 6, e27155. [Google Scholar] [CrossRef] [PubMed]
- Dimonte, A.; Adamatzky, A.; Erokhin, V.; Levin, M. On chirality of slime mould. Biosystems 2016, 140, 23–27. [Google Scholar] [CrossRef]
- Aldrich, H. Cell Biology of Physarum and Didymium V1: Organisms, Nucleus, and Cell Cycle; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Reid, C.R. Thoughts from the forest floor: A review of cognition in the slime mould Physarum polycephalum. Anim. Cogn. 2023, 26, 1783–1797. [Google Scholar] [CrossRef]
- Nakagaki, T.; Yamada, H.; Tóth, Á. Maze-solving by an amoeboid organism. Nature 2000, 407, 470. [Google Scholar] [CrossRef]
- Dussutour, A.; Latty, T.; Beekman, M.; Simpson, S.J. Amoeboid organism solves complex nutritional challenges. Proc. Natl. Acad. Sci. USA 2010, 107, 4607–4611. [Google Scholar] [CrossRef] [PubMed]
- Boisseau, R.P.; Vogel, D.; Dussutour, A. Habituation in non-neural organisms: Evidence from slime moulds. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160446. [Google Scholar] [CrossRef]
- Frankel, J. Intracellular Handedness in Ciliates. In Ciba Foundation Symposium 162—Biological Asymmetry and Handedness; John Wiley & Sons, Ltd.: Hoboken, NJ, USA; pp. 73–93.
- Brugger, P.; Macas, E.; Ihlemann, J. Do sperm cells remember? Behav. Brain Res. 2002, 136, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Hato, M.; Ueda, T.; Kurihara, K.; Kobatake, Y. Phototaxis in True Slime Mold Physarum polycephalum. Cell Struct. Funct. 1976, 1, 269–278. [Google Scholar] [CrossRef]
- Wolf, R.; Niemuth, J.; Sauer, H. Thermotaxis and protoplasmic oscillations in Physarum plasmodia analysed in a novel device generating stable linear temperature gradients. Protoplasma 1997, 197, 121–131. [Google Scholar] [CrossRef]
- Wolke, A.; Niemeyer, F.; Achenbach, F. Geotactic behavior of the acellular myxomycete Physarum polycephalum. Cell Biol. Int. Rep. 1987, 11, 525–528. [Google Scholar] [CrossRef]
- Shirakawa, T.; Konagano, R.; Inoue, K. Novel taxis of the Physarum plasmodium and a taxis-based simulation of Physarum swarm. In Proceedings of the 6th International Conference on Soft Computing and Intelligent Systems, and the 13th International Symposium on Advanced Intelligence Systems, Kobe, Japan, 20–24 November 2012; pp. 296–300. [Google Scholar]
- Winsett, K.E.; Stephenson, S.L. Global distribution and molecular diversity of Didymium difforme. Mycosphere 2011, 2, 135–146. [Google Scholar]
- Nakagaki, T.; Umemura, S.; Kakiuchi, Y.; Ueda, T. Action Spectrum for Sporulation and Photoavoidance in the Plasmodium of Physarum polycephalum, as Modified Differentially by Temperature and Starvation. Photochem. Photobiol. 1996, 64, 859–862. [Google Scholar] [CrossRef]
- Mayne, R. Biology of the Physarum polycephalum Plasmodium: Preliminaries for Unconventional Computing. In Advances in Physarum Machines: Sensing and Computing with Slime Mould; Adamatzky, A., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–22. [Google Scholar]
- Dussutour, A.; Ma, Q.; Sumpter, D. Phenotypic variability predicts decision accuracy in unicellular organisms. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182825. [Google Scholar] [CrossRef]
- Häder, D.-P.; Schreckenbach, T. Phototactic Orientation in Plasmodia of the Acellular Slime Mold, Physarum polycephalum. Plant Cell Physiol. 1984, 25, 55–61. [Google Scholar] [CrossRef]
- Shirakawa, T.; Sato, H.; Muro, M.; Konagano, R.; Ohno, R.; Inoue, K. Magnetotaxis of the Physarum Plasmodium and Construction of a Magnetically Controlled Physarum Logic Gate. Int. J. Unconv. Comput. 2020, 15, 245–258. [Google Scholar]
- Boussard, A.; Fessel, A.; Oettmeier, C.; Briard, L.; Döbereiner, H.-G.; Dussutour, A. Adaptive behaviour and learning in slime moulds: The role of oscillations. Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20190757. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, A.; Tanaka, L.Y.; Massucatto, R.C.; Laurindo, F.R.; Aiello, C.D. Automated 1D Helmholtz coil design for cell biology: Weak magnetic fields alter cytoskeleton dynamics. arXiv 2024, arXiv:2406.19555. [Google Scholar]
- Rojas, C.; Stephenson, S.L. Myxomycetes—Biology, Systematics, Biogeography and Ecology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Murugan, N.J.; Kaltman, D.H.; Jin, P.H.; Chien, M.; Martinez, R.; Nguyen, C.Q.; Kane, A.; Novak, R.; Ingber, D.E.; Levin, M. Mechanosensation Mediates Long-Range Spatial Decision-Making in an Aneural Organism. Adv. Mater. 2021, 33, 2008161. [Google Scholar] [CrossRef]
- Tekle, Y.I.; Williams, J.R. Cytoskeletal architecture and its evolutionary significance in amoeboid eukaryotes and their mode of locomotion. R. Soc. Open Sci. 2016, 3, 160283. [Google Scholar] [CrossRef]
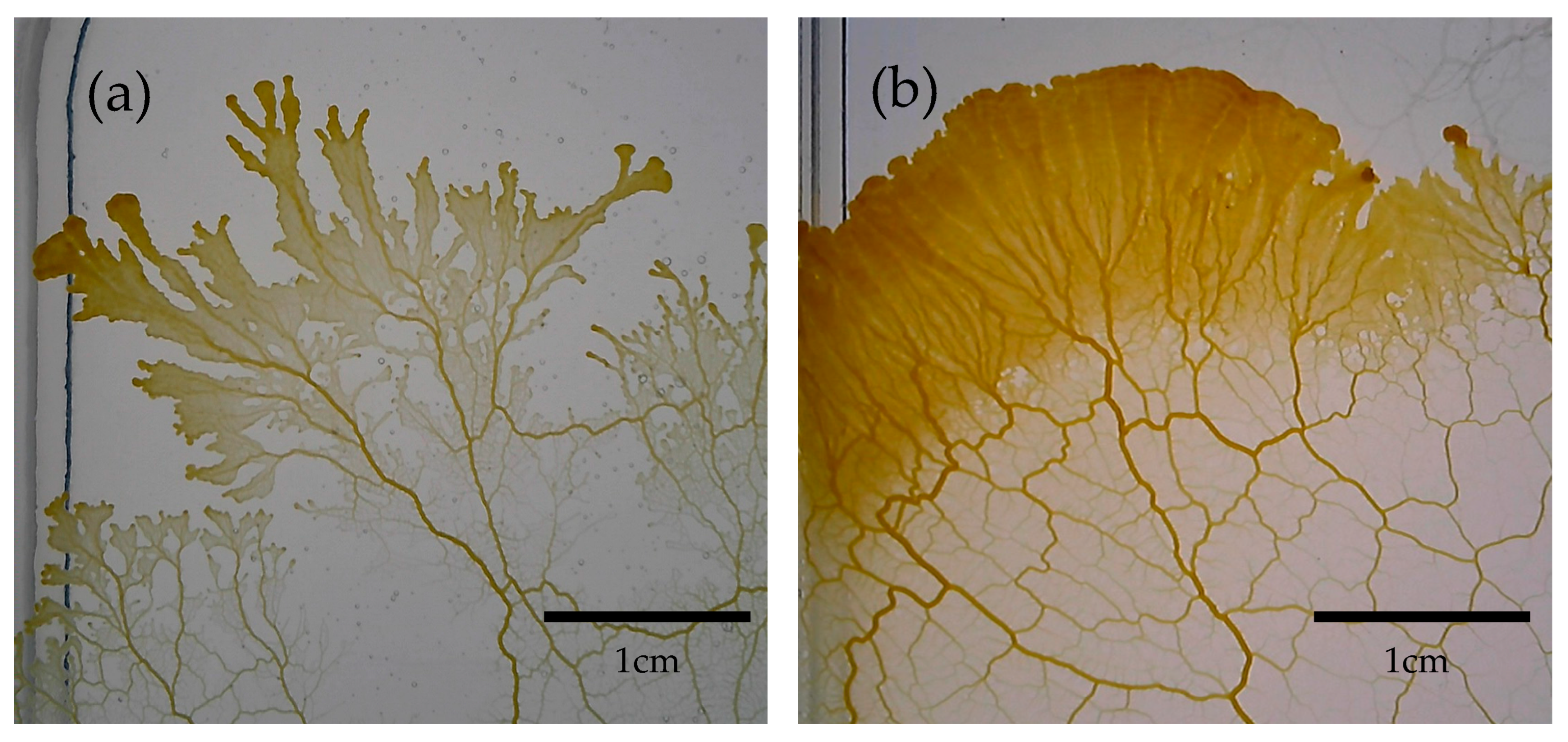
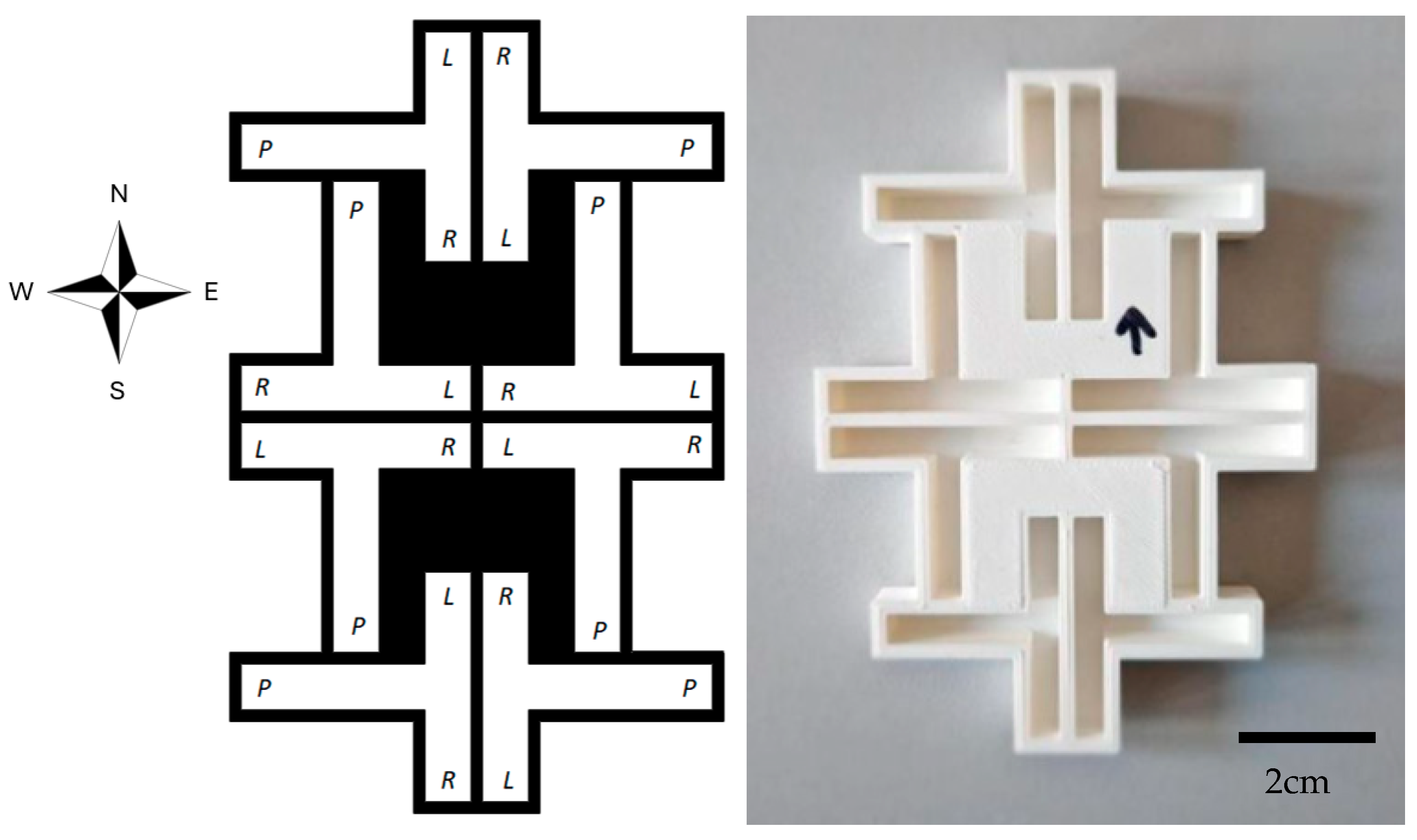

| Total | Start North | Start South | Start West | Start East | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Experiment | Right | Left | Right | Left | Right | Left | Right | Left | Right | Left | |
| 1 Replication | n | 121 | 122 | 34 | 30 | 29 | 28 | 36 | 25 | 22 | 39 |
| % | 49.8 | 50.2 | 53.1 | 46.9 | 50.9 | 49.1 | 59 | 41 | 36.1 | 63.9 | |
| p | 1 | 0.532 | 0.792 | 0.124 | 0.040 * | ||||||
| 2 Genetic variation | n | 109 | 116 | 27 | 24 | 21 | 33 | 32 | 28 | 29 | 31 |
| % | 48.4 | 51.6 | 52.9 | 47.1 | 38.9 | 61.1 | 53.3 | 46.7 | 48.3 | 51.7 | |
| p | 0.690 | 0.576 | 0.134 | 0.518 | 0.898 | ||||||
| 3 Phototaxis | n | 106 | 106 | 28 | 28 | 33 | 19 | 21 | 32 | 24 | 27 |
| (total) | % | 50 | 50 | 50 | 50 | 63.5 | 36.5 | 39.6 | 60.4 | 47.1 | 52.9 |
| p | 0.946 | 0.894 | 0.036 * | 0.168 | 0.780 | ||||||
| 3a Phototaxis | n | 58 | 53 | 12 | 15 | 17 | 11 | 15 | 13 | 14 | 14 |
| (4000 lux) | % | 52.3 | 47.7 | 44.4 | 55.6 | 60.7 | 39.3 | 53.6 | 46.4 | 50 | 50 |
| p | 0.570 | 0.702 | 0.184 | 0.572 | 0.850 | ||||||
| 3b Phototaxis | n | 48 | 53 | 16 | 13 | 16 | 8 | 6 | 19 | 10 | 13 |
| (2000 lux) | % | 47.5 | 52.5 | 55.2 | 44.8 | 66.7 | 33.3 | 24 | 76 | 43.5 | 56.5 |
| p | 0.690 | 0.458 | 0.064 | 0.014 * | 0.678 | ||||||
| 4 Magnetotaxis | n | 134 | 143 | 37 | 33 | 36 | 36 | 24 | 42 | 37 | 32 |
| (total) | % | 48.4 | 51.6 | 52.9 | 47.1 | 50 | 50 | 36.4 | 63.6 | 53.6 | 46.4 |
| p | 0.630 | 0.550 | 0.906 | 0.036 * | 0.470 | ||||||
| 4a Magnetotaxis | n | 58 | 86 | 13 | 24 | 17 | 21 | 14 | 18 | 14 | 23 |
| (2.5–3 mT) | % | 40.3 | 59.7 | 35.1 | 64.9 | 44.7 | 55.3 | 43.8 | 56.3 | 37.8 | 62.2 |
| p | 0.024 * | 0.098 | 0.628 | 0.596 | 0.188 | ||||||
| 4b Magnetotaxis | n | 76 | 57 | 24 | 9 | 19 | 15 | 10 | 24 | 23 | 9 |
| (0.3–0.5 mT) | % | 57.1 | 42.9 | 72.7 | 27.3 | 55.9 | 44.1 | 29.4 | 70.6 | 71.9 | 28.1 |
| p | 0.082 | 0.004 ** | 0.392 | 0.024 * | 0.008 ** | ||||||
| 5 Inclination | n | 110 | 101 | 27 | 29 | 31 | 23 | 35 | 17 | 17 | 32 |
| % | 52.1 | 47.9 | 48.2 | 51.8 | 57.4 | 42.6 | 67.3 | 32.7 | 34.7 | 65.3 | |
| p | 0.492 | 0.894 | 0.220 | 0.008 ** | 0.044 * | ||||||
| 6 Reverse Inclination | n | 99 | 131 | 24 | 22 | 24 | 41 | 9 | 49 | 42 | 19 |
| % | 43 | 57 | 52.2 | 47.8 | 36.9 | 63.1 | 15.5 | 84.5 | 68.9 | 31.1 | |
| p | 0.040 * | 0.658 | 0.046 * | <0.001 ** | 0.002 ** | ||||||
| 7 Vibration | n | 76 | 126 | 16 | 35 | 32 | 17 | 16 | 36 | 12 | 38 |
| % | 37.6 | 62.4 | 31.4 | 68.6 | 65.3 | 34.7 | 30.8 | 69.2 | 24 | 76 | |
| p | 0.001 ** | 0.011 * | 0.0212 * | 0.008 ** | <0.001 ** | ||||||
| Experiment | North | South | East | West | |
|---|---|---|---|---|---|
| 1 Replication | n | 75 | 47 | 62 | 59 |
| % | 0.61 | 0.39 | 0.51 | 0.49 | |
| p | 0.008 ** | 0.716 | |||
| 2 Genetic Variation | n | 63 | 57 | 60 | 45 |
| % | 0.53 | 0.48 | 0.57 | 0.43 | |
| p | 0.522 | 0.118 | |||
| 3 Phototaxis | n | 48 | 56 | 47 | 61 |
| % | 0.46 | 0.54 | 0.44 | 0.56 | |
| p | 0.493 | 0.211 | |||
| 4 Magnetotaxis | n | 56 | 79 | 73 | 69 |
| % | 0.41 | 0.59 | 0.51 | 0.49 | |
| p | 0.058 | 0.674 | |||
| 5 Inclination | n | 67 | 34 | 50 | 60 |
| % | 0.66 | 0.34 | 0.45 | 0.55 | |
| p | <0.001 ** | 0.391 | |||
| 6 Reverse Inclination | n | 28 | 91 | 65 | 46 |
| % | 0.24 | 0.76 | 0.59 | 0.41 | |
| p | <0.001 ** | 0.057 | |||
| 7 Vibration | n | 54 | 48 | 33 | 67 |
| % | 0.53 | 0.47 | 0.33 | 0.67 | |
| p | 0.488 | <0.001 ** | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gehrke, R.; Freiberg, J. Revisiting Chirality in Slime Mold: On the Emergence and Absence of Lateralized Movement in Physarum polycephalum Influenced by Various Stimuli. Symmetry 2025, 17, 756. https://doi.org/10.3390/sym17050756
Gehrke R, Freiberg J. Revisiting Chirality in Slime Mold: On the Emergence and Absence of Lateralized Movement in Physarum polycephalum Influenced by Various Stimuli. Symmetry. 2025; 17(5):756. https://doi.org/10.3390/sym17050756
Chicago/Turabian StyleGehrke, Rowena, and Jannes Freiberg. 2025. "Revisiting Chirality in Slime Mold: On the Emergence and Absence of Lateralized Movement in Physarum polycephalum Influenced by Various Stimuli" Symmetry 17, no. 5: 756. https://doi.org/10.3390/sym17050756
APA StyleGehrke, R., & Freiberg, J. (2025). Revisiting Chirality in Slime Mold: On the Emergence and Absence of Lateralized Movement in Physarum polycephalum Influenced by Various Stimuli. Symmetry, 17(5), 756. https://doi.org/10.3390/sym17050756







