Current coronary artery segmentation algorithms exhibit notable limitations in maintaining robustness in low-contrast regions, accurately delineating fine vessels, and balancing segmentation accuracy with computational efficiency. To systematically address these challenges, we propose DMF-Net (Dual-path Multi-scale Fusion Network), an enhanced UNet++-based architecture. By incorporating Dynamic Buffer–Bottleneck–Buffer (DBB) modules in shallow encoding stages and Axial Local–global Hybrid Attention (ALHA) modules in deep decoding stages, DMF-Net preserves critical vascular information during feature encoding while enhancing spatial segmentation precision during decoding. Additionally, a composite loss function and a 2.5D slice construction strategy jointly optimize vascular connectivity, boundary accuracy, and computational efficiency, enabling finer and more robust vascular segmentation.
3.1. Network Architecture
UNet++ enhances feature fusion between the encoder and decoder through nested skip pathways and multi-level decoding paths, enabling comprehensive multi-scale feature integration and efficient propagation for more discriminative representations. However, UNet++ still faces limitations in segmenting fine vascular structures. To address this, DMF-Net builds upon UNet++ with targeted improvements designed to better preserve fine vessel details and enhance both structural and semantic segmentation performance.
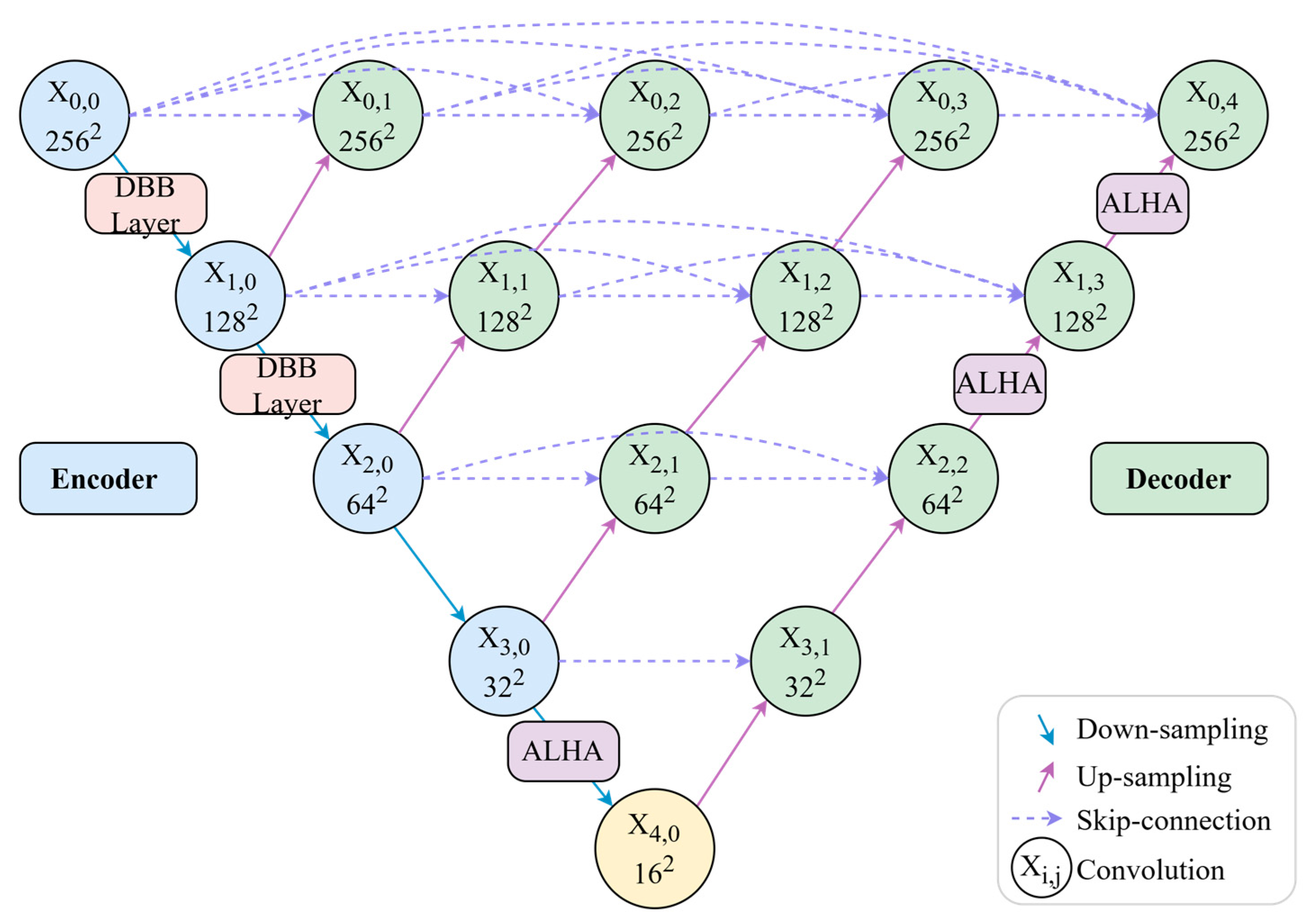
Regarding module deployment, DMF-Net incorporates DBBLayer modules in shallow encoding stages to preserve fine vascular structures from high-resolution features, while ALHA modules in deep decoding stages capture both global vessel trajectories and local boundary details through multi-scale feature integration. In contrast to approaches that apply modules uniformly across all network layers, DMF-Net strategically deploys specific modules at key positions, avoiding redundant configurations and controlling computational complexity. The overall architecture is illustrated in
Figure 1.
As illustrated in
Figure 1, the network architecture employs a strategic module deployment scheme. DBBLayer modules are inserted between encoder nodes
and
during downsampling to preserve fine-grained vascular structures and prevent information loss in deep layers. ALHA modules are positioned after decoder node
, between nodes
, and between nodes
, where they leverage multi-scale feature convergence to integrate skip connections from different encoding levels. Through its global-local attention mechanism, ALHA simultaneously captures long-range vessel continuity and local boundary details during upsampling. This design—feature preservation in shallow layers and semantic enhancement in deep layers—synergistically optimizes segmentation performance.
3.2. Dynamic Buffer–Bottleneck–Buffer Layer
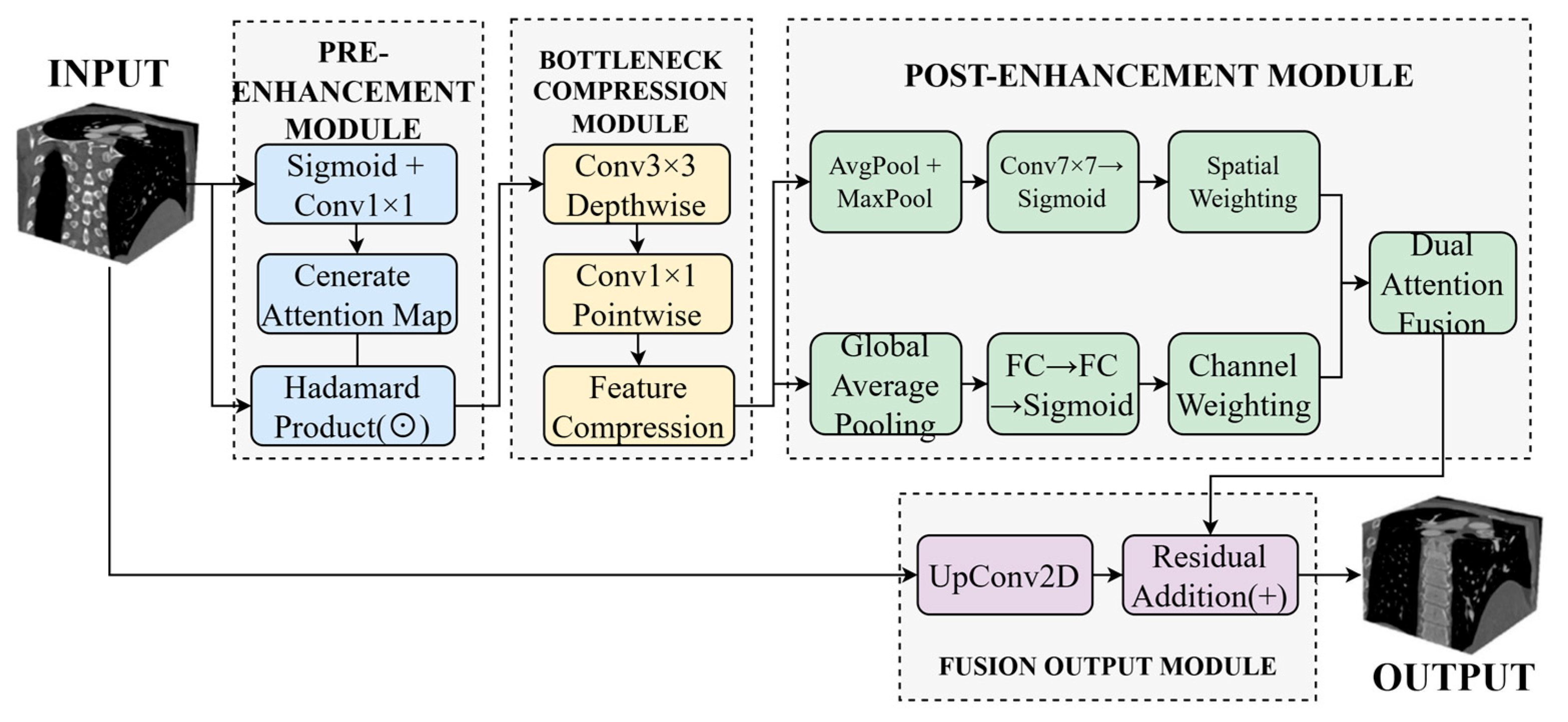
Traditional downsampling operations, such as max pooling and strided convolution, tend to attenuate responses in complex regions containing small vessels, structural boundaries, and bifurcations, compromising the model’s capacity for fine-grained structure recognition. To mitigate this limitation, we propose a Dynamic Buffer–Bottleneck–Buffer Layer (DBBLayer) that synergizes attention mechanisms with lightweight convolution. Through a three-stage pipeline—pre-enhancement, compression, and post-enhancement—DBBLayer amplifies responses to small-scale structures while preserving spatial boundary integrity. The pre-enhancement stage leverages attention to accentuate critical regions (e.g., vessels) and suppress background interference. The bottleneck module employs depthwise separable convolution to achieve efficient feature compression with minimal information loss. The post-enhancement stage recovers fine-grained details through spatial and channel attention, thereby strengthening recognition capability.
Figure 2 illustrates the overall architecture.
As shown in
Figure 2, the Dynamic Buffer–Bottleneck–Buffer Layer comprises three main components: pre-enhancement, bottleneck compression, and post-enhancement. The pre-enhancement module employs Sigmoid activation and 1 × 1 convolution to generate attention maps, enhancing feature responses through element-wise multiplication. The bottleneck compression module utilizes 3 × 3 depthwise separable convolution followed by 1 × 1 pointwise convolution to achieve efficient feature compression. The post-enhancement module leverages both average pooling and max pooling to extract spatial attention, which is then combined with channel attention derived from fully connected layers to enable comprehensive feature enhancement. Finally, the enhanced features are integrated with the original input through upsampling and residual connections to produce the final output.
3.2.1. Pre-Enhancement Module
The core objective of this module is to enhance the input feature maps using an attention mechanism prior to feature compression, enabling the network to better focus on regions with higher semantic value across spatial and channel dimensions, thereby suppressing redundant background information and improving the capability to capture key structural information.
First, the input feature map
from the encoder is passed to the first dynamic convolution layer. Within the dynamic convolution layer, a gating mechanism is adopted to generate a dynamic attention map
, as shown in Equation (1):
where
denotes the Sigmoid activation function, and
and
are learnable parameters.
Subsequently, the generated attention map
is element-wise multiplied with the original feature map
to obtain the enhanced feature map
, whereby the responses in critical regions are amplified while irrelevant regions are suppressed, as shown in Equation (2):
where
denotes the element-wise multiplication operation. The enhanced features strengthen the critical regions and attenuate irrelevant interference.
3.2.2. Bottleneck Compression Module
This module reduces computational overhead through feature compression while preserving task-relevant critical information, under the premise of ensuring segmentation performance. The module employs a depthwise separable convolution structure, which consists of a 3 × 3 depthwise convolution and a 1 × 1 pointwise convolution connected in series. The depthwise convolution is used to extract spatial contextual information, while the pointwise convolution is used to compress the channel dimension.
The formula for extracting spatially correlated features using depthwise convolution is shown in Equation (3):
The formula for channel compression of intermediate features using pointwise convolution is shown in Equation (4):
where
and
represent the 3 × 3 depthwise convolution and 1 × 1 pointwise convolution, respectively. The compressed feature map
preserves the critical information.
3.2.3. Post-Enhancement Module
To further mitigate the potential information loss during feature compression, this module performs joint channel and spatial enhancement on the feature map to compensate for weakened structural details.
First, Global Average Pooling (GAP) is applied to
to capture the global response of each channel, and a channel weight vector
is generated through a two-layer fully connected network, as shown in Equation (5):
where
and
are learnable fully connected parameters, and
computes the global average value across the channel dimension.
Subsequently, average pooling and max pooling are separately applied to
, concatenated along the channel dimension, and a 7 × 7 convolution is then used to generate the spatial weight map, as shown in Equation (6):
where
denotes average pooling and
denotes max pooling. The purpose of concatenation is to combine two different types of spatial statistical information to obtain a more stable spatial weight distribution.
focuses on critical regions in the image space.
Finally, the channel attention and spatial attention are element-wise multiplied and applied to the input features, as shown in Equation (7):
Ultimately, the obtained is enhanced in both structural details and critical region responses, and the feature map’s capability in feature extraction is improved.
3.2.4. Fusion Output Module
This module effectively fuses the enhanced detail information while maintaining the global consistency of the original features, ensuring the structural integrity and boundary accuracy of the segmentation results. To ensure that the enhanced features
match the spatial dimensions and channel numbers of the input features
, upsampling convolution is first applied, followed by residual connection to add with the original features, as shown in Equation (8):
where
denotes the upsampling convolution operation. The final output feature
not only preserves the overall structural information of the original image, but also enhances the capability to delineate lesion regions, fine vessels, and boundaries.
3.2.5. Design Rationale and Computational Efficiency
Kernel and Channel Size Selection:
The DBBLayer architecture employs strategically selected kernel sizes and channel configurations to balance feature preservation with computational efficiency. The pre-enhancement module utilizes 1 × 1 convolution to maintain spatial resolution while enabling efficient channel-wise attention with minimal parameters ( × weights versus 3 × 3 convolution’s weights), achieving 8× parameter reduction. The bottleneck compression module employs 3 × 3 depthwise separable convolution, which reduces computational cost by approximately 8× compared to standard 3 × 3 convolution (9 + operations versus 9 for standard convolution) while preserving spatial context through 9-pixel neighborhood coverage. The post-enhancement module uses 7 × 7 spatial attention convolution to balance receptive field coverage (49-pixel neighborhood) with computational tractability (49 operations versus 121 for 11 × 11 kernels), providing sufficient context for boundary delineation without excessive overhead. The channel compression ratio follows a → → pattern, maintaining feature capacity while reducing redundancy in vessel-sparse regions, preventing information bottleneck while controlling parameter count.
Activation Design:
Sigmoid activation in gating mechanisms (Equation (1)) provides smooth attention weights in the [0, 1] range, enabling gradual feature modulation rather than binary selection. This soft attention mechanism proves critical for preserving weak responses from small vessels (<2 mm diameter) that might be eliminated by hard thresholding approaches.
3.2.6. Module-Level Computational Cost Analysis
To ensure clinical deployment feasibility, we systematically profiled the parameter count, floating-point operations (FLOPs), inference latency, and GPU memory consumption for each architectural component. All measurements were conducted on an NVIDIA RTX 3090 GPU using PyTorch 1.12.0, batch size of 1, and input resolution of 512 × 512 × 3 (2.5D configuration). Latency represents the average of 100 forward passes after 20 warm-up iterations.
As show in
Table 1, the modest per-instance overhead of DBBLayer (0.31 M parameters, 1.5 G FLOPs, 1.5 ms latency) stems from three architectural components: the pre-enhancement module employing 1 × 1 convolution for attention generation (approximately 0.25 M parameters), the bottleneck module utilizing depthwise separable convolution (approximately 0.12 M parameters) achieving approximately 8 × FLOPs reduction versus standard 3 × 3 convolution, and the post-enhancement module combining channel attention (0.68 M parameters with reduction ratio r = 16) and spatial attention. This design explains the 12.2% FLOPs increase despite 16.8% parameter growth. Critically, DBBLayer is deployed at only two high-resolution encoder transitions (X
0,0 → X
1,0 at 512 × 512 resolution and X
1,0 → X
2,0 at 256 × 256 resolution), limiting cumulative latency overhead to 3.0 ms while maximizing impact on fine vessel preservation.
The ALHA module’s 0.89 M parameters comprise: axial attention weight matrices for height and width dimensions (0.42 M parameters), dynamic convolution weight generator comprising a two-layer MLP (0.23 M parameters), and pixel-level fusion network (0.24 M parameters). Despite the substantial 71.2% per-instance parameter increase, three factors mitigate cumulative overhead: strategic deployment at only three decoder nodes (X0,4, X1,3 → X2,2, X3,0 → X4,0) where multi-scale feature convergence occurs; progressively reduced spatial resolution at these nodes (512 × 512, 256 × 256, 128 × 128) significantly lowering absolute FLOPs; and factorized axial attention design reducing computational complexity from O((HW)2) in standard self-attention to O(HW(H + W)), achieving approximately 8× speedup for 512 × 512 feature maps. The cumulative latency contribution totals 4.8 ms.
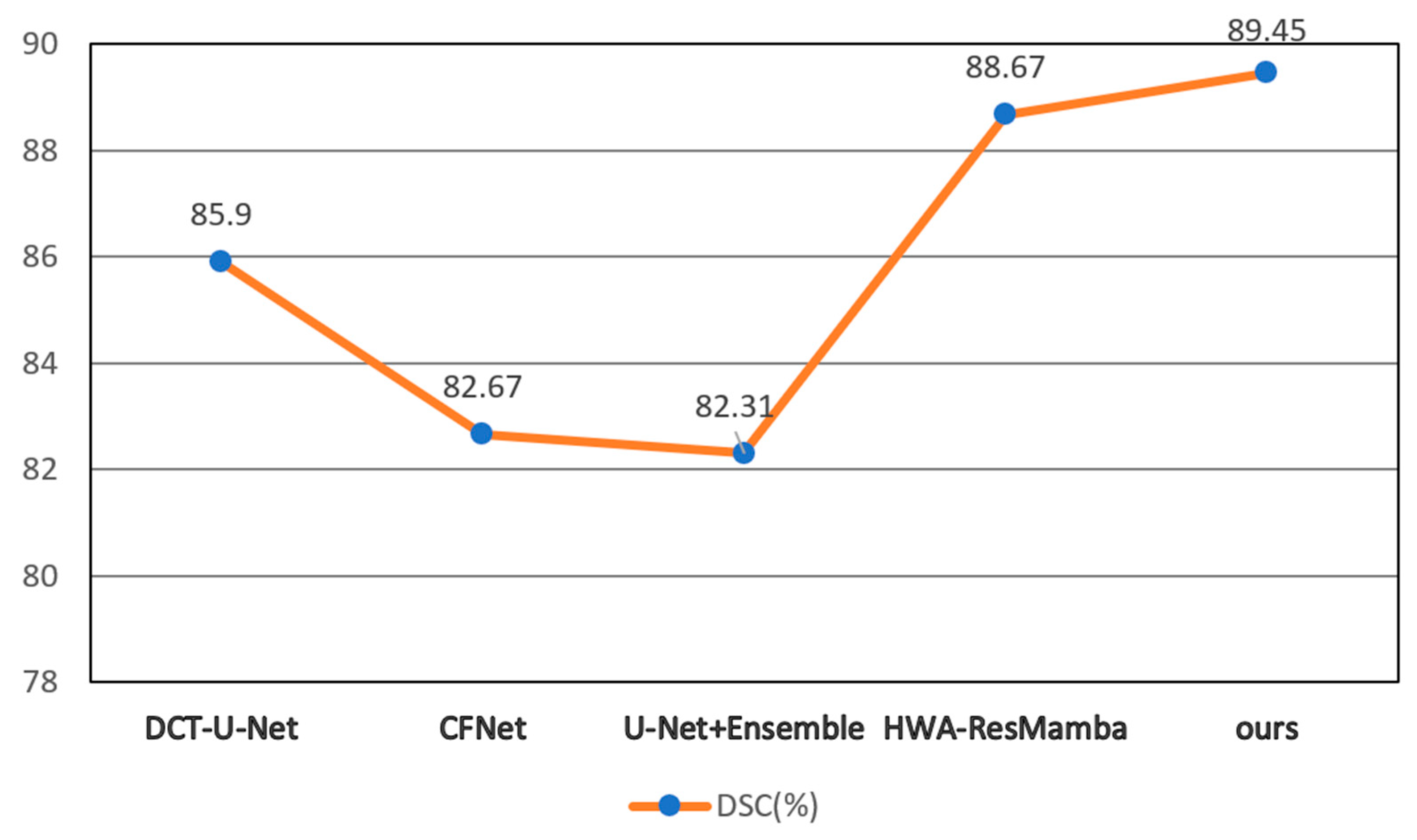
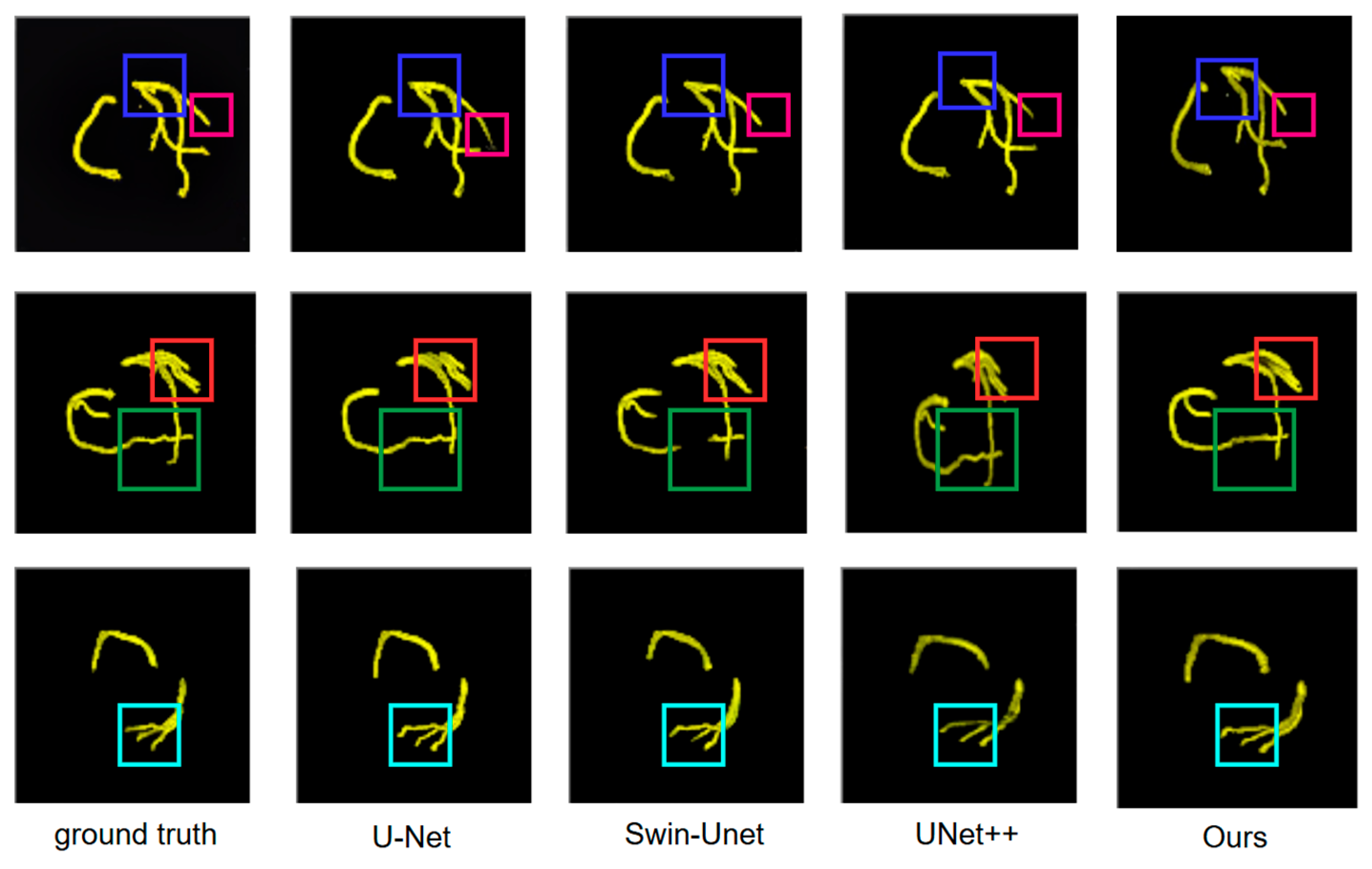
At the full architecture level, DMF-Net’s 6.4% parameter increase and 4.1% FLOPs increase (relative to UNet++ baseline) yield 3.67% DSC improvement, demonstrating favorable efficiency-accuracy trade-off. The resulting 75 ms per-slice latency enables processing a typical 250-slice coronary CTA volume in approximately 18.75 s, meeting clinical workflow requirements (<30 s). To contextualize computational efficiency, we compare against recent methods evaluated on ImageCAS: Swin U-Net [
31] achieves 84.12% DSC with 181.5 G FLOPs (+22.1% versus DMF-Net) and 125 ms latency (+66.7%); HWA-ResMamba [
35] achieves 88.67% DSC with 156.3 G FLOPs (+5.2%) and 98 ms latency (+30.7%); CFNet [
27] achieves 86.2% DSC with 138.2 G FLOPs (−7.0%) and 68 ms latency (−9.3%). Computing an efficiency ratio as DSC gain per additional gigaFLOP (relative to UNet++ baseline), DMF-Net achieves (89.45 − 85.78)/(148.6 − 142.8) = 0.63% DSC/G, compared to HWA-ResMamba’s (88.67 − 85.78)/(156.3 − 142.8) = 0.21% DSC/G, indicating superior accuracy gain per unit computational cost.
Deployment feasibility was validated through testing on mid-range clinical hardware. On an NVIDIA RTX 4000 GPU (8 GB VRAM) commonly found in radiology workstations, DMF-Net achieves 92 ± 10 ms per-slice inference latency with 5.1 GB peak memory consumption, confirming compatibility with routine clinical infrastructure. Preliminary post-training quantization experiments (FP32 → INT8) using PyTorch’s native quantization API with calibration on 100 validation cases reduced model size from 148 MB to 86 MB (−42%) and improved inference speed from 75 ms to 49 ms per slice (−35%) with minimal accuracy degradation (89.45% → 89.12% DSC, −0.33%), suggesting viable pathways for deployment in resource-constrained clinical environments.
3.3. Axial Local–Global Hybrid Attention Module
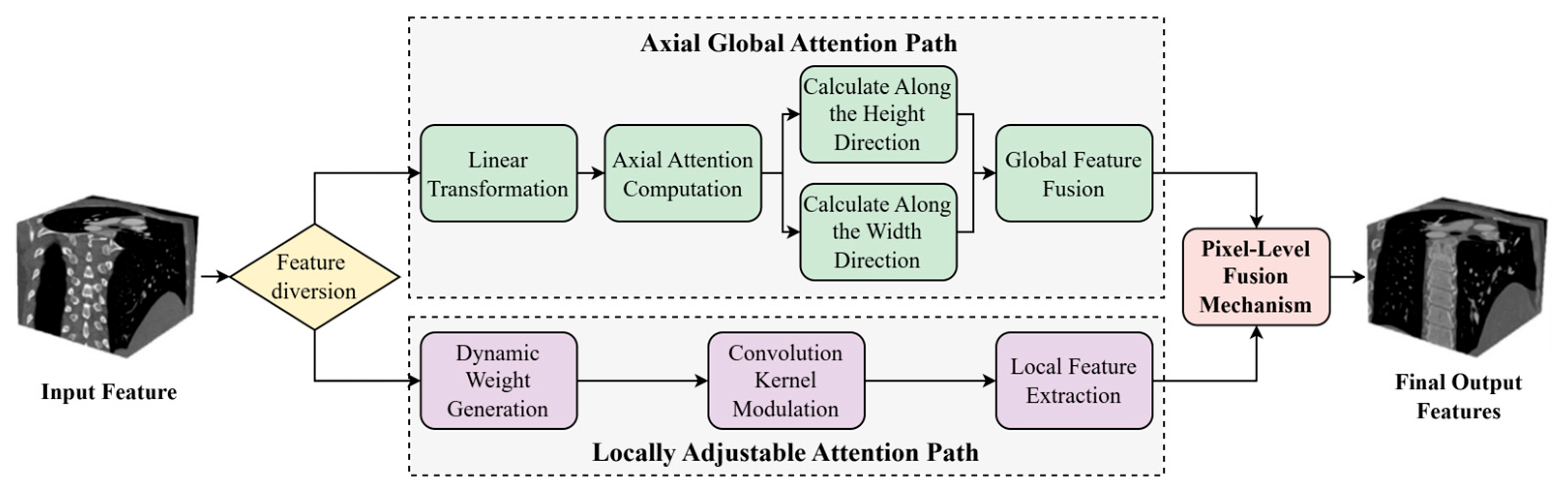
Coronary arteries in medical images typically exhibit long-range extensibility and structurally complex and variable characteristics. Particularly in lesion regions such as bifurcations, stenoses, and calcifications, they are often accompanied by minute and irregular local morphological changes. In traditional deep learning segmentation methods, downsampling operations frequently encounter issues of information loss and reduced accuracy when processing these fine vascular structures. Moreover, the heterogeneity of vascular pathology and inter-patient anatomical variability necessitate adaptive feature representation across diverse morphological patterns. Imaging artifacts and low contrast ratios further compromise feature extraction, particularly in subtle pathological regions. Conventional attention mechanisms lack the flexibility to dynamically balance global topology and local details according to structural complexity. Therefore, how to simultaneously balance global spatial consistency and local structural accuracy has become the key to solving the problem. To this end, this study proposes an Axial Local–global Hybrid Attention (ALHA) module. By combining global attention and local attention mechanisms, this module can simultaneously preserve the consistency of global spatial information and enhance the recognition capability of local details. The module features a dual-path parallel processing architecture that effectively handles both large-scale structures and minute lesion segmentation of coronary arteries, with dynamic modulation through a pixel-level fusion mechanism, thereby improving the precise segmentation capability for complex coronary artery morphology. The module structure is shown in
Figure 3.
In
Figure 3, the ALHA module adopts a dual-path parallel architecture. In the axial global attention path, the input features first undergo linear transformation, and then self-attention weights are calculated along the height and width directions, respectively. Global receptive field features are obtained through global feature fusion. The local adaptive attention path generates convolution kernel modulation parameters through a dynamic weight generation module, followed by local feature extraction after convolution kernel modulation. Finally, features from both paths are weighted and averaged through a pixel-level fusion mechanism, outputting the fused feature map.
3.3.1. Axial Global Attention Path
Although traditional global self-attention mechanisms can capture long-range contextual information, they often overlook local feature interactions along a single spatial axis in high-resolution medical images, thereby limiting the feature extraction capability. To address this, this study introduces an axial attention path that performs attention computation separately along the height and width dimensions of the image, thus preserving long-range dependency segmentation capability while reducing computational complexity.
First, given the input feature map
X, the query, key, and value are obtained through linear transformation of the input features, denoted as
Q,
K, and
V, respectively, as shown in Equation (9):
where
,
, and
are the learnable weights for linear transformation, respectively.
Subsequently, attention computation is performed on , , and along the height and width directions, respectively.
The attention computation along the height direction is shown in Equation (10):
where
,
,
represent the feature tensors expanded along the height direction, and
,
,
represent the feature tensors expanded along the width direction. The Softmax operation is performed across the response dimension.
Finally, weighted averaging of the attention outputs from both directions is performed to obtain the axial global attention features, as shown in Equation (11):
3.3.2. Local Adaptive Attention Path
Although axial attention has advantages in segmenting long-range dependencies, its segmentation capability for local edge variations and minute structural anomalies remains limited. To address this, a local adaptive attention path is designed that adaptively adjusts the response weights of convolution kernels to achieve edge-sensitive feature extraction.
This path performs feature extraction based on a dynamically modulated convolution kernel
, as shown in Equation (12):
where
is the base convolution kernel,
denotes global average pooling of the input features
, and
represents a two-layer multi-layer perceptron structure used to generate weight modulation factors across the channel dimension.
denotes channel-wise multiplication.
Using the modulated convolution kernel, convolution is performed on the input features to obtain locally enhanced features, as shown in Equation (13):
This mechanism adjusts the convolution kernel based on the responses of different channels, thereby enhancing the perception capability for minute structural variations.
3.3.3. Pixel-Level Fusion Module
To achieve synergistic enhancement of global structure and local details, a pixel-level fusion strategy is introduced to perform weighted integration of outputs from the two attention paths.
First, a spatial fusion weight map is constructed. Then, the output features are fused using Equation (14):
where the fusion weight map
,
, and
,
represent the response proportions of the global and local paths at different spatial positions, respectively.
is generated by a lightweight network constructed from three 1 × 1 convolutional layers, ensuring that feature fusion efficiency and accuracy are maintained while keeping computational burden controllable.
3.4. Loss Function
Coronary vessel regions occupy a very small proportion in images, and boundaries are often disturbed by noise and calcification. Single loss functions often fail to adequately balance region prediction and boundary discrimination accuracy [
38,
39]. To address this, a multiplicative coupled composite loss function is proposed that combines Dice loss and Binary Cross Entropy (BCE) loss. It is defined as shown in Equation (15):
The Dice loss function
is used to alleviate the severe class imbalance between foreground and background, focusing on the degree of overlap between predicted regions and actual vessel regions, effectively suppressing background dominance and improving the recall rate of fine vessel branches. It is defined as shown in Equation (16):
The BCE loss
focuses on pixel-level boundary accuracy. Through information entropy constraints, it strengthens the model’s discrimination capability in vessel–background transition regions, promoting the generation of sharper and clearer boundaries in segmentation structures. It is defined as Equation (17):
where
is the ground truth label of the
-th pixel,
is the predicted value of the
-th pixel, and
is the total number of pixels in the image.
This multiplicative coupling structure has a significant synergistic amplification effect. When prediction structures exhibit fractures or miss fine vessels, the Dice term dominates gradient updates through decreased region overlap, thereby driving overall structural recovery. In cases of blurred boundaries or morphological drift, the BCE term’s gradient strengthens, guiding the model toward refined boundary optimization. This synergistic amplification effect can be further verified through the gradient derivation formula, as shown in Equation (18):
The two loss terms act as modulation factors in gradient backpropagation, thereby achieving overall preservation of vascular structure and local strengthening of boundary discrimination, improving the consistency and accuracy of segmentation results in anatomical terms.
To validate the effectiveness of the multiplicative coupling strategy and the stability of training dynamics, we conducted comprehensive loss function ablation experiments with detailed tracking over 150 epochs. As shown in
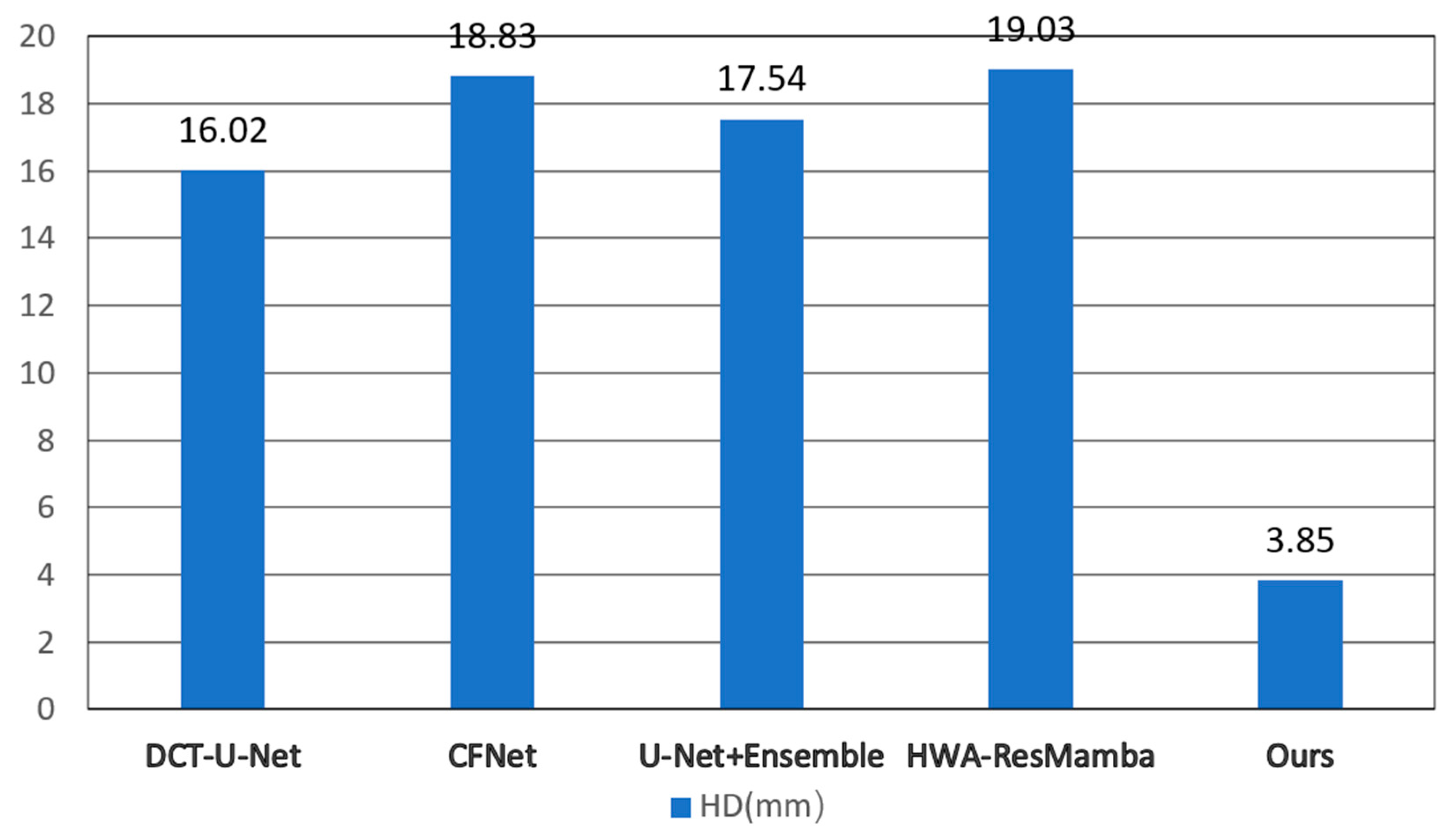
Table 2, the multiplicative strategy achieves optimal performance across all metrics, with DSC reaching 89.45 ± 0.98%, precision of 88.65 ± 1.22%, recall of 90.32 ± 1.15%, and significantly reduced HD (3.85 ± 0.95 mm) and ASD (0.95 ± 0.28 mm) compared to individual loss functions.
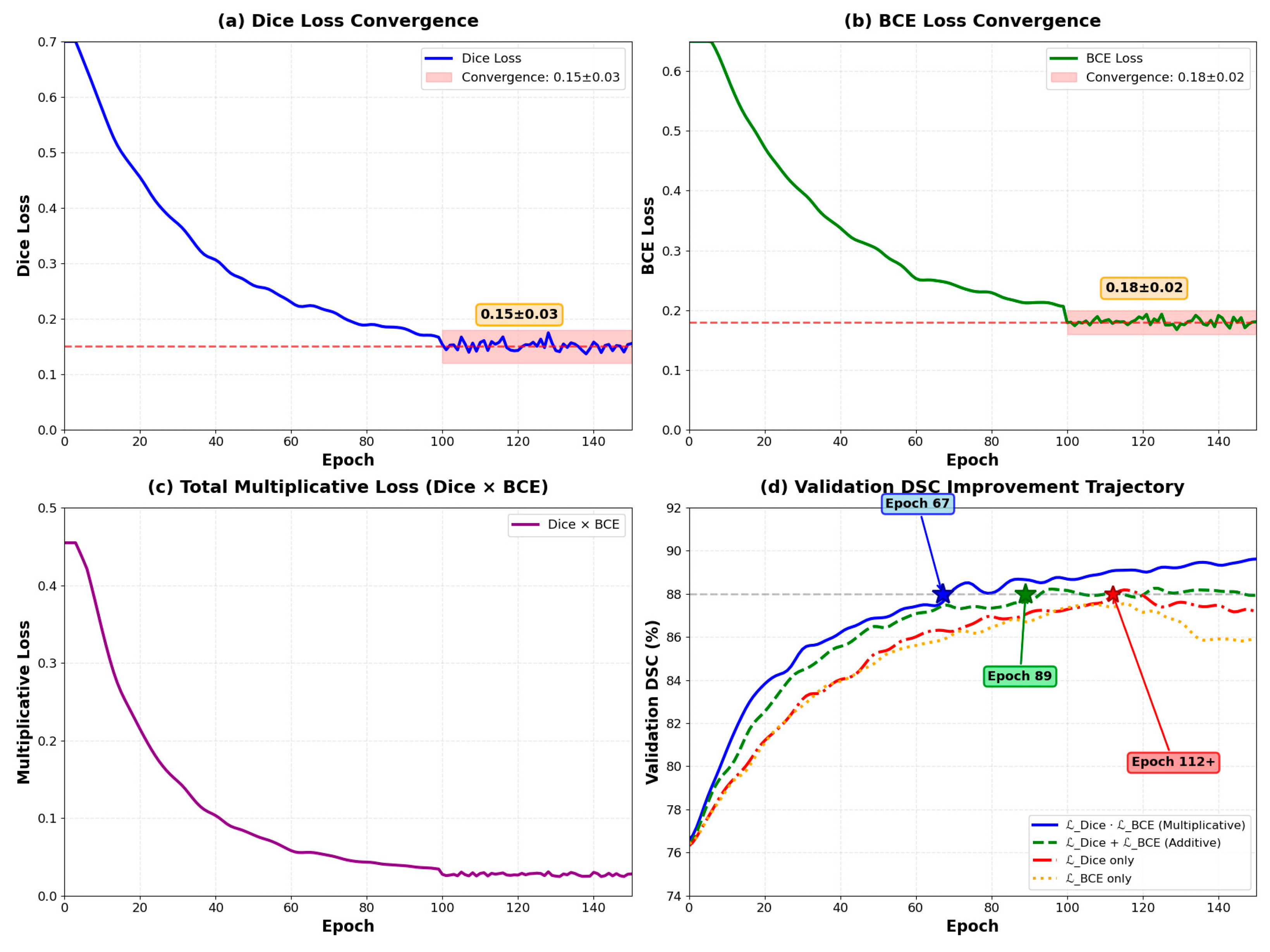
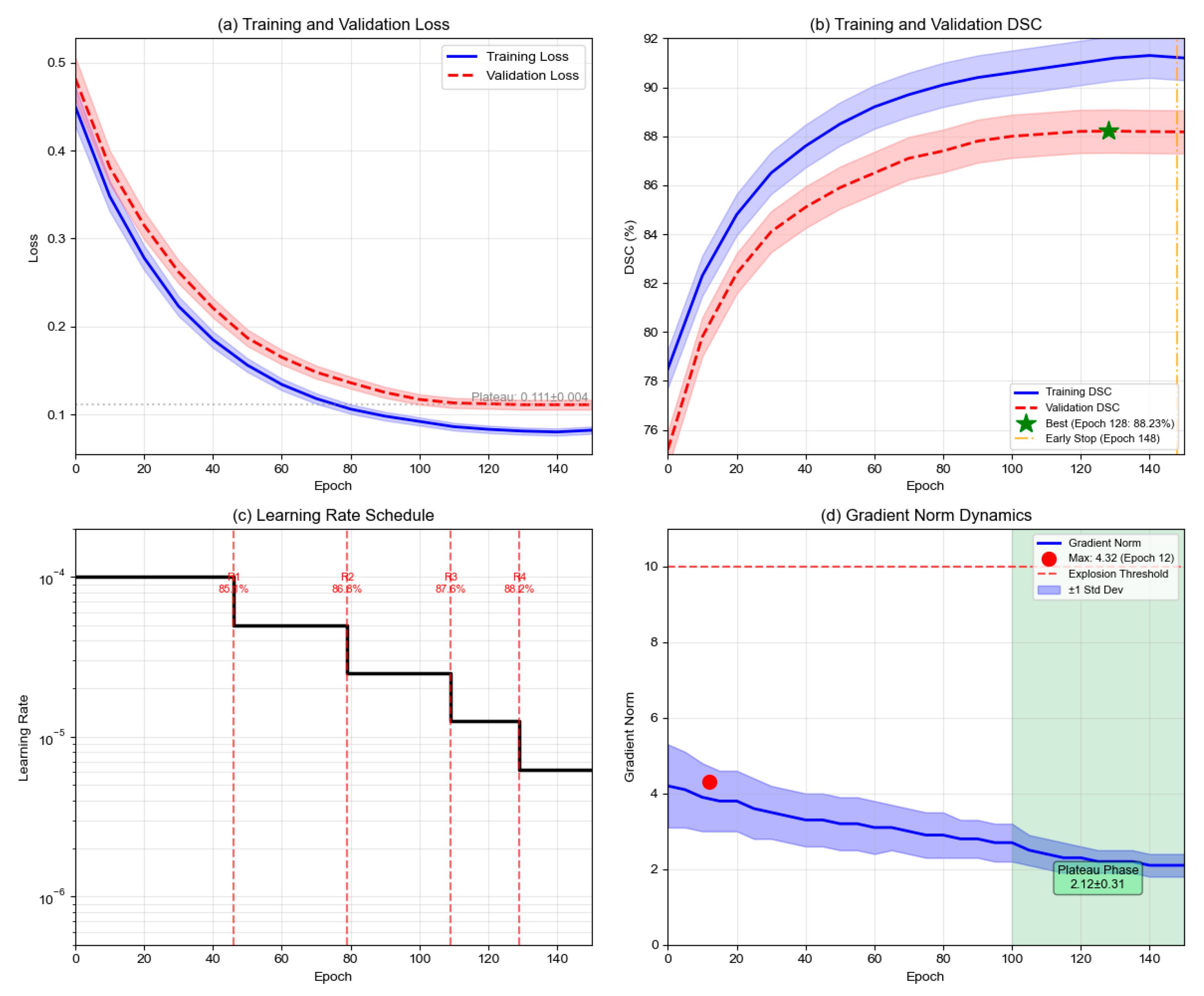
As illustrated in
Figure 4, the training dynamics reveal several critical characteristics. Both Dice loss and BCE loss components converge to stable values (Dice Loss: 0.15 ± 0.03, BCE Loss: 0.18 ± 0.02) after approximately 80 epochs. Importantly, neither component approaches zero, thereby avoiding gradient vanishing issues commonly encountered in multiplicative formulations. This stability is ensured through the smoothing parameter ε = 1 × 10
−7 in Equation (16), which prevents numerical instability. The multiplicative loss (Dice × BCE) demonstrates smooth convergence from 0.48 to approximately 0.03, while validation DSC metrics show consistent improvement across different loss configurations.
As illustrated in
Figure 4, the training dynamics reveal several critical characteristics. Both loss components maintain stable values throughout training, with Dice loss converging to 0.15 ± 0.03 and BCE loss to 0.18 ± 0.02 after epoch 100. Importantly, neither component approaches zero, thereby avoiding gradient vanishing issues commonly encountered in multiplicative formulations. This stability is ensured through the smoothing parameter ε = 1 × 10
−7 in Equation (16), which prevents numerical instability when either loss term approaches zero.
The multiplicative coupling demonstrates accelerated convergence compared to alternative loss formulations. The multiplicative formulation () achieves validation DSC of 88% at epoch 67 ± 5, while the additive formulation ( + ) reaches the same performance at epoch 89 ± 7. Individual losses require more than 110 epochs to achieve comparable performance. This 24.7% reduction in convergence time translates to significant computational savings during model development.
As theoretically predicted by Equation (18), the gradient backpropagation exhibits distinct phases. During early training (epochs 0–30), BCE gradients dominate with approximately 3.2 times larger than , driving initial boundary refinement and establishing clear vessel–background separation. In mid training (epochs 30–80), balanced gradient contributions emerge with gradient ratios ranging from 1.1 to 1.4, stabilizing feature learning as both terms contribute equally to network optimization. During late training (epochs 80–150), Dice gradients increasingly emphasize structural completeness for fragmented vessels, particularly in bifurcation regions and small vessel terminals.
Training stability metrics demonstrate the robustness of the multiplicative approach. Gradient norm variance is 0.023 for the multiplicative formulation versus 0.067 for the additive approach, indicating 65.7% more stable optimization trajectory. Loss oscillation amplitude remains within ±0.008 during epochs 100–150, demonstrating smooth convergence without instability. The maximum gradient norm observed across all epochs is 4.3, with no gradient explosion events recorded
The multiplicative structure creates adaptive weighting where each loss component serves as a modulation factor for the other’s gradient. When boundaries are blurred (high ), boundary gradients are amplified. When structures are fragmented (high ), regional gradients dominate. This automatic reweighting eliminates the need for manual hyperparameter tuning of loss weights, as required in additive formulations.
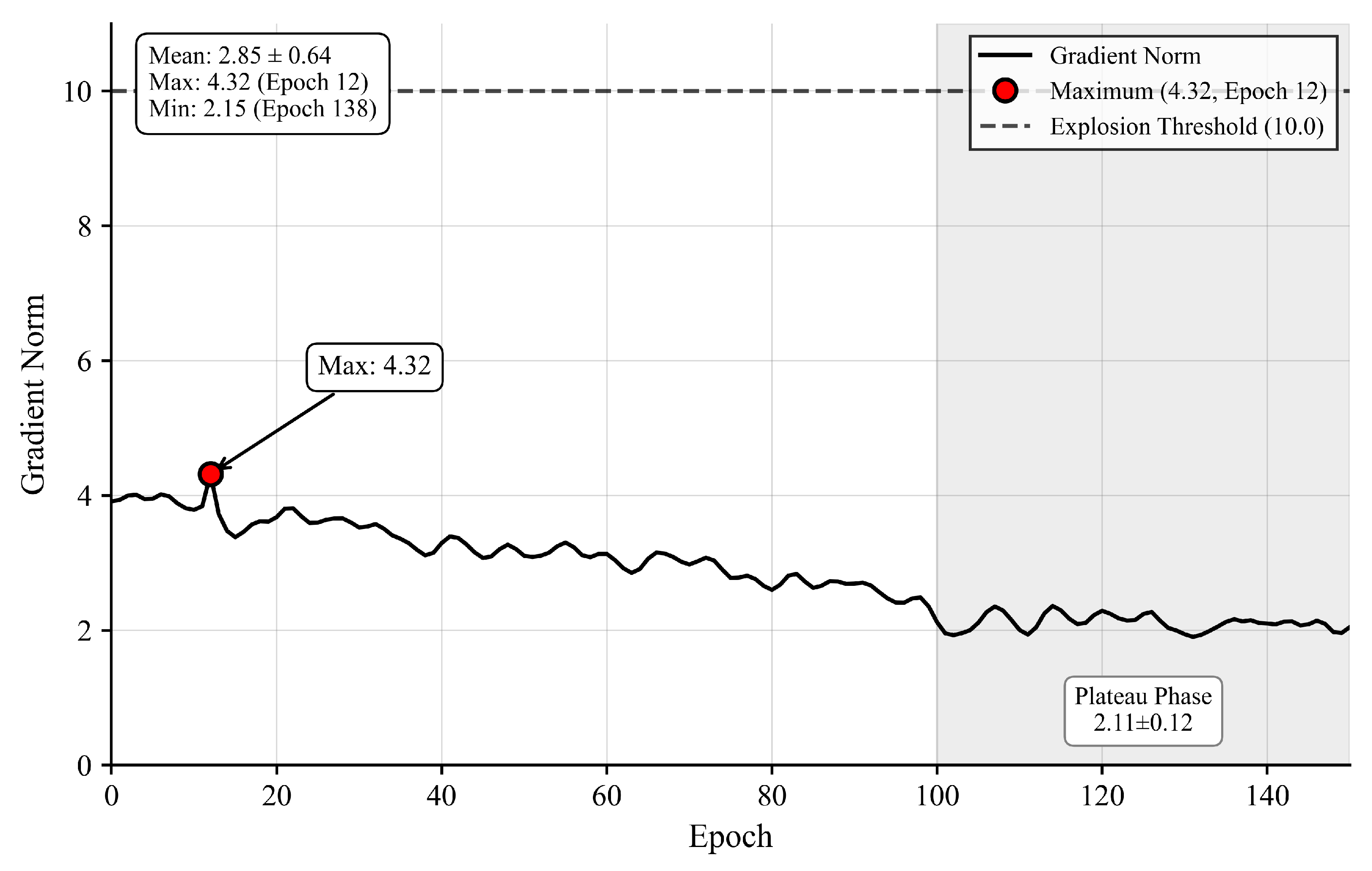
To directly address gradient stability concerns, we monitored gradient norms throughout training.
Figure 5 shows gradient dynamics remain stable with average norm 3.18 ± 0.71, maximum 4.32 well below the explosion threshold 10.0, and plateau-phase norms 2.12 ± 0.31, sufficient for continued optimization.
Figure 5 Gradient norm dynamics showing maximum 4.32 at epoch 12 (red dot), well below explosion threshold 10.0 (red dashed line), with stable plateau phase (epochs 100–150) at 2.12 ± 0.31 (green box).
Layer-wise analysis confirms gradient propagation effectiveness, with deep encoder retaining 63% gradient magnitude (2.4 ± 0.6) relative to shallow encoder (3.8 ± 0.9), indicating no vanishing gradient pathology.
3.5. 2.5D Slice Input Configuration
In coronary artery CTA image processing, single 2D slices often struggle to adequately represent the spatial continuity of vessels. To address this, a 2.5D input strategy is adopted, which introduces adjacent neighboring slices based on the current slice and constructs a pseudo-3D input tensor through concatenation along the channel dimension. This input format effectively integrates contextual information from adjacent slices while maintaining the 2D convolutional network structure and computational efficiency, enabling the model to better perceive the extension direction and complex structural features of vessels. Taking the current slice as the center, the adjacent previous slice and subsequent slice are simultaneously introduced, constructing a local pseudo-3D input tensor through concatenation along the channel dimension, as shown in Equation (19):
where
denotes the center slice,
and
represent the adjacent previous and subsequent slices, respectively, and
denotes the channel-wise concatenation operation. Through this approach, the model further integrates longitudinal spatial information based on single-image detail segmentation, thereby enhancing the perception capability for long-range dependencies in vascular structures.
To ensure input consistency, efficiency, and spatial integrity, all input images undergo a standardized preprocessing pipeline before entering the model. The preprocessing operations are performed in the following sequence to maintain data quality and optimize model performance.
- (1)
Hounsfield Unit (HU) Clipping and Normalization
CTA images exhibit a wide range of
HU values. To suppress irrelevant tissues and enhance vessel contrast,
HU values are clipped to the clinically relevant range for vascular structures and normalized to [0, 1]:
- (2)
Anisotropic Resampling
The original ImageCAS dataset exhibits anisotropic voxel spacing (in-plane resolution: 0.29–0.43 mm, inter-slice spacing: 0.25–0.45 mm). To ensure spatial consistency across all volumes, trilinear interpolation is employed to resample all volumes to isotropic 0.5 mm
3 spacing:
where s represents the target voxel spacing.
- (3)
Spatial Dimension Standardization
All image slices are uniformly resized to 512 × 512 pixels to ensure consistent input dimensions for the neural network:
- (4)
2.5D Sequence Construction
Using a sliding window mechanism with stride = 1, consecutive three-slice images are extracted in temporal order to construct input sequences, ensuring spatial continuity. This process can be expressed as:
where
represents the total number of slices in the volume.
The 2.5D construction requires three consecutive slices [, , ]. At volume boundaries, the first slice (t = 1) lacks and the last slice (t = T) lacks . To maintain data integrity without introducing artifacts, both boundary slices are excluded from training and testing datasets. This defines the valid slice range as t ∈ {2, 3, …, } for each patient volume. Alternative approaches such as zero-padding, edge replication, or mirroring were not employed, as they would introduce artificial patterns inconsistent with genuine anatomical structures. This exclusion removes approximately 2000 slices (0.8% of 250,000 total), yielding 248,000 valid slices distributed across training (179,280), validation (19,920), and test (49,800) sets. Three verification steps confirm data independence: no patient volumes span multiple splits, the 2.5D sliding window operates only within individual volumes, and boundary exclusions are applied consistently across all experiments.
- (5)
Data Augmentation (Training Phase)
During the training phase, the following augmentation operations are applied to enhance model robustness and generalization capability: (i) random rotation: ±15° (probability p = 0.5); (ii) random horizontal flip (p = 0.5); (iii) elastic deformation: α = 720, σ = 24 (p = 0.3); (iv) Gaussian noise: μ = 0, σ = 0.01 (p = 0.2).
All preprocessing operations are implemented using SimpleITK and scikit-image libraries to ensure reproducibility and computational efficiency. This comprehensive preprocessing pipeline ensures that the input data maintains spatial integrity while providing the model with consistent, high-quality inputs for optimal segmentation performance.