Enhanced Skin Lesion Segmentation via Attentive Reverse-Attention U-Net
Abstract
1. Introduction
1.1. Related Works
- Difficulty in accurately distinguishing between different types of skin lesions.
- While high-resolution images yield better performance, a significant decline in accuracy is observed for low-resolution images. Additionally, inconsistencies in image quality and resolution across various datasets further complicate classification and segmentation tasks.
- The limited availability of clinical information in datasets restricts the development of more robust computer-aided diagnostic models.
1.2. Motivation and Contribution
- Are the datasets balanced? If not, how does this imbalance affect performance?
- Which loss function is most suitable for skin lesion segmentation?
- To what extent do attention mechanisms enhance segmentation performance?
- How does the performance of deep learning models vary across different epochs?
- The impact of dataset imbalances and the generalizability of the proposed model across different data distributions were analyzed using two publicly available datasets.
- The effect of various loss functions on model performance was evaluated through a comparative analysis of four different loss functions.
- A reverse-attention-based U-Net model was introduced for skin lesion segmentation, offering a novel approach to improving segmentation accuracy.
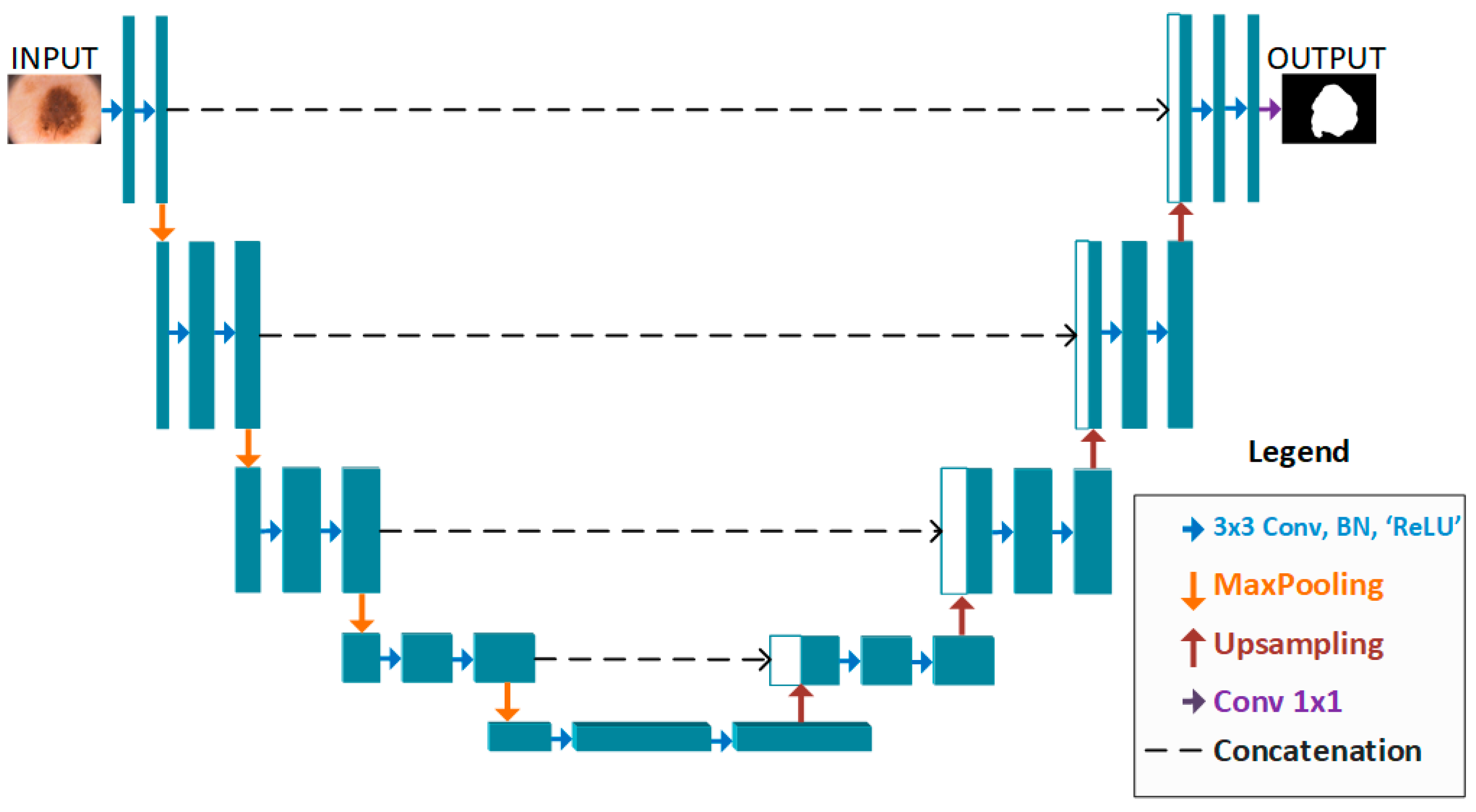
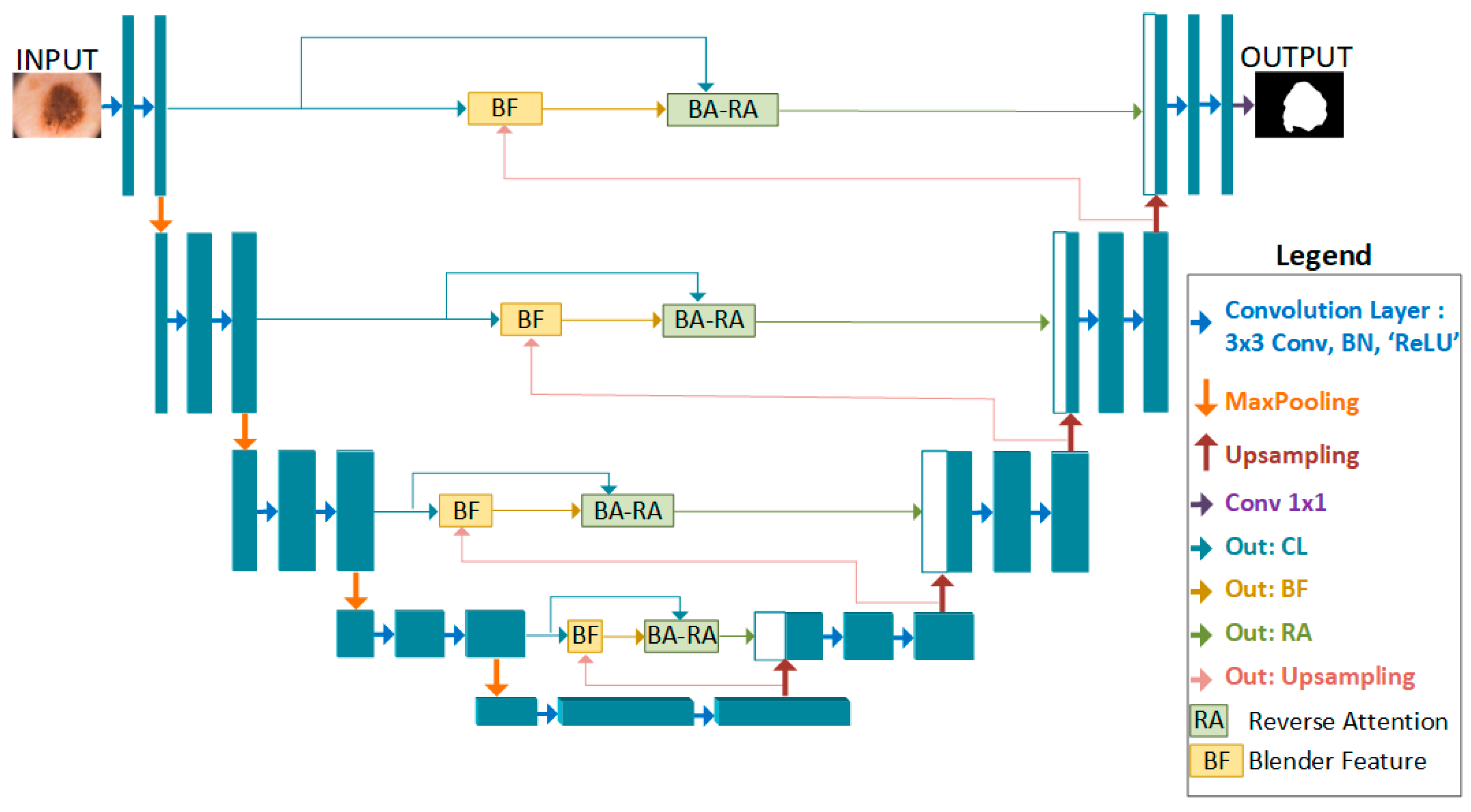
2. Proposed Method
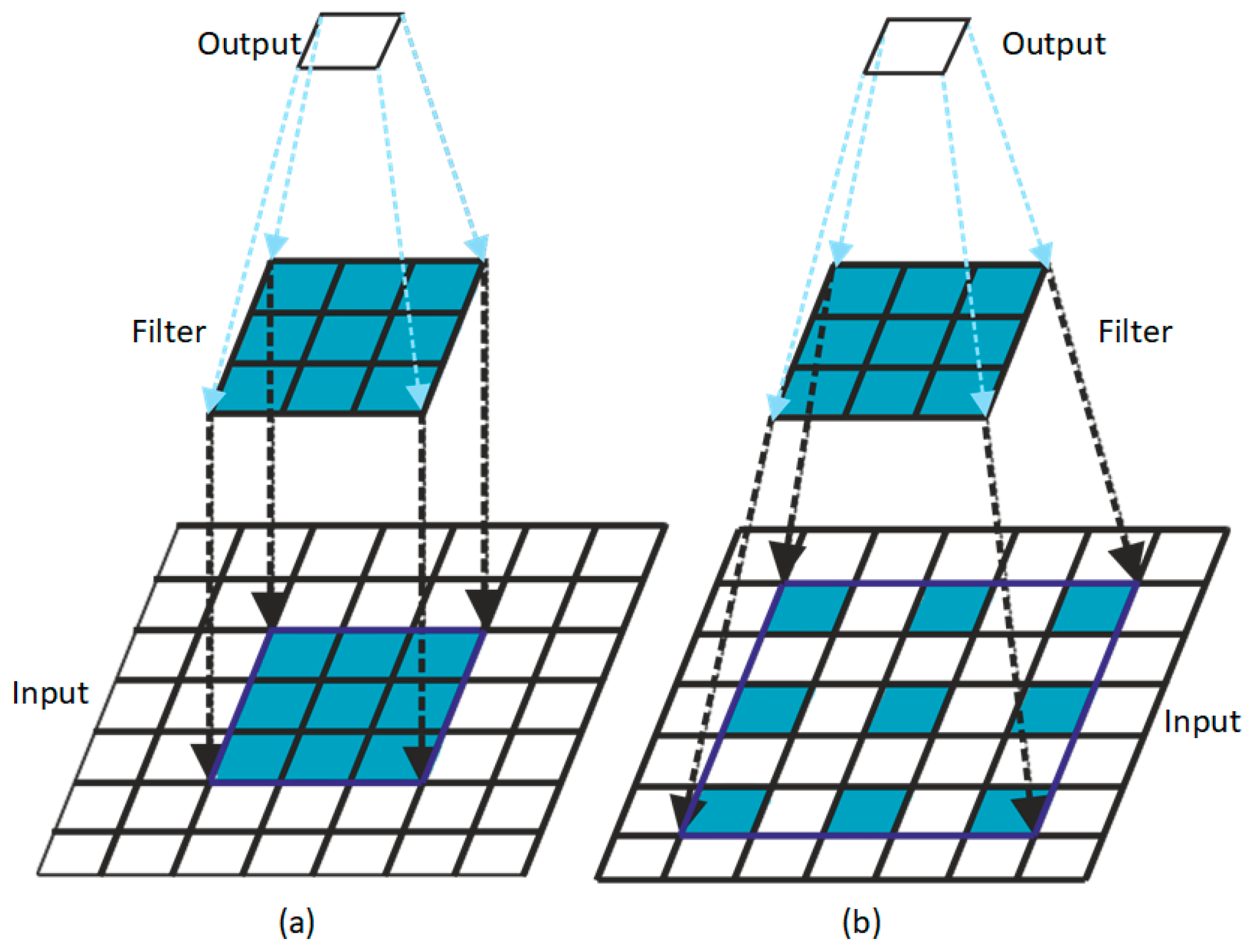
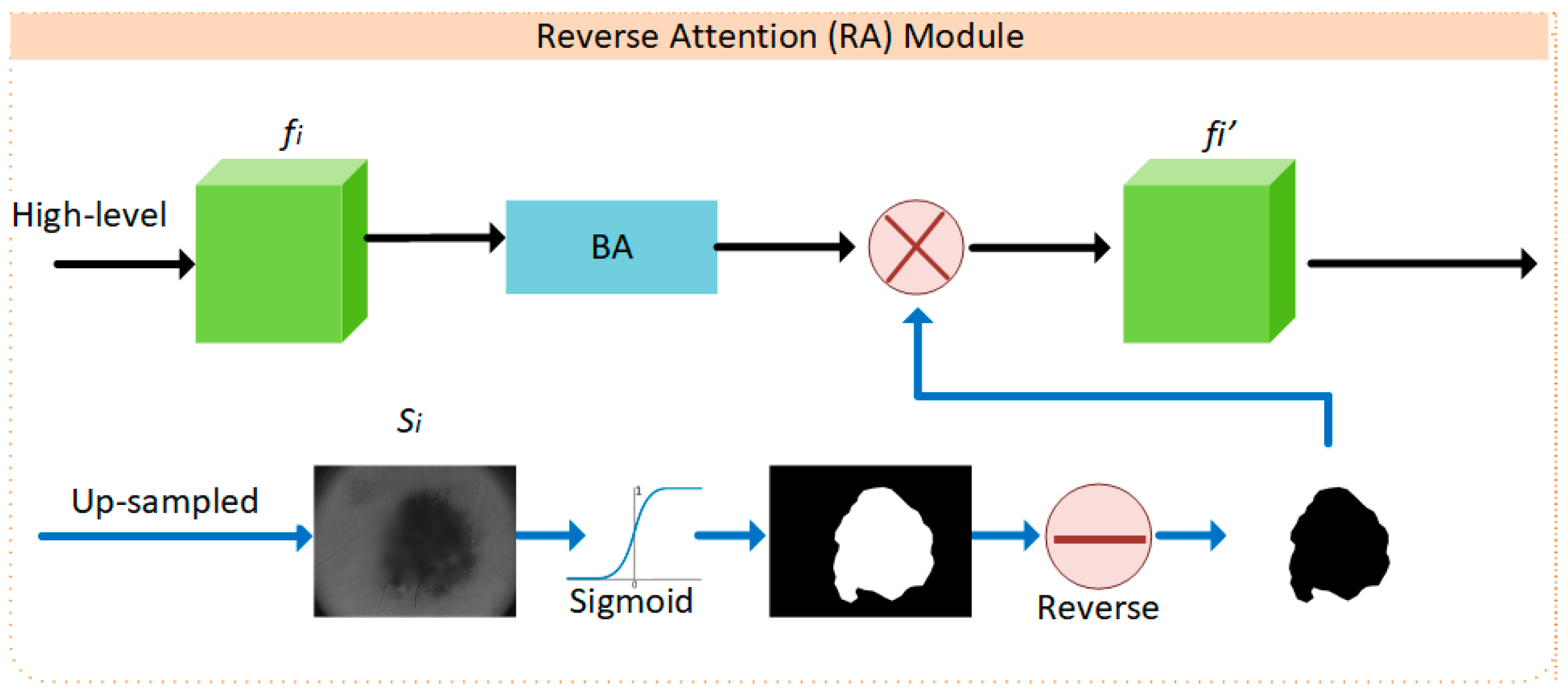
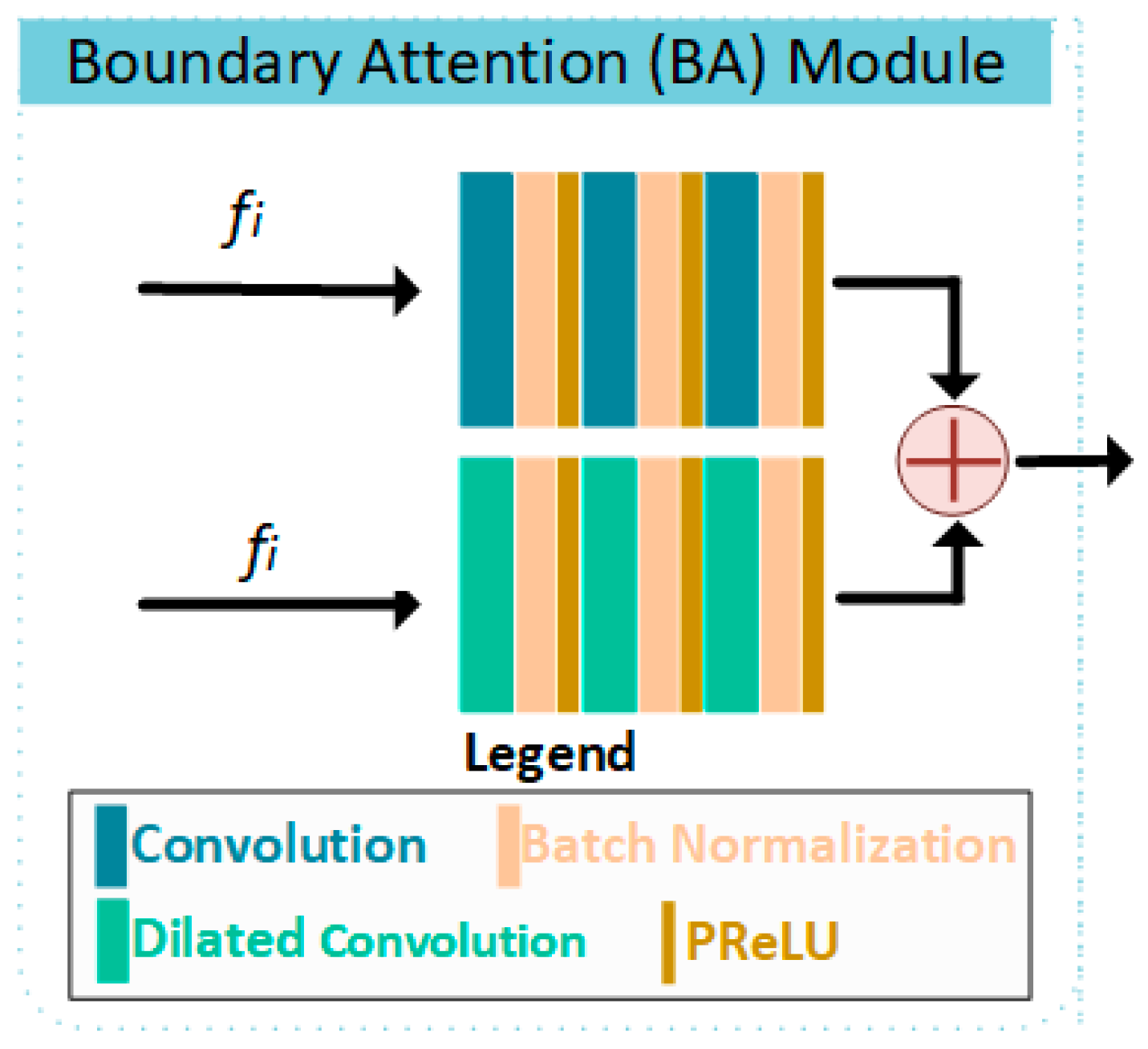
2.1. BA-RA Module
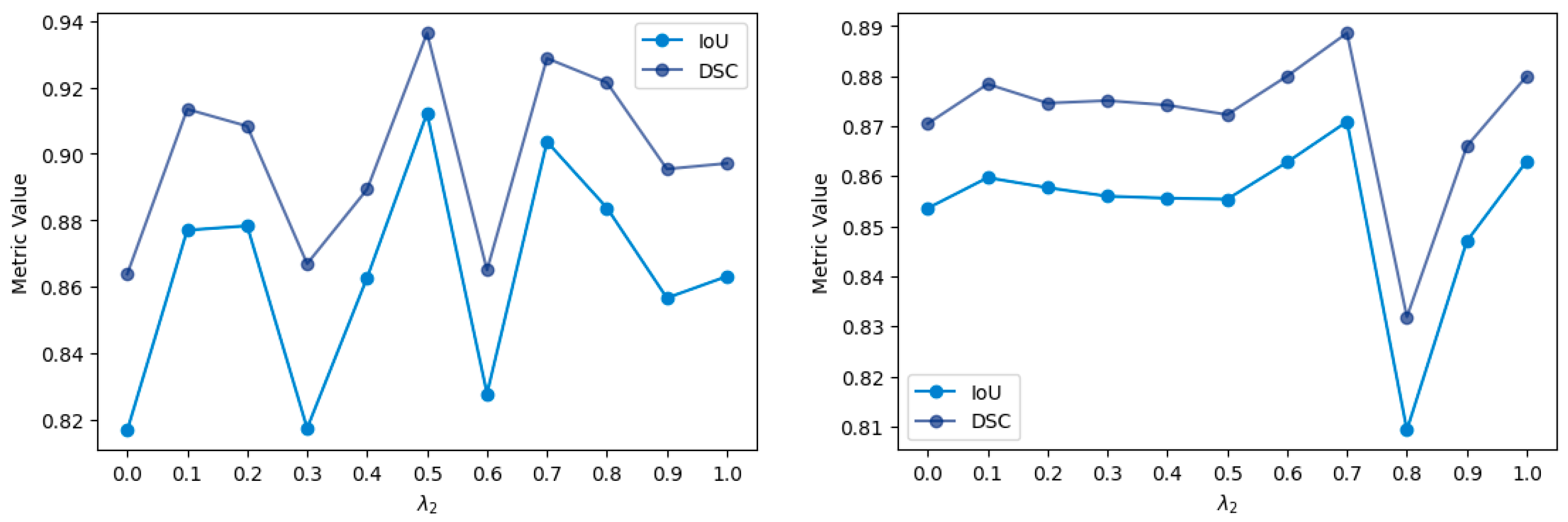
2.2. Loss Function
3. Experimental Studies
3.1. Datasets
3.2. Evaluation Metrics
3.3. Implementation Details
4. Quantitative Results
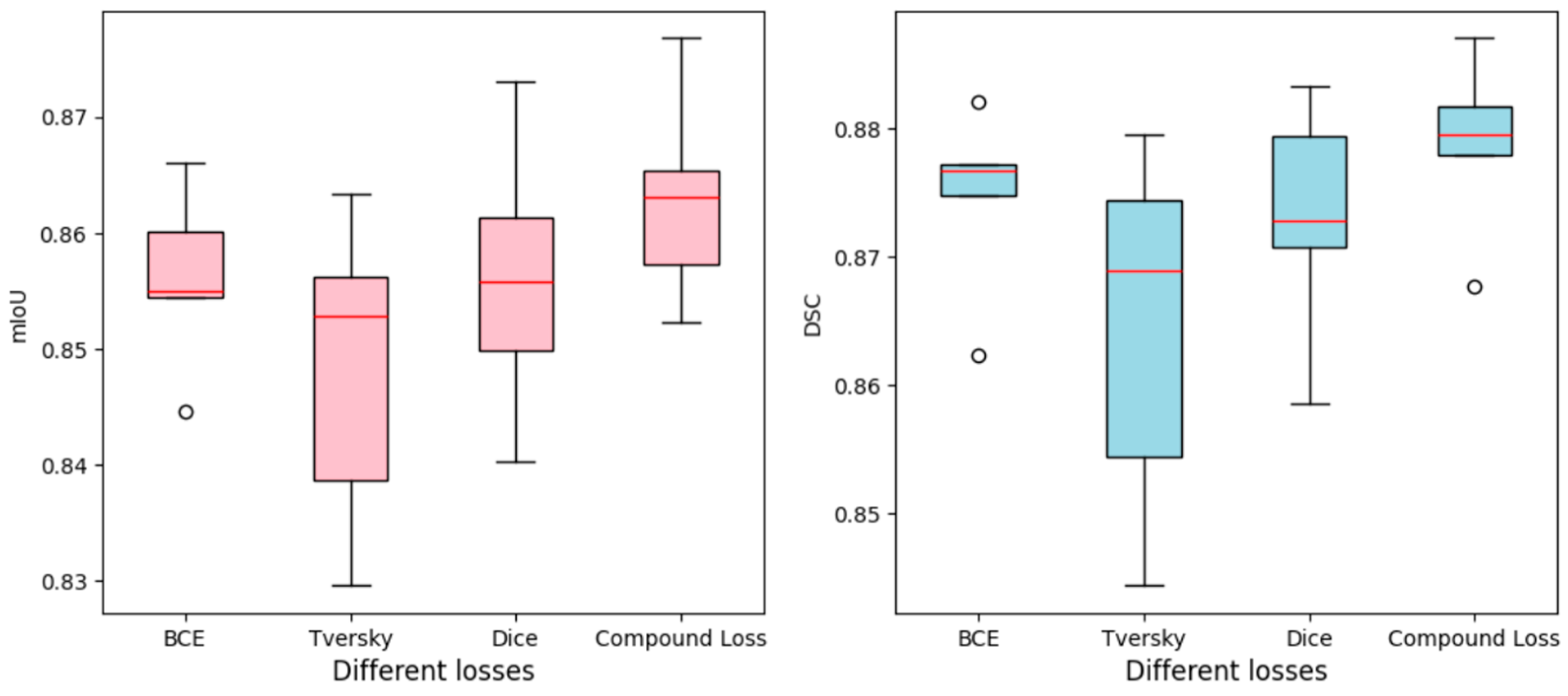
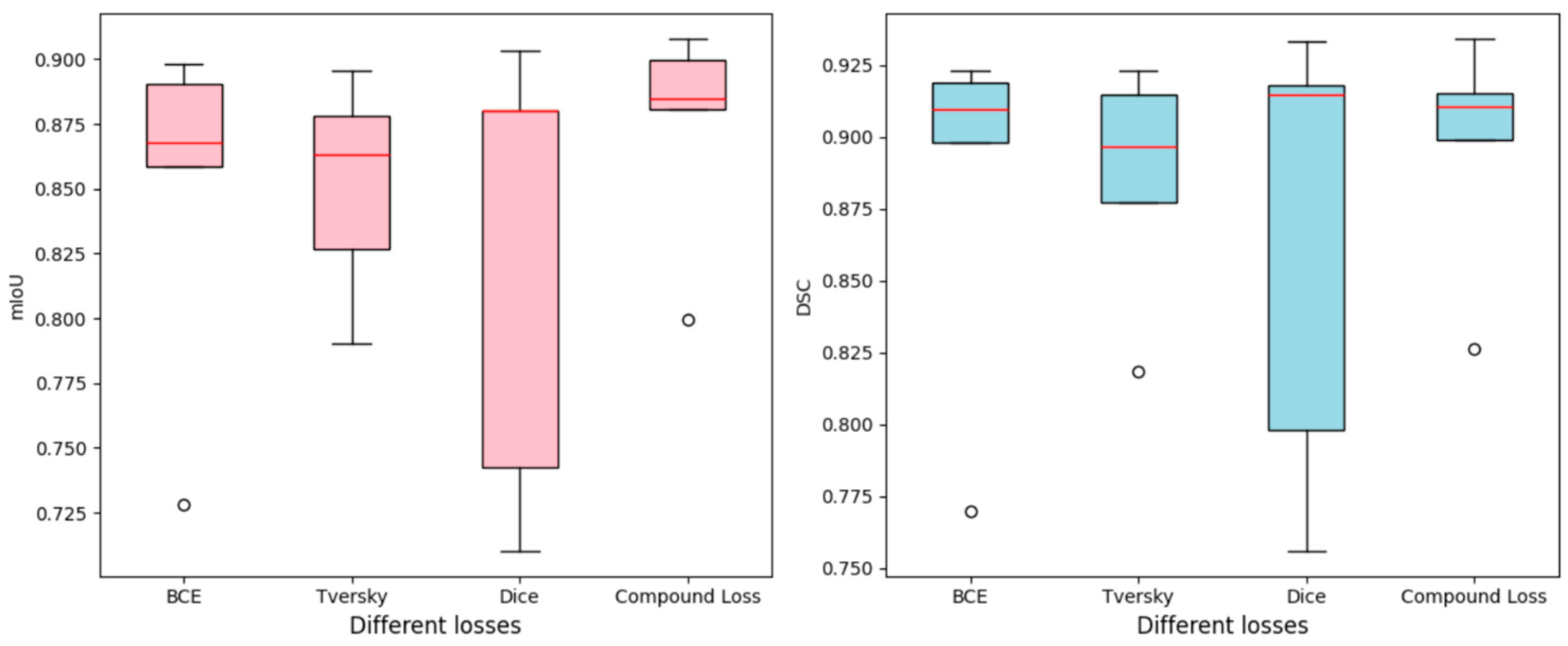
4.1. Comparison with Loss Functions
4.2. Comparison with State-of-the-Art Methods
| Networks (Ref.) | Params (M) ↓ | DSC ↑ | mIoU ↑ |
|---|---|---|---|
| U-Net ([30]) | 32.9 | 0.8417 | 0.8269 |
| Attention-U-Net ([50]) | 33.3 | 0.8253 | 0.8151 |
| Ms-RED ([44]) | 3.8 | 0.8345 | 0.8999 |
| CPFNet ([45]) | 43.3 | 0.8292 | 0.8963 |
| LeaNet ([46]) | 0.11 | 0.8825 | 0.7839 |
| AS-Net ([47]) | 24.9 | 0.8955 | 0.8309 |
| ACCPG-Net ([12]) | 11.8 | 0.9081 | 0.8352 |
| MCGFF-Net ([51]) | 39.67 | 0.8907 | 0.8179 |
| Ours | 48 | 0.8871 | 0.8768 |
| Networks (Ref.) | Params (M) ↓ | DSC ↑ | mIoU ↑ |
|---|---|---|---|
| U-Net ([30]) | 32.9 | 0.9293 | 0.9012 |
| Attention-U-Net ([50]) | 33.3 | 0.8992 | 0.8673 |
| AS-Net ([47]) | 24.9 | 0.9305 | 0.8760 |
| ICL-Net ([49]) | N.A | 0.9280 | 0.8725 |
| ACCPG-Net ([12]) | 11.8 | 0.9133 | 0.8427 |
| MCGFF-Net ([51]) | 39.67 | 0.9307 | 0.8752 |
| Ours | 48 | 0.9341 | 0.9078 |
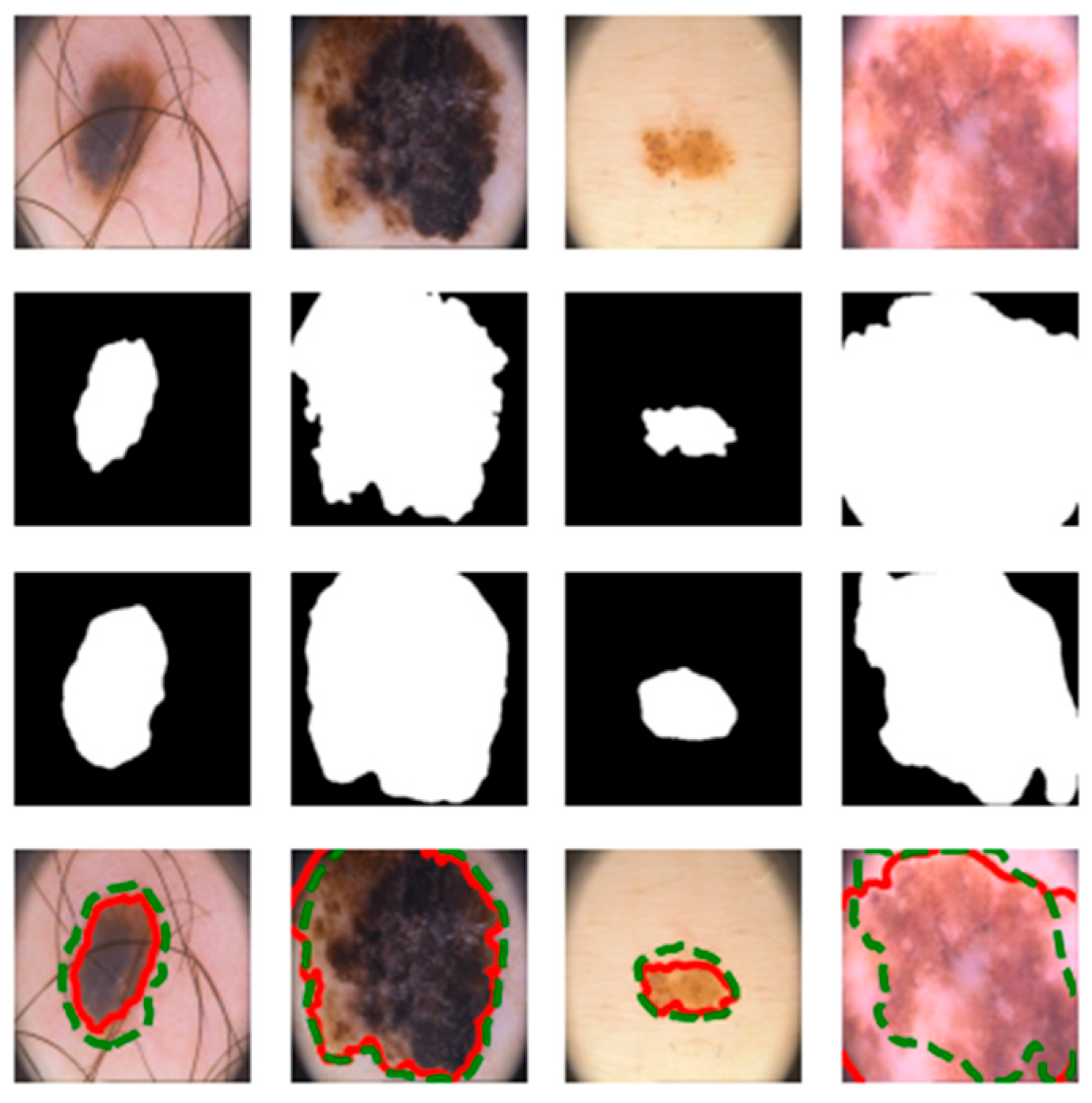
4.3. Visualization Results
5. Conclusions, Limitation and Future Work
Funding
Data Availability Statement
Conflicts of Interest
References
- Woo, Y.R.; Cho, S.H.; Lee, J.D.; Kim, H.S. The Human Microbiota and Skin Cancer. Int. J. Mol. Sci. 2022, 23, 1813. [Google Scholar] [CrossRef]
- Jiminez, V.; Yusuf, N. An update on clinical trials for chemoprevention of human skin cancer. J. Cancer Metastasis Treat. 2023, 9, 4. [Google Scholar] [CrossRef]
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global burden of cutaneous melanoma in 2020 and projections to 2040. JAMA Dermatol. 2022, 158, 495–503. [Google Scholar] [CrossRef]
- Khan, M.Q.; Hussain, A.; Rehman, S.U.; Khan, U.; Maqsood, M.; Mehmood, K.; Khan, M.A. Classification of Melanoma and Nevus in Digital Images for Diagnosis of Skin Cancer. IEEE Access 2019, 7, 90132–90144. [Google Scholar] [CrossRef]
- Harrison, K. The accuracy of skin cancer detection rates with the implementation of dermoscopy among dermatology clinicians: A scoping review. J. Clin. Aesthetic Dermatol. 2024, 17 (Suppl. S1), S18. [Google Scholar]
- Mendonca, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.S.; Rozeira, J. PH2-A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar] [CrossRef]
- Karthik, B.; Muthupandi, G. SVM and CNN based skin tumour classification using WLS smoothing filter. Optik 2023, 272, 170337. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Koundal, D.; Singh, K. Fusion of U-Net and CNN model for segmentation and classification of skin lesion from dermoscopy images. Expert Syst. Appl. 2023, 213, 119230. [Google Scholar] [CrossRef]
- Song, L.; Wang, H.; Wang, Z.J. Decoupling multi-task causality for improved skin lesion segmentation and classification. Pattern Recognit. 2023, 133, 108995. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Cai, J.; Lee, T.K.; Miao, C.; Wang, Z.J. SSD-KD: A self-supervised diverse knowledge distillation method for lightweight skin lesion classification using dermoscopic images. Med. Image Anal. 2023, 84, 102693. [Google Scholar] [CrossRef]
- He, X.; Wang, Y.; Zhao, S.; Chen, X. Joint segmentation and classification of skin lesions via a multi-task learning convolutional neural network. Expert Syst. Appl. 2023, 230, 120174. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, F.; Zhao, W.; Hu, Y.; Su, H.; Yuan, M. ACCPG-Net: A skin lesion segmentation network with Adaptive Channel-Context-Aware Pyramid Attention and Global Feature Fusion. Comput. Biol. Med. 2023, 154, 106580. [Google Scholar] [CrossRef]
- Qiu, S.; Li, C.; Feng, Y.; Zuo, S.; Liang, H.; Xu, A. GFANet: Gated Fusion Attention Network for skin lesion segmentation. Comput. Biol. Med. 2023, 155, 106462. [Google Scholar] [CrossRef]
- Karri, M.; Annavarapu, C.S.R.; Acharya, U.R. Skin lesion segmentation using two-phase cross-domain transfer learning framework. Comput. Methods Programs Biomed. 2023, 231, 107408. [Google Scholar] [CrossRef]
- Tembhurne, J.V.; Hebbar, N.; Patil, H.Y.; Diwan, T. Skin cancer detection using ensemble of machine learning and deep learning techniques. Multimed. Tools Appl. 2023, 82, 27501–27524. [Google Scholar] [CrossRef]
- Gilani, S.Q.; Syed, T.; Umair, M.; Marques, O. Skin Cancer Classification Using Deep Spiking Neural Network. J. Digit. Imaging 2023, 36, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Golnoori, F.; Boroujeni, F.Z.; Monadjemi, A. Metaheuristic algorithm based hyper-parameters optimization for skin lesion classification. Multimed. Tools Appl. 2023, 82, 25677–25709. [Google Scholar] [CrossRef]
- Shukla, M.M.; Tripathi, B.K.; Dwivedi, T.; Tripathi, A.; Chaurasia, B.K. A hybrid CNN with transfer learning for skin cancer disease detection. Med. Biol. Eng. Comput. 2024, 62, 3057–3071. [Google Scholar] [CrossRef]
- Natha, P.; RajaRajeswari, P. Advancing Skin Cancer Prediction Using Ensemble Models. Computers 2024, 13, 157. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, Y.; Zhu, X. Transfer Contrastive Learning for Raman Spectroscopy Skin Cancer Tissue Classification. IEEE J. Biomed. Health Inform. 2024, 28, 7332–7344. [Google Scholar] [CrossRef]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A simple framework for contrastive learning of visual representations. In Proceedings of the 37th International Conference on Machine Learning ICML 2020, Vienna, Austria, 13–18 July 2020; Volume Part F16814. pp. 1575–1585. [Google Scholar]
- Pandurangan, V.; Sarojam, S.P.; Narayanan, P.; Velayutham, M. Hybrid deep learning-based skin cancer classification with RPO-SegNet for skin lesion segmentation. Netw. Comput. Neural Syst. 2024, 36, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xie, C.; Zeng, N. RP-Net: A 3D Convolutional Neural Network for Brain Segmentation From Magnetic Resonance Imaging. IEEE Access 2019, 7, 39670–39679. [Google Scholar] [CrossRef]
- Eerapu, K.K.; Lal, S.; Narasimhadhan, A.V. O-SegNet: Robust Encoder and Decoder Architecture for Objects Segmentation From Aerial Imagery Data. IEEE Trans. Emerg. Top. Comput. Intell. 2022, 6, 556–567. [Google Scholar] [CrossRef]
- Ren, J.S.; Xu, L.; Yan, Q.; Sun, W. Shepard convolutional neural networks. Adv. Neural Inf. Process. Syst. 2015, 2015, 901–909. [Google Scholar]
- Sun, W.; Su, F.; Wang, L. Improving deep neural networks with multi-layer maxout networks and a novel initialization method. Neurocomputing 2018, 278, 34–40. [Google Scholar] [CrossRef]
- Kumar, M.; Mehta, U. Enhancing the performance of CNN models for pneumonia and skin cancer detection using novel fractional activation function. Appl. Soft Comput. 2025, 168, 112500. [Google Scholar] [CrossRef]
- Yang, B.; Zhang, R.; Peng, H.; Guo, C.; Luo, X.; Wang, J.; Long, X. SLP-Net:An efficient lightweight network for segmentation of skin lesions. Biomed. Signal Process. Control 2025, 101, 107242. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Wei, J.; Yuan, X.; Wu, M. A Review of Non-Fully Supervised Deep Learning for Medical Image Segmentation. Information 2025, 16, 433. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, Miccai; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Yu, Y.; Chen, S.; Wei, H. Modified UNet with attention gate and dense skip connection for flow field information prediction with porous media. Flow Meas. Instrum. 2023, 89, 102300. [Google Scholar] [CrossRef]
- Ambesange, S.; Annappa, B.; Koolagudi, S.G. Simulating Federated Transfer Learning for Lung Segmentation using Modified UNet Model. Procedia Comput. Sci. 2022, 218, 1485–1496. [Google Scholar] [CrossRef]
- Fan, D.-P.; Ji, G.-P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. PraNet: Parallel Reverse Attention Network for Polyp Segmentation. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2020; Volume 12266 LNCS, pp. 263–273. [Google Scholar] [CrossRef]
- Du Nguyen, Q.; Thai, H.-T. Crack segmentation of imbalanced data: The role of loss functions. Eng. Struct. 2023, 297, 116988. [Google Scholar] [CrossRef]
- Xu, H.; He, H.; Zhang, Y.; Ma, L.; Li, J. A comparative study of loss functions for road segmentation in remotely sensed road datasets. Int. J. Appl. Earth Obs. Geoinf. 2023, 116, 103159. [Google Scholar] [CrossRef]
- Ma, J.; Chen, J.; Ng, M.; Huang, R.; Li, Y.; Li, C.; Yang, X.; Martel, A.L. Loss odyssey in medical image segmentation. Med. Image Anal. 2021, 71, 102035. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2017; Volume 10541 LNCS, pp. 379–387. [Google Scholar] [CrossRef]
- Yeung, M.; Sala, E.; Schönlieb, C.-B.; Rundo, L. Unified Focal loss: Generalising Dice and cross entropy-based losses to handle class imbalanced medical image segmentation. Comput. Med. Imaging Graph. 2022, 95, 102026. [Google Scholar] [CrossRef]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2019, arXiv:1902.03368. [Google Scholar] [CrossRef]
- Yin, W.; Zhou, D.; Nie, R. DI-UNet: Dual-branch interactive U-Net for skin cancer image segmentation. J. Cancer Res. Clin. Oncol. 2023, 149, 15511–15524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, C.; Zuo, S.; Cai, Y.; Xu, A.; Huang, H.; Zhou, S. The impact of multi-class information decoupling in latent space on skin lesion segmentation. Neurocomputing 2025, 617, 128962. [Google Scholar] [CrossRef]
- Dai, D.; Dong, C.; Xu, S.; Yan, Q.; Li, Z.; Zhang, C.; Luo, N. Ms RED: A novel multi-scale residual encoding and decoding network for skin lesion segmentation. Med. Image Anal. 2022, 75, 102293. [Google Scholar] [CrossRef]
- Feng, S.; Zhao, H.; Shi, F.; Cheng, X.; Wang, M.; Ma, Y.; Xiang, D.; Zhu, W.; Chen, X. Cpfnet: Context pyramid fusion network for medical image segmentation. IEEE Trans. Med. Imaging 2020, 39, 3008–3018. [Google Scholar] [CrossRef]
- Hu, B.; Zhou, P.; Yu, H.; Dai, Y.; Wang, M.; Tan, S.; Sun, Y. LeaNet: Lightweight U-shaped architecture for high-performance skin cancer image segmentation. Comput. Biol. Med. 2024, 169, 107919. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Lu, J.; Lee, D.; Xiong, D.; Chen, Z. AS-Net: Attention Synergy Network for skin lesion segmentation. Expert Syst. Appl. 2022, 201, 117112. [Google Scholar] [CrossRef]
- Bertels, J.; Eelbode, T.; Berman, M.; Vandermeulen, D.; Maes, F.; Bisschops, R.; Blaschko, M.B. Optimizing the Dice Score and Jaccard Index for Medical Image Segmentation: Theory and Practice. In International Conference on Medical Image Computing and Computer Assisted Intervention—MICCAI; Springer: Cham, Switzerland, 2019; pp. 92–100. [Google Scholar]
- Cao, W.; Yuan, G.; Liu, Q.; Peng, C.; Xie, J.; Yang, X.; Ni, X.; Zheng, J. ICL-Net: Global and Local Inter-Pixel Correlations Learning Network for Skin Lesion Segmentation. IEEE J. Biomed. Health Inform. 2023, 27, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Li, Y.; Meng, W.; Ma, D.; Xu, S.; Zhu, X. MCGFF-Net: A multi-scale context-aware and global feature fusion network for enhanced polyp and skin lesion segmentation. Vis. Comput. 2024, 41, 5267–5282. [Google Scholar] [CrossRef]
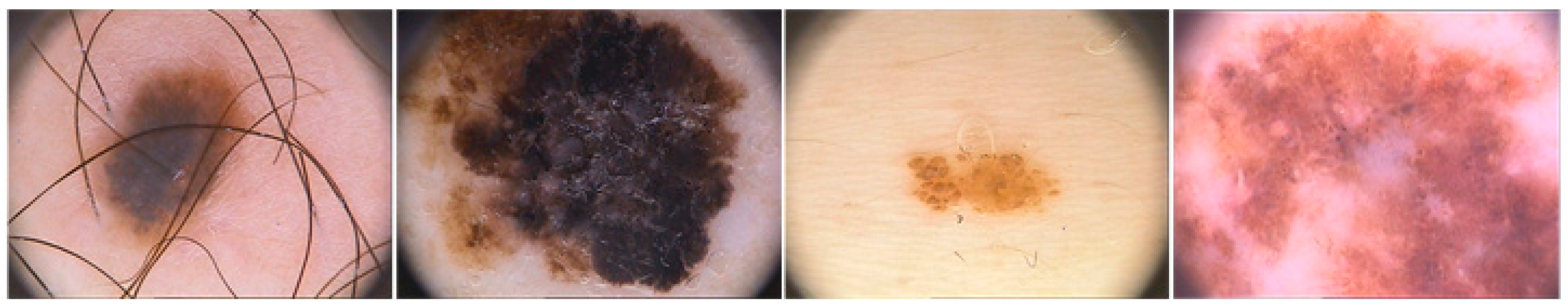


| Ref. (Year) | Method(s) | Description | Result |
|---|---|---|---|
| [7] (2022) | Feature extraction + Feature classification using SVM and CNN | Classification | Enhancement ratio 1.5 has average classification rate around 98% |
| [8] (2023) | A fusion model is proposed with the integration of U-Net and Convolution Neural Network model. | Segmentation and Classification | HAM10000: 0.979 |
| [9] (2023) | Segmentation using U-Net and classification with ResNet/DenseNet | Segmentation and Classification | PH2: Acc: 0.965 (segmentation) PH2: Acc: 0.933 (classification) ISIC2017: 0.956 (segmentation) |
| [10] (2023) | A self-supervised diverse knowledge distillation method, that is SSD-KD | Classification | ISIC2019: Acc: 0.846 (Teacher: Resnet50-Student: MobileNetV2) |
| [11] (2023) | Multi- Task Learning Convolutional Neural Network (Mtl-Cnn) | Segmentation and Classification | Xiangya-Clinic dataset: 0.959 (Melanoma Classification) ISIC2016: 0.885 (Skin lesion Classification) ISIC2017: 0.940 (Seborrheic Keratosis Classification) 0.873 (Melanoma Classification) |
| [12] (2023) | Adaptive Channel-Context-Aware Pyramid Attention and Global Feature Fusion (ACCPG-Net) | Segmentation | ISIC2016: 0.9662 ISIC2017: 0.9561 ISIC2018: 0.9613 |
| [13] (2023) | Gated Fusion Attention Network | Segmentation | ISIC2016: 0.9604 ISIC2017: 0.9397 ISIC2018: 0.9629 |
| [14] (2023) | Two-phase cross-domain transfer learning framework | HAM10000: 0.9912 MoleMap: 0.9701 | |
| [15] (2023) | Feature extraction based on deep learning and machine learning | Classification | ISIC Archive: 0.93 |
| [16] (2023) | Deep spiking Neural network | Classification | ISIC2019: 0.8957 |
| [17] (2023) | Hyperparameter tuning based on optimization | Classification | ISIC2017: 0.816 ISIC2018: 0.901 |
| [18] (2024) | A hybrid CNN with transfer learning | Classification | ISIC2019: 0.901 |
| [19] (2024) | Max Voting Model | Classification | ISIC2018 and HAM10000: 0.9580 |
| [20] (2024) | Transfer learning and contrastive learning | Classification | Private dataset |
| [22] (2024) | Hybrid deep learning method | Segmentation and Classification | ISIC2019: 0.9174 |
| [27] (2025) | Novel activation function | Classification | ISIC2018: 0.9212 |
| [28] (2025) | Segmentation | ISIC2016: 0.9570, ISIC2018: 0.9387 PH2: 0.9162 |
| Loss Function | DSC | mIoU | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Epoch | Epoch | |||||||||
| 20 | 40 | 60 | 80 | 100 | 20 | 40 | 60 | 80 | 100 | |
| BCE | 0.8624 | 0.8821 | 0.8748 | 0.8767 | 0.8772 | 0.8446 | 0.8661 | 0.8544 | 0.8550 | 0.8601 |
| Tversky | 0.8544 | 0.8444 | 0.8690 | 0.8744 | 0.8796 | 0.8386 | 0.8296 | 0.8528 | 0.8562 | 0.8633 |
| Dice | 0.8708 | 0.8586 | 0.8795 | 0.8834 | 0.8728 | 0.8499 | 0.8403 | 0.8613 | 0.8731 | 0.8558 |
| Compound | 0.8677 | 0.8818 | 0.8796 | 0.8871 | 0.8780 | 0.8523 | 0.8631 | 0.8653 | 0.8768 | 0.8573 |
| Loss Function | DSC | mIoU | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Epoch | Epoch | |||||||||
| 20 | 40 | 60 | 80 | 100 | 20 | 40 | 60 | 80 | 100 | |
| BCE | 0.7697 | 0.9095 | 0.9233 | 0.8981 | 0.9190 | 0.7280 | 0.8680 | 0.8983 | 0.8587 | 0.8905 |
| Tversky | 0.8183 | 0.8771 | 0.9149 | 0.8965 | 0.9232 | 0.7903 | 0.8266 | 0.8779 | 0.8630 | 0.8954 |
| Dice | 0.7562 | 0.7979 | 0.9304 | 0.9146 | 0.9181 | 0.7426 | 0.7104 | 0.9033 | 0.8800 | 0.8803 |
| Compound | 0.8265 | 0.8988 | 0.9341 | 0.9150 | 0.9106 | 0.7994 | 0.8996 | 0.9078 | 0.8846 | 0.8809 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toptaş, B. Enhanced Skin Lesion Segmentation via Attentive Reverse-Attention U-Net. Symmetry 2025, 17, 2002. https://doi.org/10.3390/sym17112002
Toptaş B. Enhanced Skin Lesion Segmentation via Attentive Reverse-Attention U-Net. Symmetry. 2025; 17(11):2002. https://doi.org/10.3390/sym17112002
Chicago/Turabian StyleToptaş, Buket. 2025. "Enhanced Skin Lesion Segmentation via Attentive Reverse-Attention U-Net" Symmetry 17, no. 11: 2002. https://doi.org/10.3390/sym17112002
APA StyleToptaş, B. (2025). Enhanced Skin Lesion Segmentation via Attentive Reverse-Attention U-Net. Symmetry, 17(11), 2002. https://doi.org/10.3390/sym17112002




