Multiple Alignments of Protein Families with Weak Sequence Similarity Within the Family
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview of the MAHDS Algorithm
2.2. The Algorithm for Creating MSAr and PWMr
2.3. PWMr Optimization Using Set Q
2.4. Calculation of F(Li, L) for Sequence S of Set Q with Length Li and PWM with Length L
2.5. Calculation of MSAQ
2.6. Assessment of the Statistical Significance of MSAs
2.7. Software Used
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| MSA | Multiple sequence alignment |
| PWM | Position–weight matrix |
References
- Thompson, J.D.; Linard, B.; Lecompte, O.; Poch, O. A Comprehensive Benchmark Study of Multiple Sequence Alignment Methods: Current Challenges and Future Perspectives. PLoS ONE 2011, 6, e18093. [Google Scholar] [CrossRef] [PubMed]
- Kemena, C.; Notredame, C. Upcoming Challenges for Multiple Sequence Alignment Methods in the High-Throughput Era. Bioinformatics 2009, 25, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Chatzou, M.; Magis, C.; Chang, J.-M.; Kemena, C.; Bussotti, G.; Erb, I.; Notredame, C. Multiple Sequence Alignment Modeling: Methods and Applications. Brief. Bioinform. 2016, 17, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Chowdhury, B.; Garai, G. A Review on Multiple Sequence Alignment from the Perspective of Genetic Algorithm. Genomics 2017, 109, 419–431. [Google Scholar] [CrossRef]
- Paruchuri, T.; Kancharla, G.R.; Dara, S.; Yadav, R.K.; Jadav, S.S.; Dhamercherla, S.; Vidyarthi, A. Nature Inspired Algorithms for Solving Multiple Sequence Alignment Problem: A Review. Arch. Computat Methods Eng. 2022, 29, 5237–5258. [Google Scholar] [CrossRef]
- Ibrahim, M.; Yusof, U.; Eisa, T.; Nasser, M. Bioinspired Algorithms for Multiple Sequence Alignment: A Systematic Review and Roadmap. Appl. Sci. 2024, 14, 2433. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A Novel Method for Fast and Accurate Multiple Sequence Alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef]
- Higgins, D.G.; Sharp, P.M. CLUSTAL: A Package for Performing Multiple Sequence Alignment on a Microcomputer. Gene 1988, 73, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Lassmann, T.; Sonnhammer, E.L. Kalign—An Accurate and Fast Multiple Sequence Alignment Algorithm. BMC Bioinform. 2005, 6, 298. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, T. On the Complexity of Multiple Sequence Alignment. J. Comput. Biol. 1994, 1, 337–348. [Google Scholar] [CrossRef]
- Feng, D.-F.; Doolittle, R.F. Progressive Sequence Alignment as a Prerequisitetto Correct Phylogenetic Trees. J. Mol. Evol. 1987, 25, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Hogeweg, P.; Hesper, B. The Alignment of Sets of Sequences and the Construction of Phyletic Trees: An Integrated Method. J. Mol. Evol. 1984, 20, 175–186. [Google Scholar] [CrossRef]
- Hirosawa, M.; Totoki, Y.; Hoshida, M.; Ishikawa, M. Comprehensive Study on Iterative Algorithms of Multiple Sequence Alignment. Comput. Appl. Biosci. 1995, 11, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Notredame, C.; O’Brien, E.A.; Higgins, D.G. RAGA: RNA Sequence Alignment by Genetic Algorithm. Nucleic Acids Res. 1997, 25, 4570–4580. [Google Scholar] [CrossRef]
- Kim, J.; Pramanik, S.; Chung, M.J. Multiple Sequence Alignment Using Simulated Annealing. Comput. Appl. Biosci. 1994, 10, 419–426. [Google Scholar] [CrossRef]
- Althaus, E.; Caprara, A.; Lenhof, H.-P.; Knut, R. A Branch-and-Cut Algorithm for Multiple Sequence Alignment. Math. Program. 2006, 105, 387–425. [Google Scholar] [CrossRef]
- Korotkov, E.V.; Kostenko, D.O. Application of the MAHDS Method for Multiple Alignment of Highly Diverged Amino Acid Sequences. Int. J. Mol. Sci. 2022, 23, 3764. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Korotkov, E.V.; Suvorova, Y.M.; Kostenko, D.O.; Korotkova, M.A. Multiple Alignment of Promoter Sequences from the Arabidopsis Thaliana L. Genome. Genes 2021, 12, 135. [Google Scholar] [CrossRef] [PubMed]
- Pugacheva, V.; Korotkov, A.; Korotkov, E. Search of Latent Periodicity in Amino Acid Sequences by Means of Genetic Algorithm and Dynamic Programming. Stat. Appl. Genet. Mol. Biol. 2016, 15, 381–400. [Google Scholar] [CrossRef]
- Korotkov, E.; Suvorova, Y.; Kostenko, D.; Korotkova, M. Search for Dispersed Repeats in Bacterial Genomes Using an Iterative Procedure. Int. J. Mol. Sci. 2023, 24, 10964. [Google Scholar] [CrossRef] [PubMed]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bairoch, A. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007, 406, 89–112. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Comet, J.P.; Aude, J.C.; Glémet, E.; Risler, J.L.; Hénaut, A.; Slonimski, P.P.; Codani, J.J. Significance of Z-Value Statistics of Smith-Waterman Scores for Protein Alignments. Comput. Chem. 1999, 23, 317–331. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Schneider, T.D.; Stephens, R.M. Sequence Logos: A New Way to Display Consensus Sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef]
- Thompson, J.D.; Plewniak, F.; Poch, O. BAliBASE: A Benchmark Alignment Database for the Evaluation of Multiple Alignment Programs. Bioinformatics 1999, 15, 87–88. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Koehl, P.; Ripp, R.; Poch, O. BAliBASE 3.0: Latest Developments of the Multiple Sequence Alignment Benchmark. Proteins 2005, 61, 127–136. [Google Scholar] [CrossRef] [PubMed]

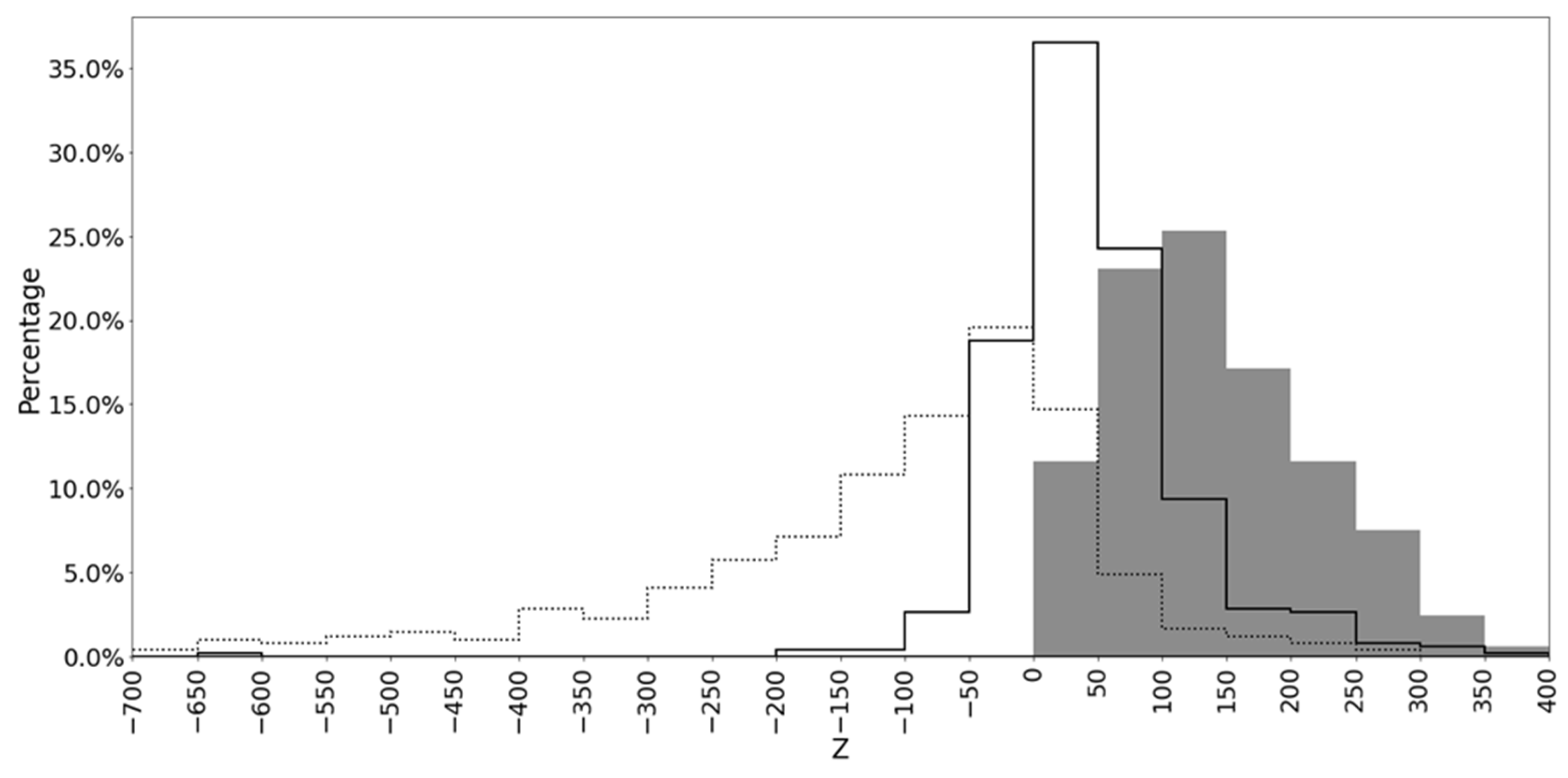
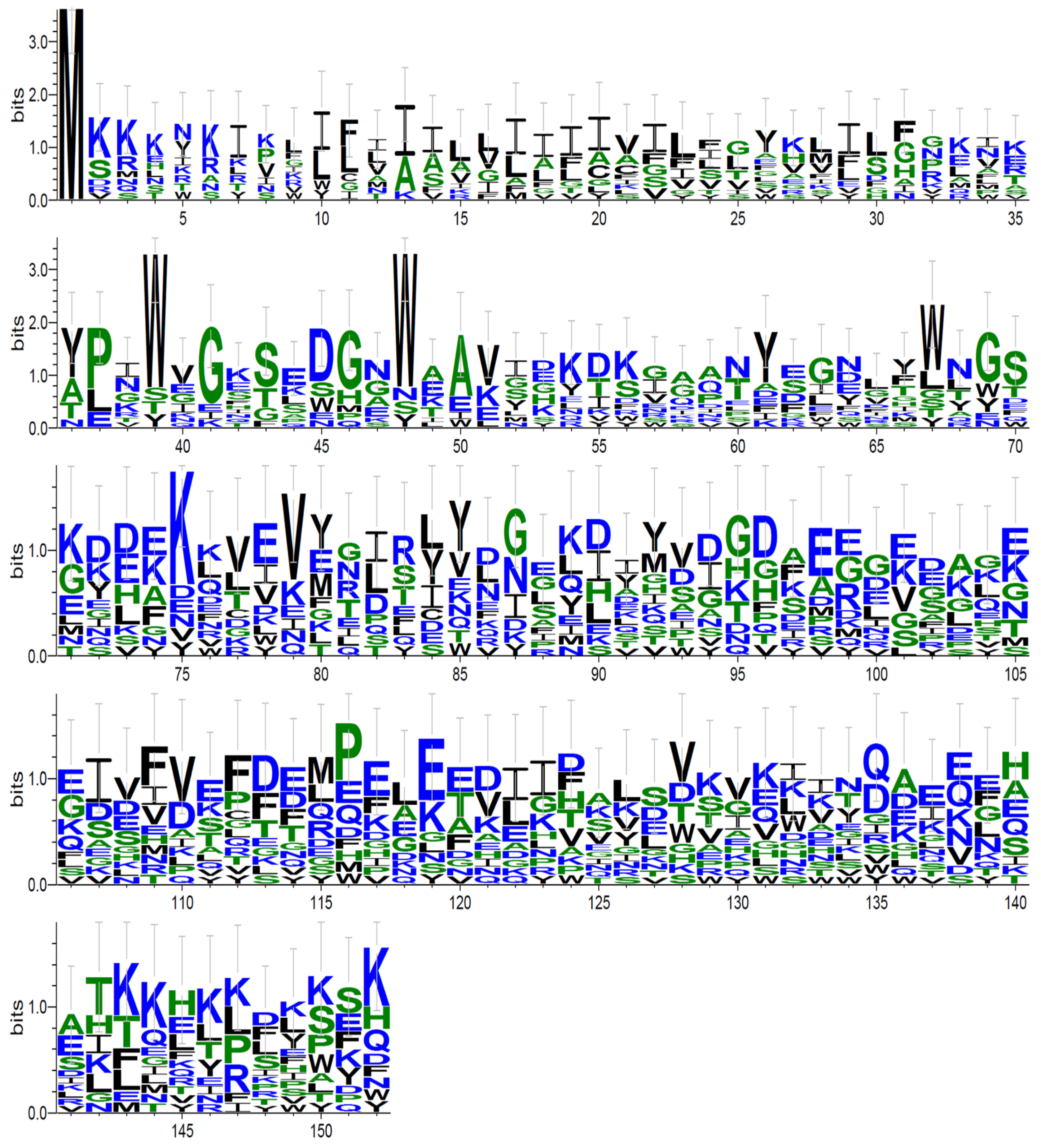
| Methods | Mean 1 | Significant Alignments 2 | Most Significant Alignments 3 | Unique Significant Alignments 4 |
|---|---|---|---|---|
| MAHDS | 141.99 | 480 | 476 | 138 |
| MUSCLE | 47.81 | 344 | 6 | 2 |
| T-Coffee | −136.16 | 96 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostenko, D.; Korotkova, M.; Korotkov, E. Multiple Alignments of Protein Families with Weak Sequence Similarity Within the Family. Symmetry 2025, 17, 408. https://doi.org/10.3390/sym17030408
Kostenko D, Korotkova M, Korotkov E. Multiple Alignments of Protein Families with Weak Sequence Similarity Within the Family. Symmetry. 2025; 17(3):408. https://doi.org/10.3390/sym17030408
Chicago/Turabian StyleKostenko, Dmitrii, Maria Korotkova, and Eugene Korotkov. 2025. "Multiple Alignments of Protein Families with Weak Sequence Similarity Within the Family" Symmetry 17, no. 3: 408. https://doi.org/10.3390/sym17030408
APA StyleKostenko, D., Korotkova, M., & Korotkov, E. (2025). Multiple Alignments of Protein Families with Weak Sequence Similarity Within the Family. Symmetry, 17(3), 408. https://doi.org/10.3390/sym17030408






