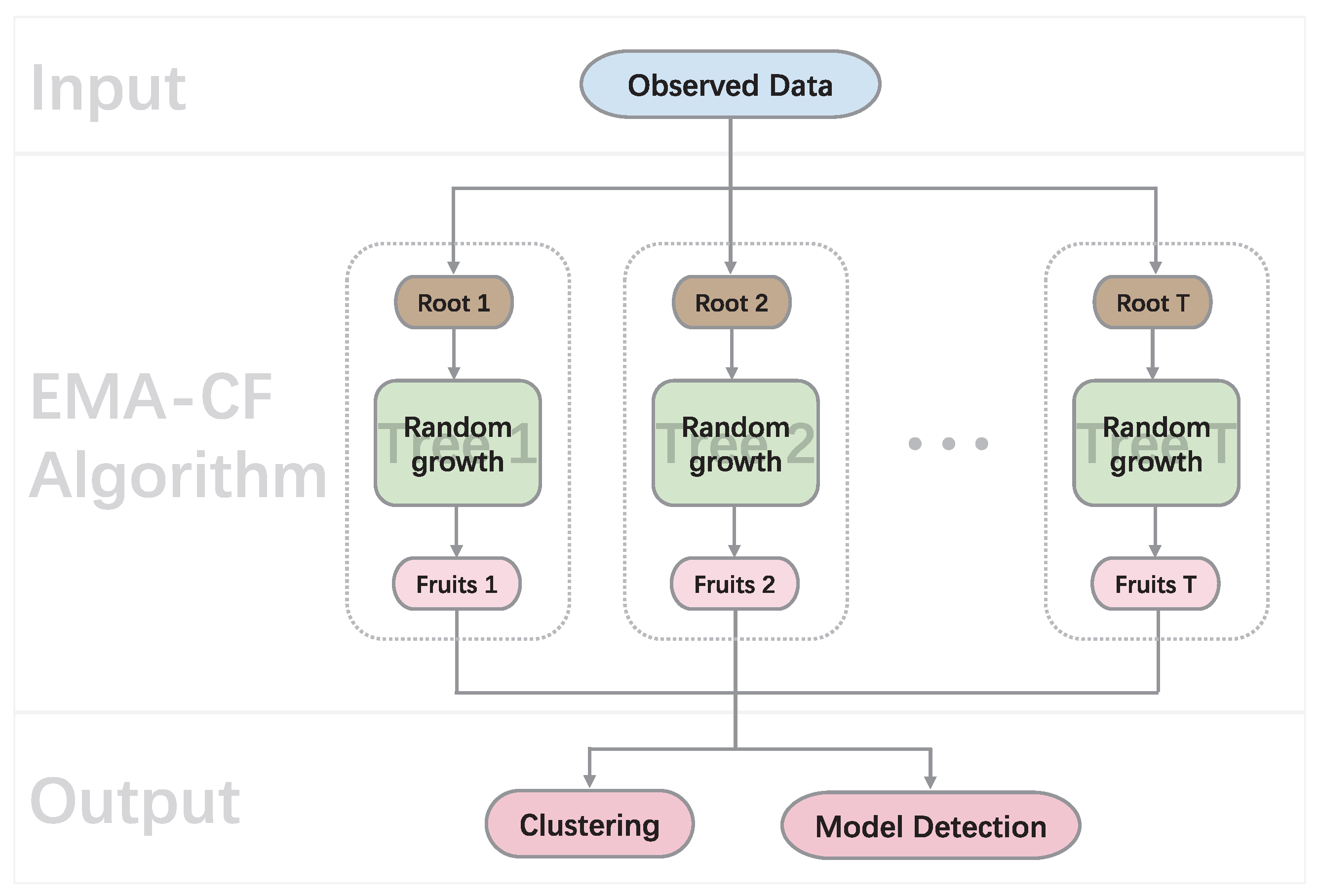
6.1. Experiments on Synthetic Data
In this section, we assess the proposed parsimonious modeling and approximated BMA approach for model-based clustering through different simulated data scenarios. The objective is to evaluate the ability of this framework to recover the original grouping of the data, as well as its ability to select features and detect covariance structures. We refer to the proposed method for clustering and model detection as EMA-CF.
Eight state-of-the-art model-based clustering methods were chosen for comparison. They are closely related to the proposed one and summarized in
Table 1. To the best of our knowledge, except the proposed method, there are few approaches to performing clustering, feature selection, and covariance structure detection simultaneously. The MBIC method suggested in [
49] uses the structural EM algorithm combined with the BIC to realize simultaneous feature selection and clustering in Gaussian mixture models. The MICL method proposed in [
21] further tackles parameter uncertainty by considering the ICL, where an alternating optimization algorithm has been designed to find the data separation and distinctive features. Both MBIC and MICL are established on the local independence assumption. They were implemented using the R package “VarSelLCM”. The mcgStep method [
15] directly seeks the sparse pattern of the component covariance matrices using the structural EM algorithm. Stepwise searching is required in each iteration to refine the component covariance graphs. The mcgStep method was implemented using the R package “mixggm” with the BIC model selection criterion. The mclust approach [
50] in the R package “mclust” is based on the Gaussian parsimonious clustering model (GPCM) family, which is composed of 14 component covariance matrix structures under different levels of constraints after eigenvalue decomposition. mclust-BIC conducts model selection based on the BIC value after the fitting of each model. mclust-RMA and mclust-PMA are approximated BMA methods proposed in [
27] based on the 14 model structures. While mclust-RMA performs averaging on the response matrices, mclust-PMA takes averaging on the estimated model parameters. When conducting model averaging, the model weights are approximated by normalizing the BIC values. In addition, we included the EMA algorithm in comparison, which takes BMA over only the naïve Bayes structures to investigate the effects of accounting for the within-class associations with the CF architecture. We denote the method as EMA-naïve. (In the
Supplementary Materials, by viewing the EMA algorithm as an approximated CVB approach, we provide comparison results with two additional related clustering methods VarFnMS [
51] and VarFnMST [
3], which are both VB methods based on the naïve Bayes assumption.)
The MBIC, MICL and mcgStep algorithms require initialization of the cluster allocations as well as the model structure. In the R package “VarSelLCM” for MBIC and MICL, each feature is initialized as discriminant or not discriminant via random sampling. The initial class allocations are then provided by the EM algorithm associated with the initial feature discrimination pattern. We set the number of random initializations in the two algorithms as 100. The output was the result with the maximum BIC value for MBIC and maximum ICL for MICL across the 100 initializations. In the packages “mixggm” and “mclust”, the Gaussian model-based hierarchical clustering approach in [
52] is used to initialize the class allocations. Initialization of the component covariance graphs for mcgStep is provided by filtering the sample correlation matrix in each class. The EMA-CF and EMA-naïve algorithms only need initialization of the cluster allocations. Like the competitive algorithms, a poor choice of the starting values may make the convergence very slow and obtain a local optimal. Thus, the choice of a good starting class allocation for EMA-CF or MEA-naïve is important. Or, multiple starting points should be tried as in MBIC and MICL. For overall efficiency of the algorithm, we chose to use the result from the
k-means clustering algorithm. To implement the EMA-CF algorithm, the number of maximum allowed times of iterations
in the EMA algorithm was fixed at 100 when growing a clustering vector and set as 500 for the final EMA algorithm with the obtained clustering vector. We set the number of paralleled cluster instances as 100. The maximal allowed times of tree growth
and the maximal allowed times of consecutive unsuccessful attempts
were both set as 5. Further explorations on the CF controllable parameters are given in the
Supplementary Materials. The values specified for these parameters are adequate to ensure a stable and good performance of the algorithm in the following simulation settings.
We considered the synthetic data from a bi-component Gaussian mixture model with mixing proportions
, composed of ten features. The component mean of class one was
, where
were generated randomly from the uniform distribution
. The mean of class two was given by
. The value
permits us to tune the class overlaps. Six scenarios differentiated by the covariance structures were considered. In Scenarios 1–4, the covariance structures were well-specified with two, three, five, and ten feature blocks from the LAN family. Scenario 4 corresponds to the naïve Bayes configuration. We also took into account the scenarios where the covariance structures were mis-specified as the Toeplitz type matrix and the Erdos–Renyi model [
15] in Scenarios 5 and 6. The probability of two features being marginally correlated was set as 0.2 in the Erdos–Renyi specification. We used the same covariance matrix for different mixture components in each scenario. Without loss of generosity, the diagonal elements of each covariance matrix were set as one. The non-zero off-diagonal elements were randomly chosen from the uniform distribution
submitting to the symmetric and positive definite constraint.
Figure 2 exhibits the simulated covariance graphs and the corresponding Gaussian graphical models in the six scenarios. For each scenario, we generated random datasets with different combinations of sample size
and class overlap
, and we replicated each experiment twenty times.
As the true class assignment of the synthetic dataset was known, we computed the classification error rate to evaluate the quality of the clustering obtained by the competitive algorithms. The results averaged over the twenty replicates are reported in
Table 2. The first and the second smallest classification error rates in each case are marked in bold.
Table S3 provides the results of the standard deviation of the classification error rates. In general, the EMA-CF method outperforms the others and shows robustness across various simulation settings. The performance gain is substantial when the within-component correlation is strong, such as in Scenarios 1 and 2. It is noticeable that the mcgStep method also obtains relatively small classification errors in these scenarios, which emphasizes the importance of modeling the component covariance structures when presumptively high association relationships are present in the data. For small class overlap
, all the methods tend to attain an almost perfect classification of the data. The EMA-CF method improves the data separation dramatically when the class overlap is increased. While EMA-CF improves the performance of EMA-naïve in Scenarios 1 and 2 when the within-component correlation is strong, the two methods show an overall competitive performance in the remaining four scenarios. Moreover, the EMA-naïve method outperforms MBIC and MICL when the class overlap is high. While the three methods all assume the naïve Bayes network structure, EMA-naïve considers the model uncertainty by employing the BMA.
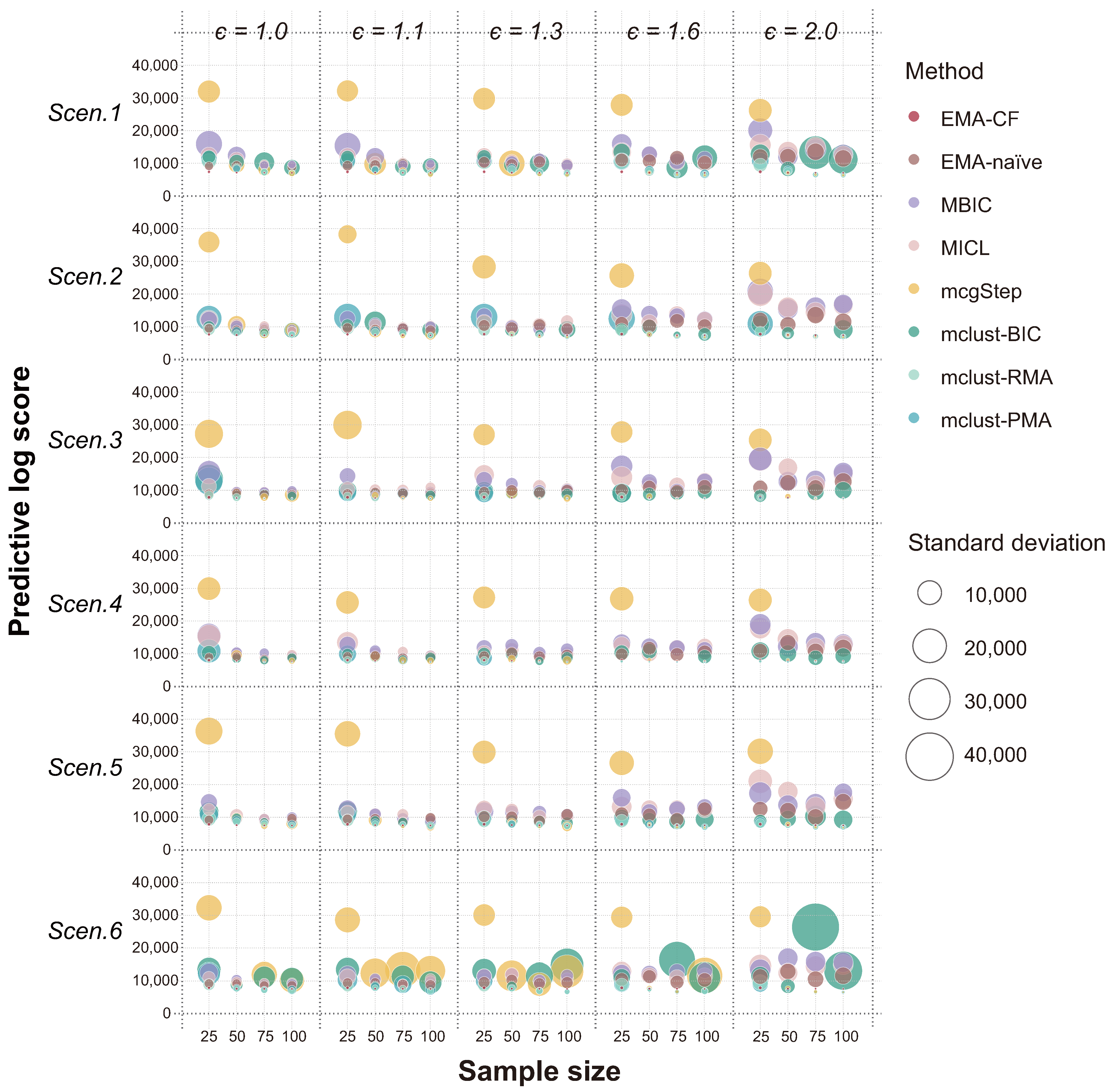
The predictive log score (PLS) [
25] is defined by
where
is the set of testing data and
denotes the predictive probability on datum
given in the target method. Following the logarithmic scoring rule of Good [
23], a better modeling strategy should consistently assign higher probabilities to the events that actually occur. Therefore, the smaller the PLS, the more reliable the method. The results of PLS obtained using the eight methods in the six scenarios are compared in the bubble plot shown in
Figure 3. The radius of each circle indicates the variation of the PLS values over the twenty replicates. The EMA-CF method shows remarkably high predicting performance across different covariance structure configurations. And, the results are robust across various simulation settings. Particularly, using EMA-CF reduces the PLS of EMA-naïve in each case, which suggests the significance of modeling the within-component correlations.
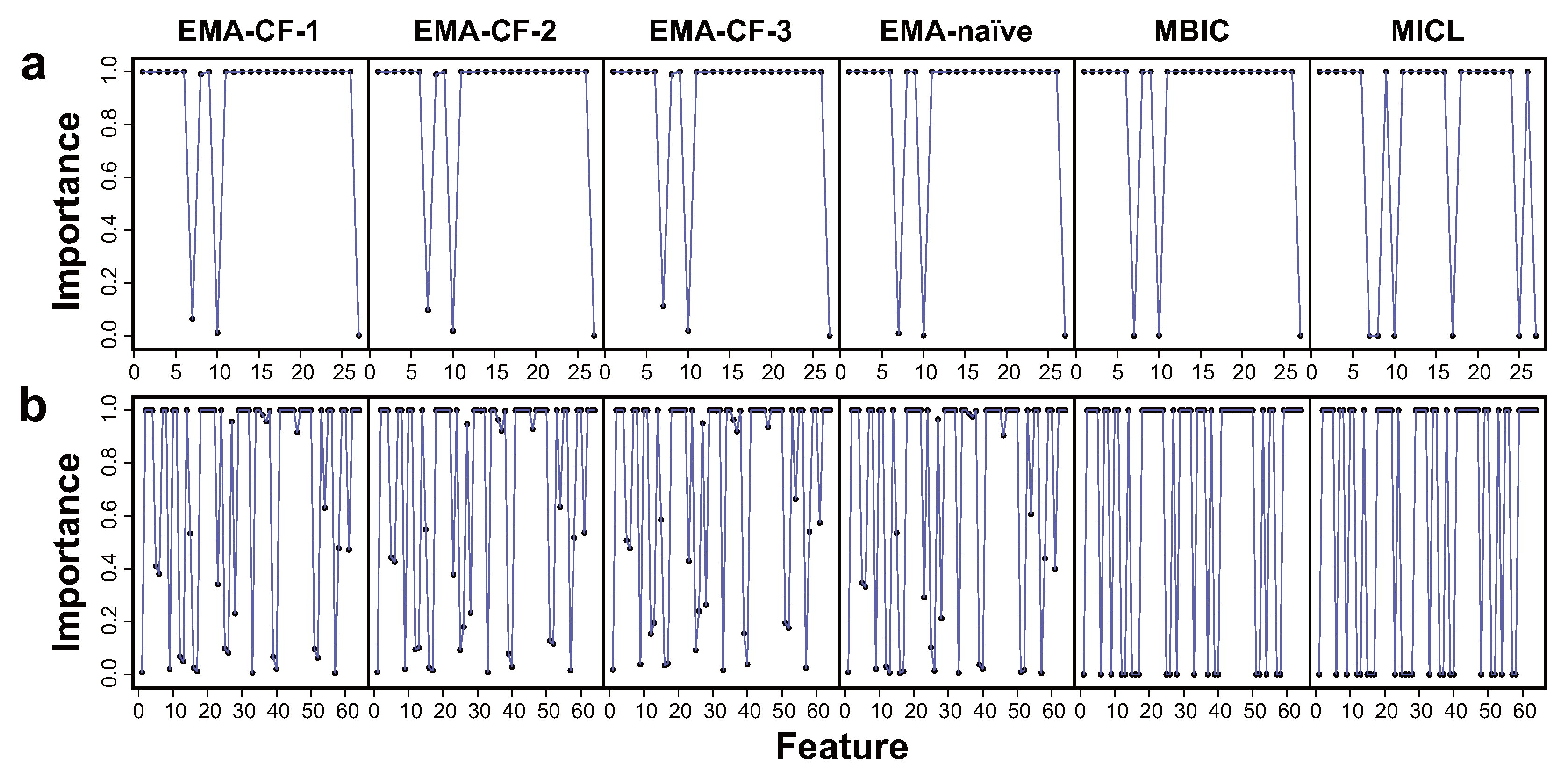
The covariance structure detection ability was compared between EMA-CF, mcgStep, and mclust-BIC, and the ability to identify the feature importance pattern was compared between EMA-CF, EMA-naïve, MBIC, and MICL.
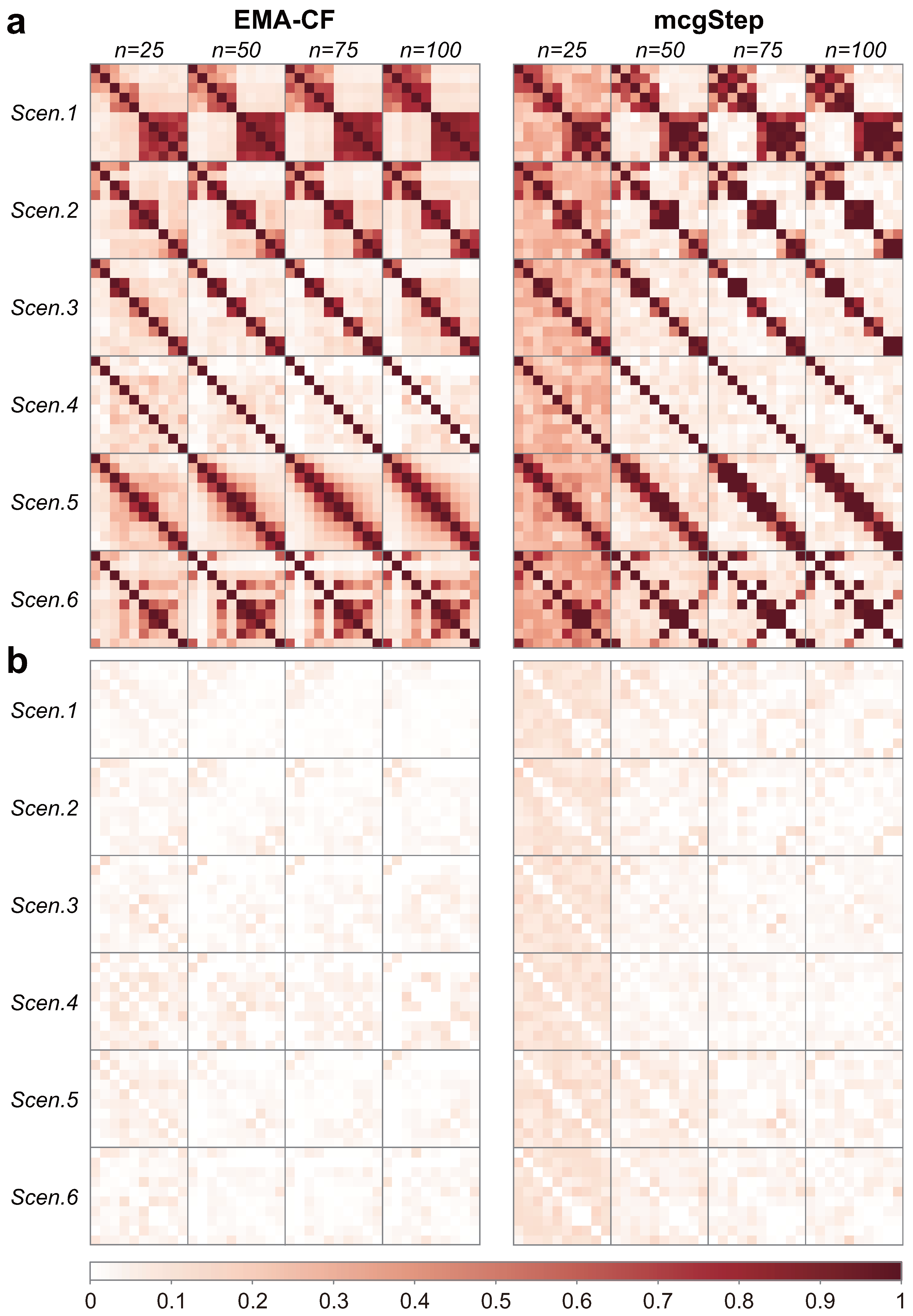
Figure 4 shows the covariance structures detected using the EMA-CF and mcgStep methods. As the results are not sensitive to the class overlap, only the cases with
are illustrated here. The remaining results are present in the
Supplementary Materials. While the mcgStep method gives an estimation of the covariance structure as the covariance graph, EMA-CF provides an estimation as the Gaussian graphical model. Both methods show good performance in detecting the underlying graph configurations. The EMA-CF method exhibits more robustness, especially when the sample size is small. The covariance structures detected most frequently over the twenty replicates by the mclust-BIC method are shown in
Table 3. The detection frequency is present within the parentheses. The EII structure indicates that the component covariance matrices are diagonal and homogeneous. EEE indicates a homogeneous full covariance structure for the components.
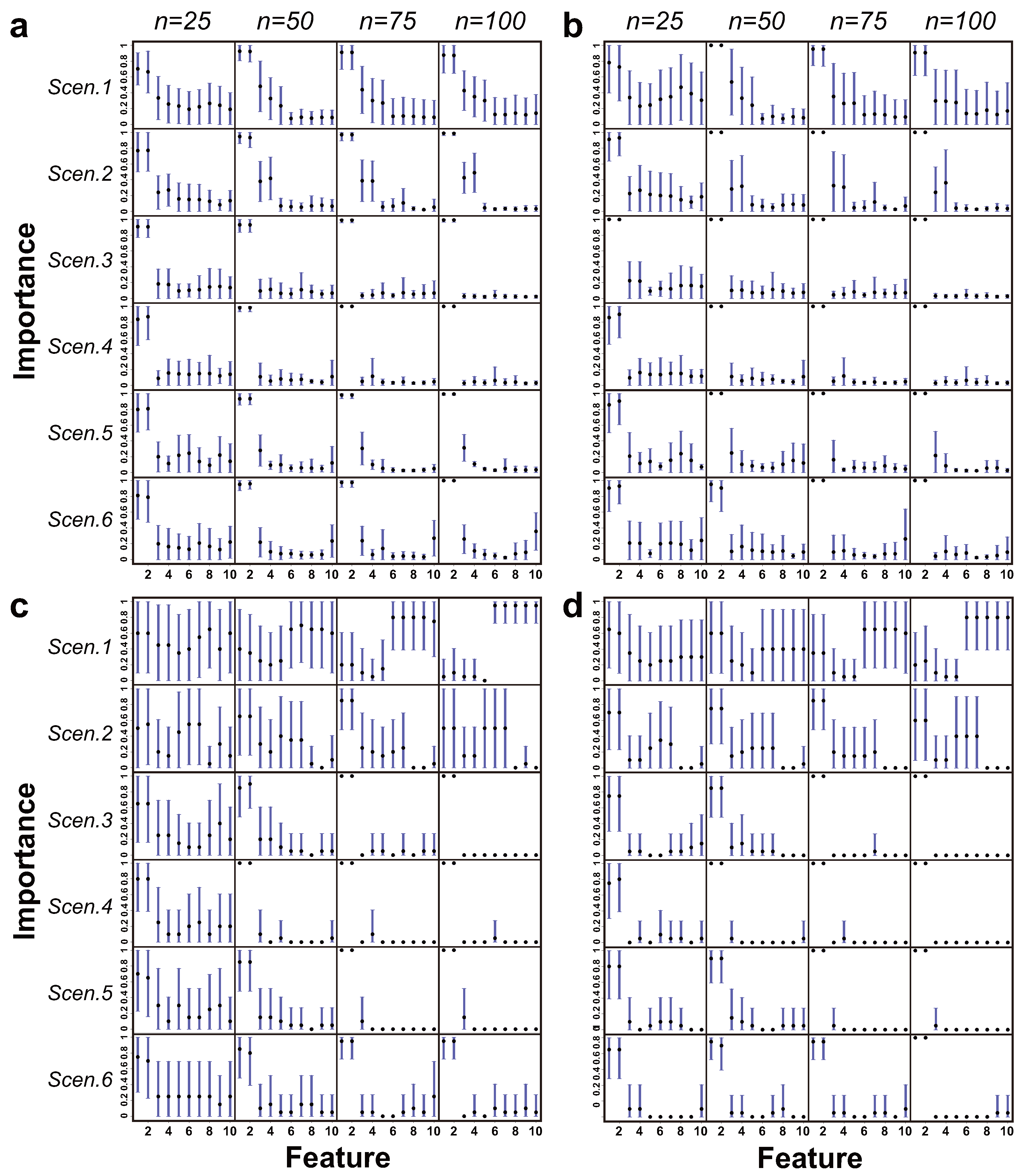
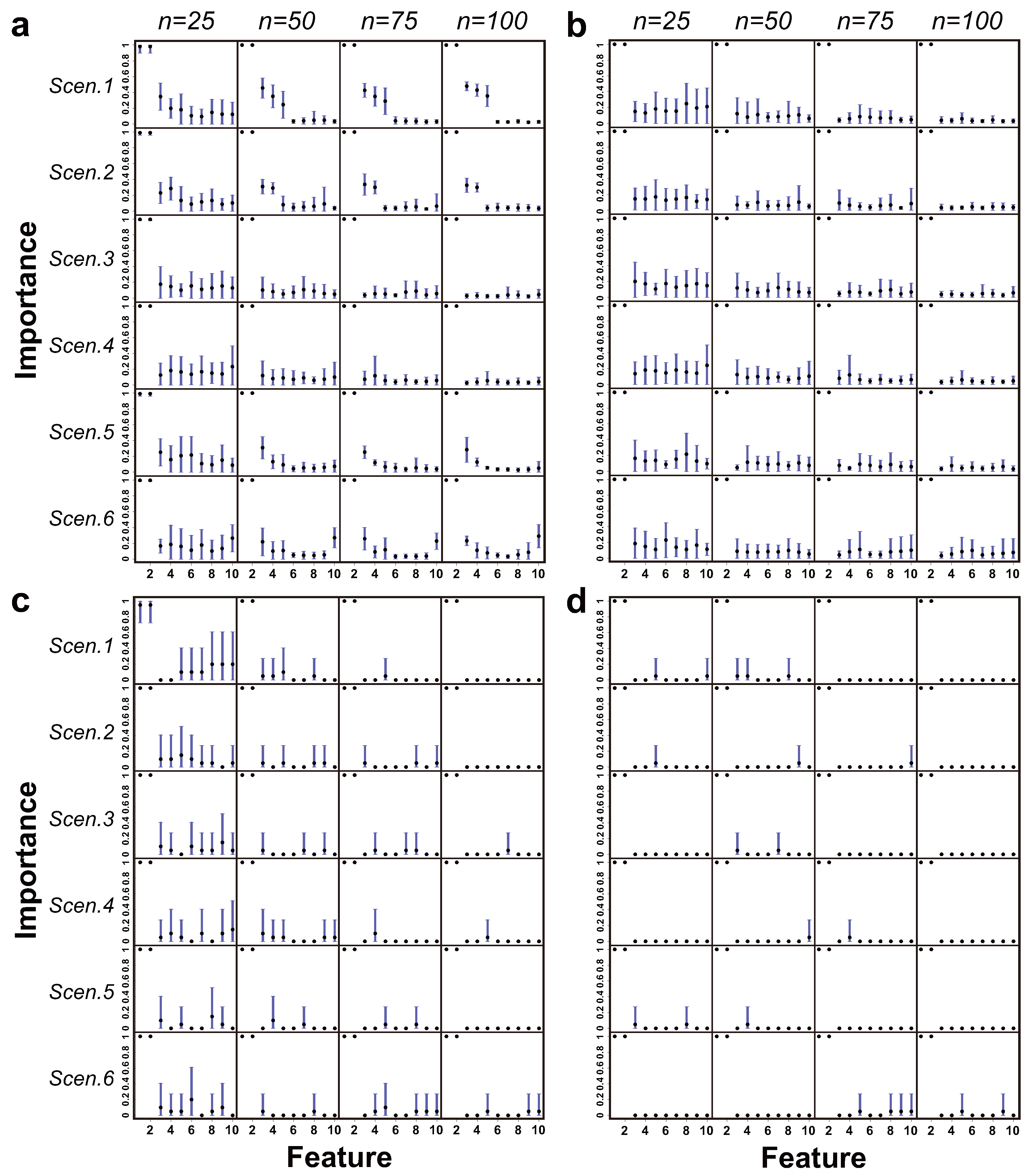
Figure 5,
Figure 6 and
Figure 7 show the patterns of feature importance estimated by the EMA-CF, EMA-naïve, MBIC, and MICL methods under class overlap settings of
, respectively. The cases for
are present in the
Supplementary Materials. Overall, the results of EMA-CF show great robustness, responsive to the corresponding association structures between features. It is noticeable that the estimated patterns of feature importance for Scenarios 1 and 2 by EMA-CF are distinguished from those estimated by EMA-naïve, MBIC, and MICL. The difference becomes evident as the class overlap decreases and the sample size increases. Indeed, the associations of the last eight features with the first two induce their indirect contributions to the classification. The MBIC and MICL methods perform erratically in Scenarios 1 and 2 when the class overlap is high. In Scenarios 3 and 4, where there is no conditional association between the first two and the remaining eight features, the four methods give similar identification results. In Scenarios 5 and 6, where the structures assumed are not from the LAN family, EMA-CF detects some latent patterns of the feature’s significance induced by the simulated association structures. Overall, EMA-CF and EMA-naïve, which are based on the BMA, show consistency behaviors as the sample size increases. Such behaviors are conformable to the principle of BMA, where the probability mass function of model structures (model weights) peaks gradually at the MAP structure as the size of the dataset increases [
31].
6.2. Experiments on Real-World Datasets
In this section, we illustrate the proposed method by applying it to some benchmark datasets: Iris, Olive, Wine, and Digit. The Iris dataset was obtained from the R package “datasets”. Olive is the Italian olive oil dataset and Wine the Italian wine dataset. They were both obtained from the R package “pgmm”. The Digit dataset was obtained from the UCI machine learning repository (
https://doi.org/10.24432/C50P49; accessed on 26 August 2025). The complete data of Digit contain more than 5000 images for the handwritten digits 0–9. They are gray-scale with a size of
pixels. We focused on separation of the 4 and 9 digits and randomly reserved 100 images for each digit. As the variability of some pixels for a digit was exactly zero, singularity problems could occur in the model-based clustering algorithms. Therefore, we put a noise mask on the data matrix (of size
). Each element of the noise mask was generated from
.
Table 4 presents the basic information for the four datasets.
To implement the EMA-CF method, as we had little information about the covariance structure of the data, we varied the setting of that controls the tree growth in the CF architecture between , and (rounded to the nearest integer) to match with different levels of sparsity. We denote the corresponding EMA-CF algorithms as EMA-CF-1, EMA-CF-2, and EMA-CF-3, respectively. The number of cluster instances was fixed at , and let . We kept the settings of the other algorithms the same as those in the simulation study.
The clustering quality was evaluated by comparing it with the original grouping of the data.
Table 5 shows the classification errors obtained by the eight competing methods. In general, the EMA-CF method gives the lowest classification errors for all the four datasets. For the Wine and Digit datasets, the best results can be achieved when
is small, and the EMA-naïve method that does not model the covariance structure performs equally well. Larger
values improve data separation for the Iris and Olive datasets. The MBIC and MICL methods based on the conditional independence assumption provide the worst results for Iris and Olive, but they show sound performance for Wine and Digit. The mcgStep method provides relatively good classification for Iris and Olive but is undesirable for Wine and Digit.
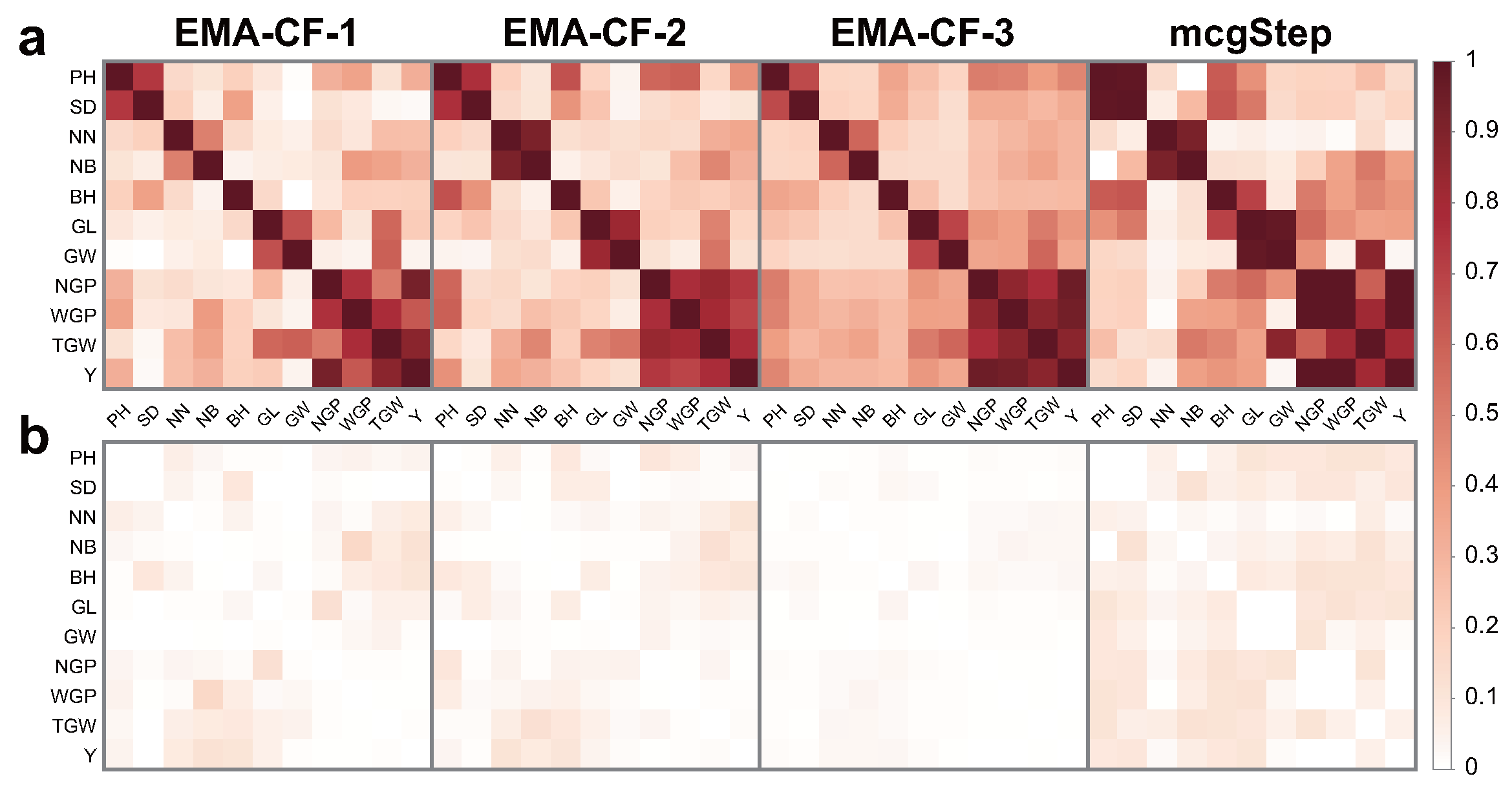
Figure 8 shows the covariance structures detected by EMA-CF and mcgStep. The two methods both advocate strong within-class correlations in Iris and Olive. Even with a small
value, the overall color of the normalized occurrence matrix given by EMA-CF is remarkably deep. In contrast, the associations in Wine and Digit are much sparser. The covariance structures given by the mclust-BIC method are VEV, EVV, EVI, and EEE for Iris, Olive, Wine, and Digit, respectively. While EVI indicates diagonal component covariance matrices, VEV, EVV, and EEE produce full covariance matrices. The combined results with the classification performance indicate that the EMA-CF method can accommodate various kinds of covariance structures and give more reliable clustering results.
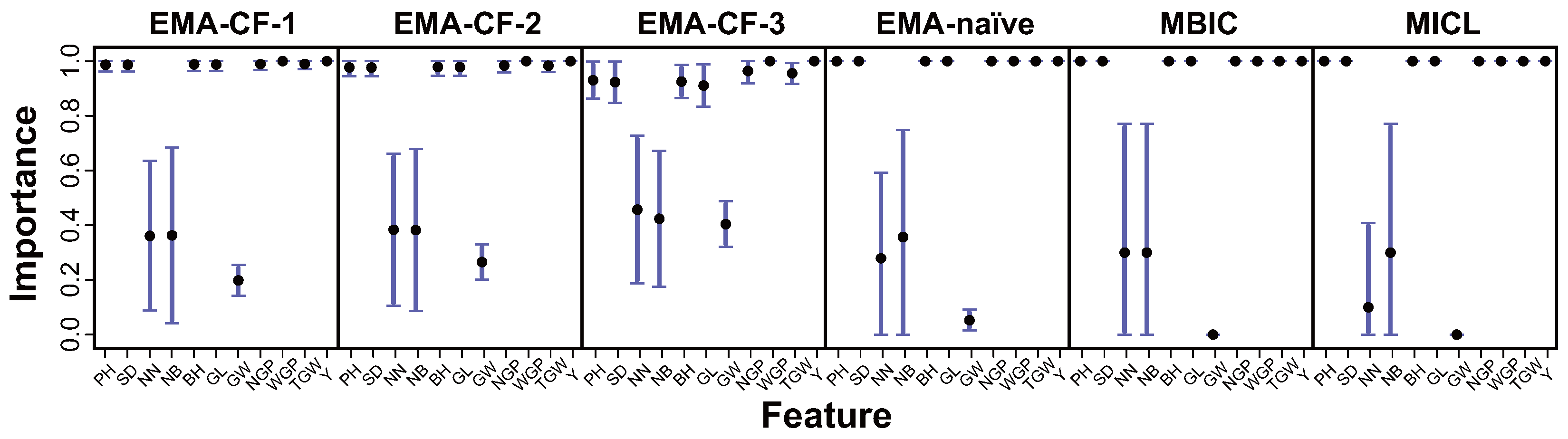
Implementation of EMA-CF, EMA-naïve, MBIC, and MICL gives the estimated patterns of feature importance. For Iris and Olive, the four methods have the same identification, where all the features are significant at the highest level (see
Figure S11 in the Supplementary Materials). The results for Wine and Digit are compared in
Figure 9. While the MBIC and MICL methods separate the features as discriminating and undiscriminating, EMA-CF and EMA-naïve give relatively conservative estimations of feature importance by taking into account model uncertainty with BMA. There are no apparent differences between the results of EMA-CF andEMA-naïve, which agrees with the sparse patterns of feature association in the two datasets, as detected in
Figure 8.
6.3. Application on Tartary Buckwheat Data
In this section, we present an application of the developed method in a real agriculture problem. The data reflect a traditional edible and medicinal crop, Tartary buckwheat. The experimental data contain a total of 200 Tartary buckwheat landraces growing in two different locations with distinct climate conditions [
53], denoted as E1 and E2, respectively. Eleven phenotypic traits of the Tartary buckwheat plant were investigated:
plant morphological traits
plant height (PH), stem diameter (SD), number of nodes (NN), number of branches (NB), and branch height (BH)
grain-related traits
grain length (GL) and grain width (GW)
yield-related traits
number of grains per plant (NGP), weight of grains per plant (WGP), 1000-grain weight (TGW), and yield per hectare (Y)
It is commonly acknowledged that changing environment has non-negligible impacts on the growth and development of crops. Our study is concerned with the influence of environmental changes on the Tartary buckwheat landraces. We mixed the phenotypic data of 100 randomly selected landraces from environments E1 and E2. The experiment was repeated twenty times. Again, in using the EMA-CF method, we set as , , and . In each of the eight competing methods, a bi-component clustering model was constructed. Therefore, major impacts of the environmental changes could be confirmed by separation of the data according to their original environments.
Table 6 shows the classification errors and the PLS values obtained using the eight methods. The class assignment given by EMA-CF with
shows the highest consistency with the original grouping of the data divided by the environments. Moreover, EMA-CF shows the highest predictive ability compared with the competitive methods.
The covariance structures detected by EMA-CF and mcgStep show strong evidence of the within-class correlations between the phenotypic traits of Tartary buckwheat. As shown in
Figure 10, there are heavy associations between the yield-related traits. Moreover, the conditional correlations are evident between PH and SD, between NN and NB, and between GL and GW, as assessed by the EMA-CF method. The phenotype TGW is found to be related to GL and GW, which agrees with the findings in a previous study [
54]. The covariance structures provided by mclust-BIC over the twenty replicates are EVE (10 times) and VVE (10 times), both of which indicate full component covariance matrices.
From
Figure 11, we can find that all the yield-related traits are discriminant between environments E1 and E2. In addition, the phenotypes PH, SD, BH, and GL are also identified as significant. These facts suggest the sensitivity of the Tartary buckwheat landraces to the environmental changes. While the significance of GW is gradually identified by EMA-CF as
increases, it is regarded as unimportant by the EMA-naïve, MBIC, or MICL methods, which have ignored the associations between GW and the yield-related traits.
While most Tartary buckwheat landraces can be separated according to their growing environments (E1 and E2), there are a small group of landraces that are easy to misclassify.
Table 7 summaries those landraces that have data misclassified into the same class ten times or more across the twenty replicates by the EMA-CF-3 algorithm. Among them, SC-8 and GZ-32 exhibit high yields in both environments, which could provide potentially excellent varieties for further investigation.