1. Introduction
Bryophytes, second only to angiosperms in terms of diversity and distribution, constitute a vital component of global biodiversity [
1]. Bryophytes are often neglected in ecological environmental research because of their small size and simple structures [
2]. However, research has shown that bryophytes play significant roles in ecological conservation, environmental monitoring, and scientific studies of biodiversity [
3]. For example, the change in heavy metal content in the same bryophyte species or genuses can indicate the air pollution situation in specific areas [
4]. Therefore, relevant research on bryophytes should be strengthened to protect their populations and communities [
5].
Plant-related research such as plant species identification, disease detection, and yield estimation typically requires professional knowledge and significant manpower resources [
6,
7]. Compared with the traditional machine learning methods, the development of deep neural networks reduces labor demand and improves work efficiency [
8]. Classical deep neural networks, such as ResNet [
9], VGG [
10], Inception [
11], and EfficientNet [
12], have been well applied in species classification, plant pest and disease identification, fruit detection, etc. These networks leverage image features of plants, which may include flowers, fruits, leaves, stems, or the entire plant, to achieve high accuracy. Rzanny et al. [
13] employed the Inception-ResNet-v2 [
14] network and fused the flower front view, flower side view, and leaf top view of herbaceous plants for training, thereby achieving a high-precision recognition of herbaceous plants. Voncarlos et al. [
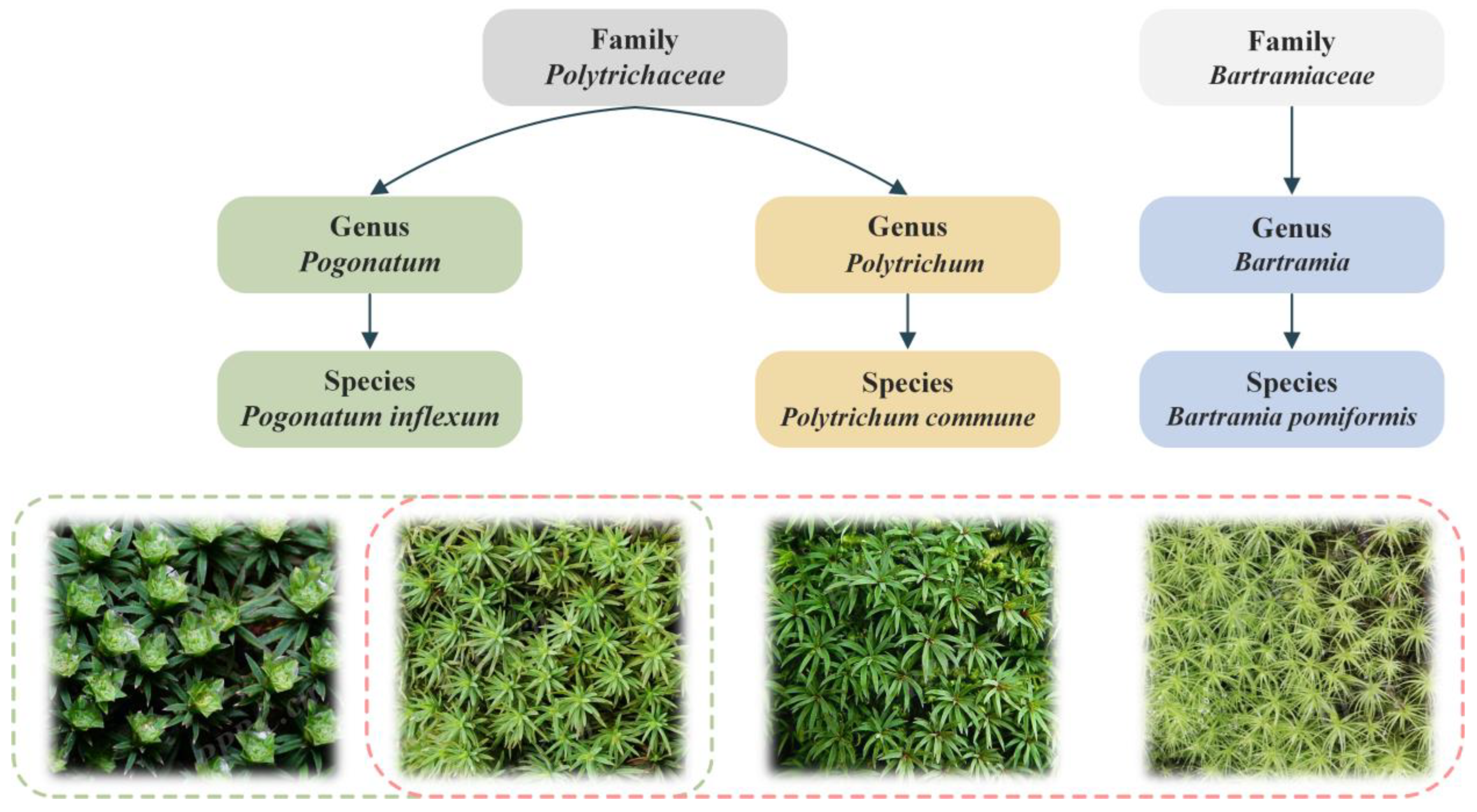
15] proposed a fine-grained plant species recognition method using a hierarchical classification strategy with global and local leaf images, employing a Siamese Convolutional Neural Network (SCNN) to identify plant genera and species from coarse to fine. Similarly, bryophytes can be classified by leaf and spore characteristics. However, species recognition in complex real scenes is a fine-grained visual classification problem that has high inter-class similarity and high intra-class variability. As shown in
Figure 1, from left to right, the images in the red box are three different bryophyte species,
Pogonatum inflexum,
Polytrichum commune, and
Bartramia pomiformis, but their image features are highly similar. Conversely, the two images in the green box both belong to
Pogonatum inflexum, but their image features are obviously different. Meanwhile, the bryophyte leaves presented in
Figure 1 exhibit local or global symmetry, such as radial symmetry. Therefore, the design of a deep learning model specifically for the fine-grained image recognition of bryophyte features is helpful to promote research on bryophyte diversity.
Publicly available datasets for bryophyte classification are limited, which makes model training challenging. Only the dataset constructed by Uzhinsky et al. [
16], which contains 599 images, covers a small number of moss species and has an insufficient data volume. Moreover, its relatively uniform collection environment may restrict the model’s generalization ability. To address this limitation, we construct BryophyteFine, a dataset dedicated to bryophyte classification. The dataset collects and organizes images of 26 bryophyte species, covering 16 families and 23 genera, and containing images under different lighting and background conditions. After data preprocessing steps such as cleaning, standardization, and augmentation, each species has 1000 images, ensuring a comprehensive and sufficient sample for model training. This work provides a solid data foundation for bryophyte classification studies. The leaves of bryophytes typically grow densely, creating a thin, lush green or yellow-green covering, and their images display various shapes, such as linear and sheet-like forms. Due to the small size of individual bryophytes, varying degrees of enlargement occur during image collection, resulting in images with multi-scale features. Meanwhile, a single enlarged image may contain several dozens of plant bodies, with intricate shapes and veins of leaves that are often difficult to identify clearly. These characteristics of bryophyte images make it challenging for existing classification methods (such as CNN and Transformer variants) to effectively extract multi-scale features and fine details in complex real-world scenarios, leading to reduced recognition accuracy. CNN methods often rely on fixed scale feature extraction, resulting in poor adaptability to bryophyte images at different magnification ratios. Although the Transformer method leverages self-attention to handle multi-scale features, it struggles to capture fine-grained local details. This limitation creates a bottleneck in Top-1 recognition accuracy while also demanding extensive computational resources, limiting its practical application. To address these challenges, deep neural network models need to focus on both local and global features while optimizing computational efficiency. Local features include details of leaf shapes and veins in bryophytes, while global features provide scale invariance, enabling the model to retain stability across different scales and effectively deal with scale changes in the image (such as distance and scaling).
Based on this concept, we propose a lightweight deep learning model that constructs modules specifically for the fine-grained features of bryophytes, improving classification accuracy while maintaining low complexity. Specifically, we propose a Dual-Branch Multi-scale (DBMS) module that integrates Partial Convolution (PConv) [
17] with Dilated Convolution Enhanced Attention (DCEA). PConv is an advanced convolution method that can extract local details more efficiently. Meanwhile, the DCEA module introduces dilated convolution based on the self-attention mechanism and overcomes the limitations of PConv by expanding the receptive field, so as to capture features of different scales more comprehensively on the premise of ensuring computational efficiency. Multiscale fusion is essentially a symmetrical processing in scale space, which helps models maintain feature consistency at different scales. To further enhance the quality of the model’s input features, we propose a Convolutional Patch Embedding (CPE) module. By performing the convolutional processing before dividing the patch, our approach retains more fine-grained details and enhances the efficiency of the self-attention mechanism while reducing the computational load. Convolution operation naturally has translational invariance, suitable for capturing local symmetric textures or structures. CPE makes the model more competent for processing the BryophyteFine dataset. By integrating the DBMS module and the CPE module, MOSSNet forms a lightweight deep learning network with a clean and efficient design that achieves high performance while reducing computation and storage requirements. On BryophyteFine’s test set, MOSSNet achieves 99.02% Top-1 accuracy. This shows that the proposed model has significant performance advantages in the fine-grained image classification of bryophytes. The main contributions of this paper are as follows:
We construct BryophyteFine, a fine-grained dataset for bryophyte classification that consists of 26,000 images covering 16 families, 23 genera, and 26 species of bryophytes and presents their diverse visual features in different environments, scales, and morphologies.
According to the characteristics of bryophyte images, we propose a DBMS module that integrates convolution with DCEA. This approach effectively extracts both local and global features while reducing computational redundancy and ensuring high recognition accuracy.
We propose a CPE module to improve the input feature quality of subsequent modules, thereby further improving the model’s ability to capture complex symmetric features in bryophyte images and reducing computational complexity.
With DBMS and CPE, the fine-grained classification model MOSSNet for bryophyte significantly reduces the number of parameters and computational complexity, achieving 99.02% Top-1 classification accuracy on the BryophyteFine test set with only 1.59 M parameters and 0.07 G FLOPs.
The structure of the paper is as follows:
Section 2 provides an overview of relevant research on plant image classification and mainstream methods of image classification;
Section 3 introduces the structure of our proposed model and the construction of the BryophyteFine dataset in detail;
Section 4 presents the model evaluation experiment as well as the analysis of experimental results;
Section 5 makes an overall summary of the paper’s work and discusses the future direction for improvement.
2. Related Work
This section provides an overview of general plant classification methods and bryophyte classification methods, as well as exploring the deep neural network model combining Vision Transformer (ViT) and Convolutional Neural Network (CNN) and the feasibility of its application to bryophyte classification.
Plant Classification Research: Plant classification research is an important branch of the computer vision field that usually relies on image recognition techniques. There are currently multiple related image datasets available to evaluate plant classification methods, such as LeafSnap [
18], Flavia [
19], Swedish [
20], MalayaKew [
21], and PlantCLEF [
22]. These datasets contain feature images of leaves, flowers, fruits, and whole plants. Much existing research on plant species classification focuses on utilizing deep learning-based image recognition techniques to improve recognition efficiency and automation levels. An effective dataset is a prerequisite for applying deep learning methods. Lee et al. [
23] proposed a deep learning model that combines CNN and ViT to achieve the automatic recognition of plant species. They employed four pre-trained models—ResNet-50, DenseNet-201, Xception, and ViT—to fully leverage the advantages of different deep learning models by concatenating and fusing their outputs, significantly improving the effectiveness of leaf feature extraction. However, this model integration method also increases the computational complexity of the classification process. Voncarlos et al. [
13] utilized SCNN and selected VGG16 as the backbone, combined with two-view leaf image representation and a hierarchical classification strategy to achieve the accurate classification of plant leaves. The method has the potential to be extended to new plant species and, to some extent, reduces the dependence of deep learning models on large numbers of training data. Lapkovskis et al. [
24] shifted the focus of plant classification research to multimodal learning and proposed a multimodal fusion architecture based on MobileNetV3. This architecture integrates images of multiple plant organs (flowers, leaves, fruits, and stems), which have stronger representation ability and robustness.
Bryophyte classification Research: The classification of bryophytes falls under fine-grained classification, a field where research is relatively limited and publicly available datasets for model evaluation are scarce. Bryophyte images have the characteristics of small and dense plant bodies, and previous studies have primarily focused on the recognition of a few bryophyte species. Ise et al. [
25] proposed a “chopped picture” method, which cuts large-sized images into multiple small-sized images to process blurry and shapeless image data of bryophytes. Using the LeNet model, they successfully identified three bryophyte species. However, this image processing method lacks global information after cutting the image, and as the number of recognition species increases, feature confusion may arise. Uzhinskiy et al. [
16] constructed a public dataset containing 599 images of bryophytes and proposed an SCNN architecture based on MobileNetV2. This architecture integrates a triplet loss function to measure image similarity, enabling the classification of five bryophyte species. Li et al. [
26] constructed a dataset covering 110 species of bryophyte for classification research and optimized the data processing method proposed by Ise et al. using the K-means clustering algorithm. Based on the Swin Transformer architecture, they introduced the Swin Routiformer Block, a feature interaction module that improved classification accuracy and image processing speed for bryophytes, achieving a Top-1 accuracy of 82.19%. Although this method can recognize more species compared with previous bryophyte classification methods, the increase in model size also results in higher computational resource demands, limiting its practical application. Similarly, to study the fine-grained classification of bryophytes, we construct a dataset named BryophyteFine. It includes images of four bryophyte species collected by Uzhinskiy et al. and 22 additional bryophyte species from the Plant Picture Bank of China (PPBC) [
27]. BryophyteFine ensures the professionalism of data labels while providing sufficient training samples for deep learning models.
ViT and CNN: Since the ViT [
28] architecture was proposed, it has made remarkable progress in the fields of computer vision tasks such as image classification, segmentation, and object detection. Transformer architecture can effectively capture global features through self-attention mechanisms, while CNN architecture can effectively extract local features through hierarchical feature learning. However, there are limitations to using these two methods alone, which leads to unsatisfactory results when dealing with complex image tasks. Therefore, how to integrate CNN and Transformer architecture has become a hot topic in current research. The Convolutional vision Transformer (CvT) [
29] model replaces the original positional linear projection in the self-attention layer with convolutional projection and employs overlapping convolution to divide patches in the embedding layer, thereby enhancing the performance of the Transformer. These improvements introduce the advantages of convolution into the Transformer, which achieved 87.7% accuracy on the ImageNet dataset. Similarly, the Compact Convolutional Transformer (CCT) [
30] model improved the performance of ViT on a small dataset by introducing a convolution module, with a design that includes overlapping convolution, ReLU activation, and max pooling. CCT achieved a new state-of-the-art (SOTA) performance on the Flower102 dataset. Both CvT and CCT models demonstrate that introducing the convolutional mechanism into the Transformer architecture can enhance the performance of the model in image classification tasks. Chakrabarty et al. [
31] input the local feature maps extracted by CNN into Transformer, capture global dependencies using Transformer, and then fuse the outputs of the two. This method effectively integrates local and global information, ensuring optimal feature extraction. Overall, with architectural enhancements, local module improvements, and module splicing, the image classification performance of the models mentioned in [
29,
30,
31] are enhanced.
The fine-grained image classification of bryophytes in complex backgrounds is a challenging task. To recognize bryophyte images efficiently and accurately, we propose a lightweight fine-grained image classification model, MOSSNet. The model utilizes a multi-stage hierarchical structure, combining CNN and Transformer architectures to effectively capture symmetric local details and global structures in bryophyte images. We first introduce the CPE to preprocess the input image through two different size convolutional layers to capture more representative features while enriching self-attention labeling and reducing the amount of computation. Next, multiple DBMS modules are employed at each stage. These modules weighed fusion the outputs generated by both the PConv and the DCEA-based self-attention mechanism, effectively extracting both fine-grained and coarse-grained features. Ultimately, MOSSNet achieved the best classification performance on our self-constructed fine-grained bryophyte dataset, BryophyteFine. In the following sections, we will provide a detailed explanation of the proposed method.
3. Method
In this section, we introduce the production of datasets and the structure of MOSSNet, which is a deep neural network designed for bryophyte recognition. We also elaborate on the CPE module, the DBMS module, and the internal self-attention mechanism, DCEA.
3.1. Dataset Construction
To ensure the professionalism and accuracy of data labels, we base our work on the standard image dataset containing four bryophyte species created by Uzhinskiy et al. and select PPBC as the data source to construct a new dataset with a larger number of bryophyte species. For the bryophyte images in PPBC, we apply a consistent screening methodology: (1) selecting images with a large proportion of plant areas and a large number and density; (2) removing blurred images and those with significant occlusions.
When processing the image data, it is necessary to perform center cropping to remove most of the background interference in the image, which can preserve the main feature information of bryophytes and enhance the feature representation ability of data. The center-cropped image needs to normalize to 256 × 256 size, which is conducive to the feature extraction of the model and the stability of training. We performed the enhancement process on the image data to enrich the bryophyte image samples: (1) Random Cropping, which is randomly cropping the regions with coordinates in the upper-left and lower-right within the range of 1/4 to 3/4 of the width and height of the original image. This enhances the robustness of the model to different viewpoints and partial occlusion. (2) Random Rotating, which is rotating around the image center and randomly selecting the rotation angle within the range of [−5°, 10°], explicitly utilizing symmetry to simulate various growth angles of bryophytes in the natural environment. (3) Random Brightness Enhancement, where the brightness adjustment factor is randomly selected within the range of [0.7, 1.3] to simulate the bryophyte images under various lighting conditions. (4) Random Contrast Enhancement, where the contrast factor is randomly selected within the range of [0.7, 1.3] to modify the difference between the bright and dark areas of the image, which allows the model to be adaptive to different background conditions. After the above processing, we finally constructed a bryophyte dataset and named it BryophyteFine. This dataset has 26,000 images, covering 16 families, 23 genera, and 26 species of bryophytes.
3.2. MOSSNet
We design the deep learning network MOSSNet for the efficient identification of bryophyte images. The network adopts a modular architecture of four-stage feature extraction, which is consistent with the pyramid feature extraction design in mainstream models like ResNet and Swin Transformer, as shown in
Figure 2. The input image has a size of
, which is initially processed by the CPE module. The CPE module comprises two convolutional layers: the first layer uses convolution with a kernel size of 2 × 2 and a stride of 2 for downsampling, while the second layer uses convolution with a kernel size of 4 × 4 and a stride of 4 for patch division. This preprocessing step reduces the size of the input image to
, thereby decreasing computational load and enhancing the efficiency of feature extraction.
The four feature extraction stages of MOSSNet all adopt a lightweight depth-wise structure, and each stage is composed of a different number of DBMS modules (1, 2, 8, 2 respectively). The design of DBMS fully considers the diversity and complexity of bryophyte images, presenting an overall horizontally symmetrical structure. MOSSNet extracts features through two parallel branches: one uses convolution operations to capture local features, and the other one uses the DCEA module to capture global features. Among them, the DCEA module uses dilated convolutions to achieve multi-scale feature fusion and integrates the global contextual information through the self-attention mechanism. The outputs of these two branches are combined by weighted fusion to effectively integrate local and global features. In addition, at the end of each of the first three stages, there is a merging layer (convolution layer with kernel size of 2 × 2 and stride of 2) expanding the number of channels. This merging not only reduces computational cost but also facilitates the layer-by-layer extraction of high-level semantic features.
At the end of the network, the classification head consists of a global average pooling layer, a 1 × 1 convolutional layer, and a fully connected (FC) layer. The global average pooling layer compresses the spatial information into a fixed-size feature vector, the 1 × 1 convolutional layer adjusts the number of feature channels, and the FC layer maps features to category labels to achieve accurate classification.
The aforementioned design enables MOSSNet to effectively extract features from bryophyte images with a concise and low-complexity architecture, thereby achieving accurate image classification. In
Section 4, we will demonstrate through experiments that the model performs well on the BryophyteFine dataset and the network in this paper is effective.
3.3. Dual-Branch Multi-Scale
The fine-grained image classification tasks focus on distinguishing visually similar subcategories in coarse-grained species, such as different species of bryophyte [
32]. Meanwhile, in the BryophyteFine dataset, the scaling degree of each image is different, and the plant body distribution in the image may be dense, so the model is required to have the ability to effectively extract detailed features and overall structure. Although some existing bryophyte image classification methods [
16,
25] can recognize a few species, their ability to extract multi-scale features gradually declines as the number of species increases. This limitation hinders the model’s capacity to fully utilize both detailed and global information, which may negatively affect its ability to recognize more species.
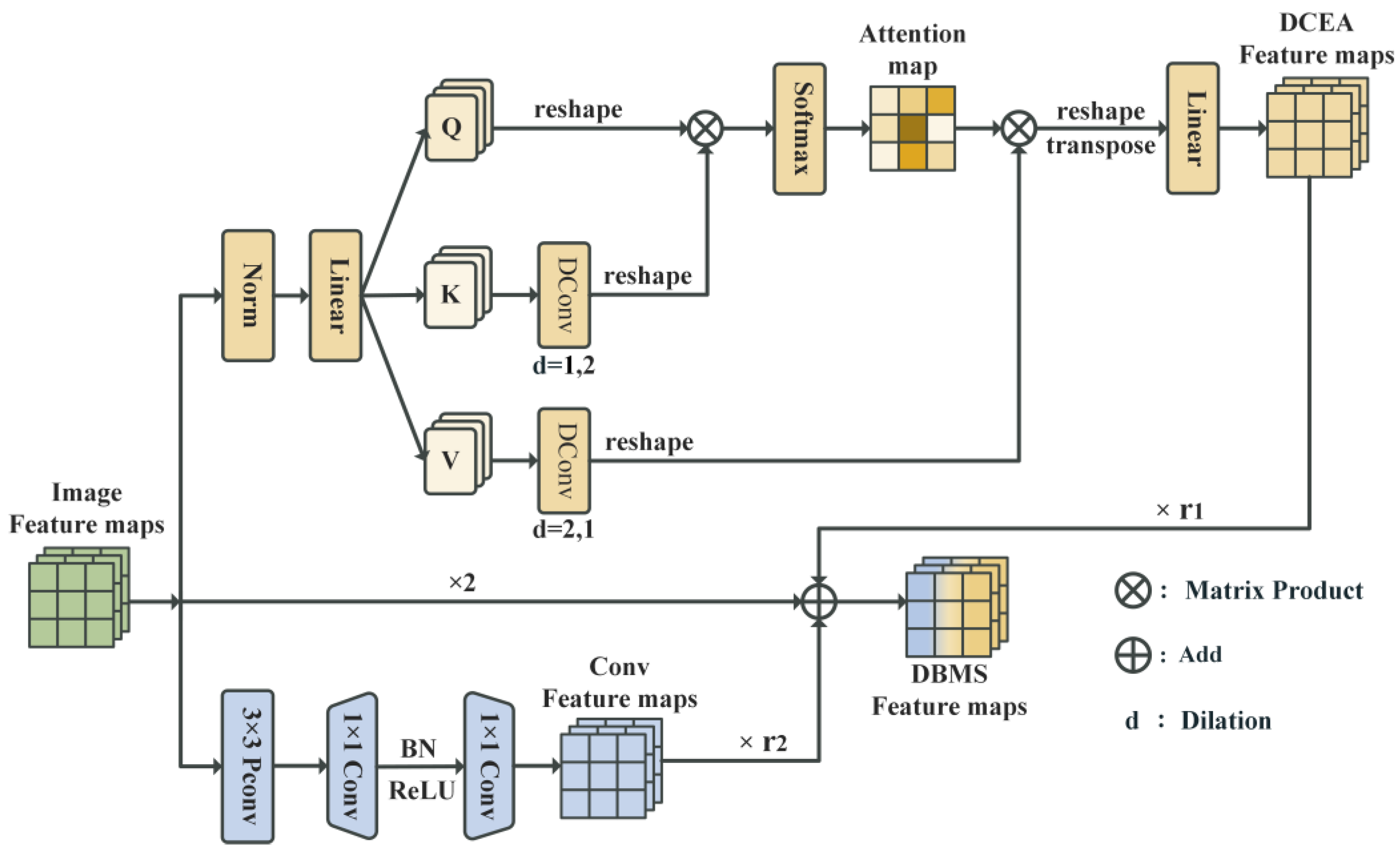
To address the challenge of bryophyte fine-grained image classification, we propose a DBMS module. This module realizes the effective fusion of multi-scale features through the dual-branch structure, thus improving the performance of the model in the bryophyte classification task. The structure of DBMS is shown in
Figure 3. In the DBMS module, the output feature map is from the upper layer, which is processed by two parallel branches. The convolutional branch (Conv Block) consists of 3 × 3 PConv and two 1 × 1 convolutional layers (also known as pointwise convolution). After the middle layer, the normalization layer and activation layer are applied to extract local features essential for fine-grained classification tasks. The PConv computers feature within the effective region, reducing computational redundancy and memory access complexity, thereby accelerating the feature extraction process. This efficient convolutional design not only improves the processing speed of the model but also maintains the accuracy of classification, so that it can effectively deal with the needs of large-scale image processing in bryophyte classification. Additionally, the attention branch (DCEA Block) employs dilated convolution to expand the receptive field, which achieves the fusion of multi-scale features that capture both fine-grained and global information. The specific implementation method of the DCEA block will be detailed in
Section 3.4. Both branches include residual connections to better preserve feature information and enhance the training stability of the network, as indicated by the ×2 arrows in the middle. The dual branch structure is a symmetrical design, in which two branches extract features of different scales and then use weighted fusion to emphasize the symmetry and asymmetry information at multiple scales. The final output,
, of the DBMS module is obtained by fusing features from two different extraction branches, calculated as follows:
where
and
represent the output features of the Conv Block and the DCEA Block, respectively, and
and
are two learnable scalars to control the weights of the two branches [
33]. This dual-branch structure can effectively fuse fine- and coarse-grained features at different scales, which enables the model to capture both detailed and overall information. For example, in bryophyte image classification tasks, the Conv Block can extract texture details of leaves, while the DCEA Block can capture a wider range of morphological features. Through the fusion mechanism of DBMS modules, the model can have a more comprehensive understanding of image information.
3.4. Dilation Convolutional Enhanced Attention
The challenge of the fine-grained image classification of bryophytes arises from their dense and multi-scale features. The convolution branch primarily extracts local features through 3 × 3 PConv, but as the depth of the model increases, its receptive field will be gradually limited, thus affecting the ability to capture overall and detailed features. To address this, the DBMS module introduces dilated convolution within the attention branch, using varying dilation rates to effectively expand the receptive field. The dilated convolution technique expands the receptive field by inserting “gaps” into the convolution kernel without adding additional parameters, which allows the model to capture broader contextual information [
34]. The implementation process of the DCEA module is shown in
Figure 3. Initially, the linear transformation layer maps the input features into a combination query (Q), key (K), and value (V). In the same attention head, K and V are processed by dilated convolution with different dilation rates. Specifically, the dilated convolution operation is performed on K and V to obtain
and
with Q as the center. The formula is as follows:
where
represents the dilated convolution operation with a 3 × 3 kernel, which expands the receptive field through sparse sampling to enable the model to focus on a wider range of regions. The dilation rates,
and
, determine how the convolution samples feature in a skipping manner. When
and
, this ensures complementary information across different scales. Based on the results of these dilated convolutions, a multi-head self-attention computation is performed to obtain attention output
, given by:
where
represents the i-th head and different heads can learn different levels of information. The
is a scaling factor used to prevent excessive gradients from affecting training stability. The
operation ensures the normalization of attention weights, making feature weighting calculations more reasonable. Finally, the attention features from dilated convolution are concatenated with different dilation rates to form the final output feature
, as follows:
Through the above self-attention calculation, the dilated convolution with different dilation rates provides multi-scale feature information for the self-attention mechanism, which enables the DCEA module to achieve effective multi-scale feature fusion in the same head. This fusion expands the depth and breadth of feature extraction, allowing the model to capture potential symmetrical features across regions in bryophyte images.
In addition, due to the locality and sparsity of the shallow attention matrix in ViT, it often exhibits low relevance when modeling the semantics of distant patches [
35]. The characteristic limits the contribution of parts that are far from the central region of the image in global feature modeling. The DCEA module utilizes the dilated convolution technique to effectively extend the receptive field, thereby enhancing the correlation between distant features and central features. At the same time, it reduces the redundancy in global attention computation to a certain extent. The 3 × 3 kernel size for dilated convolution is chosen because it achieves a balance between receptive field size, computational complexity, and detail preservation. Moreover, a 3 × 3 kernel size is a widely validated standard in deep learning. Through this strategy, the DCEA module can integrate multi-scale information more efficiently in the process of global feature modeling.
Compared with the method in [
26], our model employs the efficient and concise PConv and DCEA in the BDMS module, reducing computational redundancy and resource consumption to better adapt to the diverse requirements of bryophyte classification tasks.
3.5. Convolutional Patch Embedding
The CPE module is proposed based on the DBMS module to further enhance the efficiency of the model in capturing the complex features of bryophyte images. As shown in
Figure 2, the CPE module consists of two convolutional layers with the ReLU activation function in the middle, producing the output
from the input
as follows:
where
represents the first convolutional layer, which is used for initial feature extraction. This layer applies a convolutional kernel size of 2 × 2 with a stride of 2 to downsampling the input image, thus reducing its spatial dimensions. After the first convolutional layer, the ReLU activation function is applied to set all negative values to zero, thereby enhancing the model’s ability to express complex features and reducing the risk of gradient vanishing. The
represents the second convolutional layer, which utilizes a kernel size of 4 × 4 with a stride of 4 to extract higher-level embedded features from the downsampled image. A convolution kernel size of 4 × 4 provides a larger local receptive field than 3 × 3, making it more suitable for effectively transforming spatial information into feature space in the early stages of the model. The CPE module maps the input image to the potential feature space through convolution, thereby providing more representative initial features for the subsequent DBMS module. The experimental results in
Section 4.4 demonstrate that feature mapping enables DBMS to efficiently extract multi-scale features and improve the effectiveness of feature fusion.
In addition, the computational complexity of the self-attention mechanism is proportional to the square of the number of labels, which is equal to the resolution of the input feature map. Therefore, further downsampling can significantly reduce the computational load [
30]. Differently from the convolution module in the CCT model mentioned earlier, the CPE module proposed in this paper employs non-overlapping convolution instead of maximum pooling. While the maximum pooling performs well in reducing the size of the feature map, it compresses information within the pooling window, retaining only the maximum value. This may result in the loss of crucial local details. When dealing with bryophyte images with complex textures and small structural differences, preserving these details may be beneficial for improving classification performance. In contrast, non-overlapping convolution preserves all the information within the convolution window. This improves computational efficiency while retaining more details, so it is more suitable for the feature extraction task of complex images.
4. Experiments
We use the BryophyteFine dataset to perform experiments in the fine-grained classification of bryophytes, then evaluate and analyze the experimental results. Additionally, we also conduct ablation experiments to verify the effectiveness of the proposed network.
4.1. Dataset
The dataset utilized in the experiments is the BryophyteFine dataset, which we developed. For model training, the dataset is randomly partitioned into a training set, validation set, and test set according to the ratio of 6:2:2. The names of all bryophyte species and the corresponding number of training images used in the experiment are listed in
Table 1.
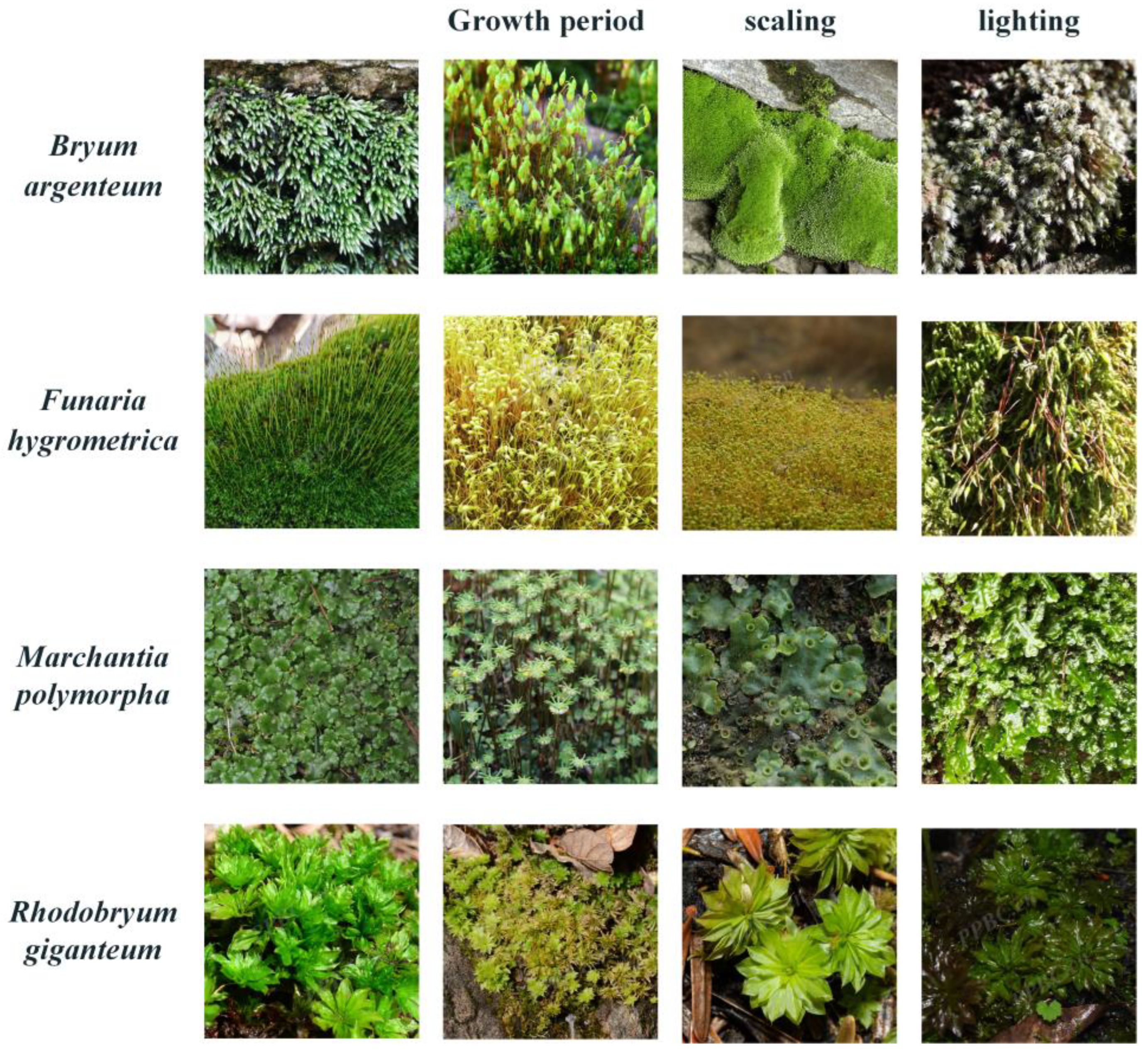
As shown in
Figure 4, the dataset exhibits several fine-grained factors, such as the morphological and environmental similarities among different bryophyte species, as well as significant variations within the same species due to different growth periods, shooting angles, and lighting conditions. These characteristics truly reflect the complexity of the natural environment, which is very consistent with the application requirements of the real-time classification of bryophytes. The images in BryophyteFine are dominated by the leafy gametophytes and erect sporophytes of bryophytes, fully showing the typical characteristics of bryophytes in different growth stages. The gametophyte images provide views of leaf structure, arrangement, and color changes in bryophytes in detail, while the sporophyte images focus on the morphology of the elongated stipe and sporocapsules and their appearance at different maturity stages [
36]. These images encompass both the overall morphology and growth status of bryophytes under varying environmental conditions, offering rich visual data for bryophyte classification studies.
4.2. Experimental Setup
The experimental hardware platform uses a single NVIDIA GTX 1080 Ti GPU (Microsoft Corporation, Shanghai, China) with 11 GB video memory. is developed based on the PyTorch 1.10.2 deep learning framework in the Python 3.9.13 programming environment, and PyCharm 2020.3.1 is used as the programming platform.
The image in the training set and verification set are adjusted to 224 × 224 pixels and normalized. Data augmentation techniques, including blur processing and color dithering, are applied to the image in the training set to enhance the generalization ability of the model.
During the model training phase, the AdamW optimizer is employed with an initial learning rate of , and the cosine annealing learning rate scheduler is used to dynamically adjust the learning rate. Training is performed for a total of 200 epochs with a batch size of 64 per round, and the training data are shuffled before the start of each epoch. The cross-entropy loss function is used to improve the efficiency and stability of training through mixed precision training.
Upon the completion of training, select the best weight to evaluate the performance of the model on the test set. The evaluation metrics include Top-1 accuracy, Top-3 accuracy, F1 score (F1), and mean average precision (mAP), as well as floating-point operations (FLOPs) and the number of parameters (Params).
measures whether the correct class is included in the Top-k predictions of the model. Here, represents the number of correctly classified samples in the Top-k predictions, and represents the total number of samples. The F1 score is used to evaluate the overall classification performance of the model, ranging from 0 to 1, with values closer to 1 indicating better overall performance. The F1 score is calculated from the combination of precision () and recall (), where , , and represent true positives, false positives, and false negatives, respectively. mAP is obtained by calculating the area under the precision-recall curve (AP) for each class and then averaging these APs, making it more suitable for evaluating the stability of the model. Here, represents the total number of classes, and denotes the average precision of the i-th class.
4.3. Performance Comparison of Models
To validate the performance of the proposed MOSSNet model in the task of bryophyte classification, we conduct model performance verification experiments based on the BryophyteFine image dataset constructed. The classification performance is compared with five different models (ResNet50 [
9], MobileNetV2 [
37], ViT-base [
28], CCT [
30], and FasterNet [
17]) where CCT and FasterNet include two model structures. These models cover a range of architectures and scales, including the pure CNN models (ResNet50 and MobileNetV2), the pure Transformer model (ViT-base), and hybrid models combining CNN and Transformer (CCT, FasterNet), with parameters ranging from 2.25 M to 53.59 M.
Experimental results for multiple metrics: All models are trained from scratch under the same experimental conditions and evaluated using the best weight on the test set. The results are shown in
Table 2. Our proposed MOSSNet network performs exceptionally well across all metrics. Compared with FasterNet-s and FasterNet-m, which only use PConv in all stages, MOSSNet with the DCEA module increases 8.1% and 7.13% in Top-1 accuracy, respectively. This significant improvement is primarily attributed to the constraints imposed by the 3 × 3 PConv on the receptive field. In contrast, the DCEA module effectively expands the receptive field by introducing dilated convolutions, making it more suitable for dealing with the densely and widely distributed plant structures in bryophyte images. Notably, CCT is also a hybrid model that combines CNN and Transformer. It has relatively low computational resource consumption. For instance, the FLOPs of CCT-7/7 × 2 are only 1.61 G. However, in the bryophyte image classification task, our model not only achieves higher classification accuracy but also reduces the FLOPs by 1.54 G compared to CCT-7/7 × 2. This further shows that the CPE and DBMS modules not only introduce multi-scale information in feature fusion but also significantly reduce computational redundancy in traditional CNN and Transformer architectures.
Confusion matrix: To demonstrate the performance of each model more intuitively in the bryophyte classification task, we calculate the confusion matrix based on the test set results and select the thirteen bryophyte species that are most easily confused. As shown in
Figure 5, the classification results for these species are visualized through a confusion matrix heatmap. The figure reveals that CNN models, such as ResNet50 and MobileNetV2, exhibit higher confusion rates in most species, while Transformer-based models, such as ViT, CCT, and FasterNet, achieve more accuracy in classifying most species. Notably, all five models tend to misclassify species 13 (
Marchantia polymorpha) as species 8 (
Reboulia hemisphaerica). The high similarity in leaf structure, color distribution, and texture features may make these two species difficult to distinguish. In contrast, with the DBMS module effectively fusing multi-scale information, MOSSNet only shows misclassification on one sample, indicating its ability to capture fine-grained differences. Additionally, MOSSNet does not exhibit misclassification in eight species, which further demonstrates its superior performance in bryophyte classification tasks.
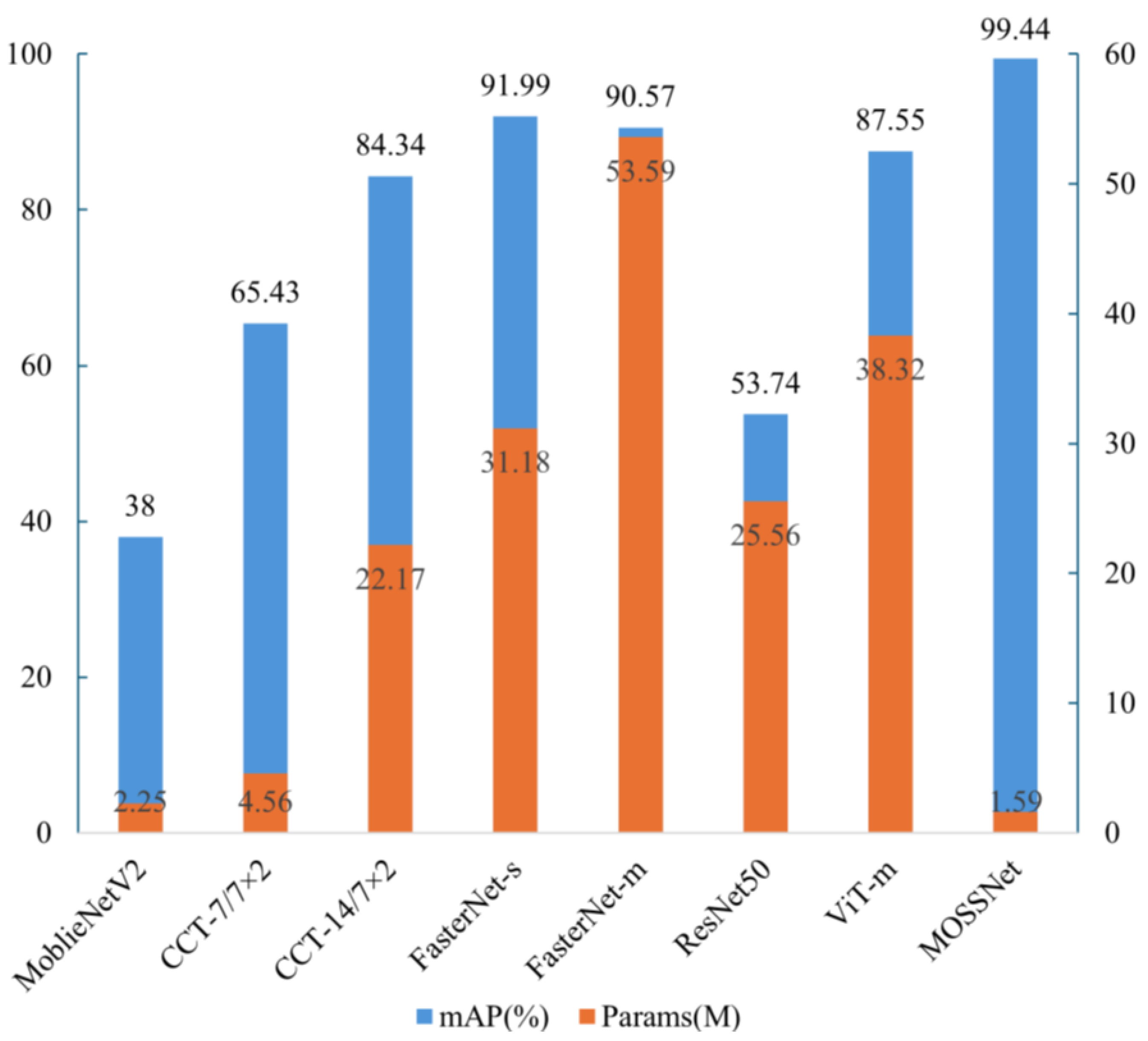
Parameter and mAP Analysis:
Figure 6 shows the columnar distribution of parameters and mAP for each model. Overall, as the number of parameters increases, mAP tends to improve. However, our model MOSSNet achieves the highest accuracy (mAP: 99.44%) while maintaining a low parameter, indicating that MOSSNet achieves a good balance between complexity and performance. In contrast, although ResNet50 has a higher parameter compared with CCT, it fails to improve mAP. A similar situation also occurs in the comparison between ViT-m and FasterNet-s. Notably, while MobileNetV2 is closest to MOSSNet in terms of parameters and FLOPs, it performs poorly in bryophyte classification tasks. This phenomenon, like that observed in ResNet50, may be attributed to the lack of effective fusion of global information in these models. This underscores the importance of integrating both global and local information in the fine-grained classification task of bryophytes. Our model achieves multi-scale feature fusion through the DBMS module, significantly improving classification performance.
In summary, compared with the classical classification models and the recent hybrid models, MOSSNet not only achieves the highest classification accuracy but also significantly reduces the consumption of computing resources in the bryophyte fine-grained classification task. The dual advantages of performance and efficiency make MOSSNet more suitable for bryophyte classification needs in practical applications.
4.4. Ablation
CPE module: To assess the effect of the CPE module on the subsequent DCEA modules in each stage, we conduct two sets of experiments at the input stage: one set includes the CPE module (DCEA+CPE), and the other set only uses a single 4 × 4 convolution layer for patch division (DCEA). The experimental results on the test set are summarized in
Table 3. As shown in
Table 3, the inclusion of the CPE module increases Top-1 accuracy, Top-3 accuracy, F1 score, and mAP by 1.52%, 0.44%, 0.015, and 1.99%, respectively. These results indicate that the CPE module can enhance the model’s capacity to capture detailed information, thereby boosting classification accuracy. Furthermore, the confusion matrix results in
Figure 5 reinforce this conclusion: for the frequently confused species 13 and 8, the number of misclassified samples decreased from 7 to 1 after adding the CPE module. While the CPE module slightly increases the parameter, it reduces FLOPs by 0.155G, demonstrating that the dual-layer convolutional structure helps to reduce computational complexity.
DCEA module dilation rate selection: To achieve multi-scale feature fusion within the same attention head, we conduct classification experiments using a 3 × 3 convolution kernel size, select different dilation rates, and configure the number of attention heads as multiples of the dilation rate in the experiment. According to the test results in
Table 4, the optimal performance is observed with dilation rates of [1, 2], where both Top-1 and Top-3 accuracies reach their maximum values. Although a larger dilation rate can provide a larger receptive field, it may also lead to computational redundancy. Meanwhile, the larger dilation rate may also lead to the loss of local detailed information, making it difficult for the model to capture fine-grained features. Thus, choosing the appropriate dilation rate is crucial. For the dilation rates of [1, 2], we also explore different configurations of attention heads. The experimental results indicate that the best performance is achieved with [4, 8, 12, 16] attention heads across the four stages. The reasonable configuration of dilation rates and number of attention heads can effectively enhance the model’s feature extraction capability, thereby improving the classification accuracy of bryophyte classification.