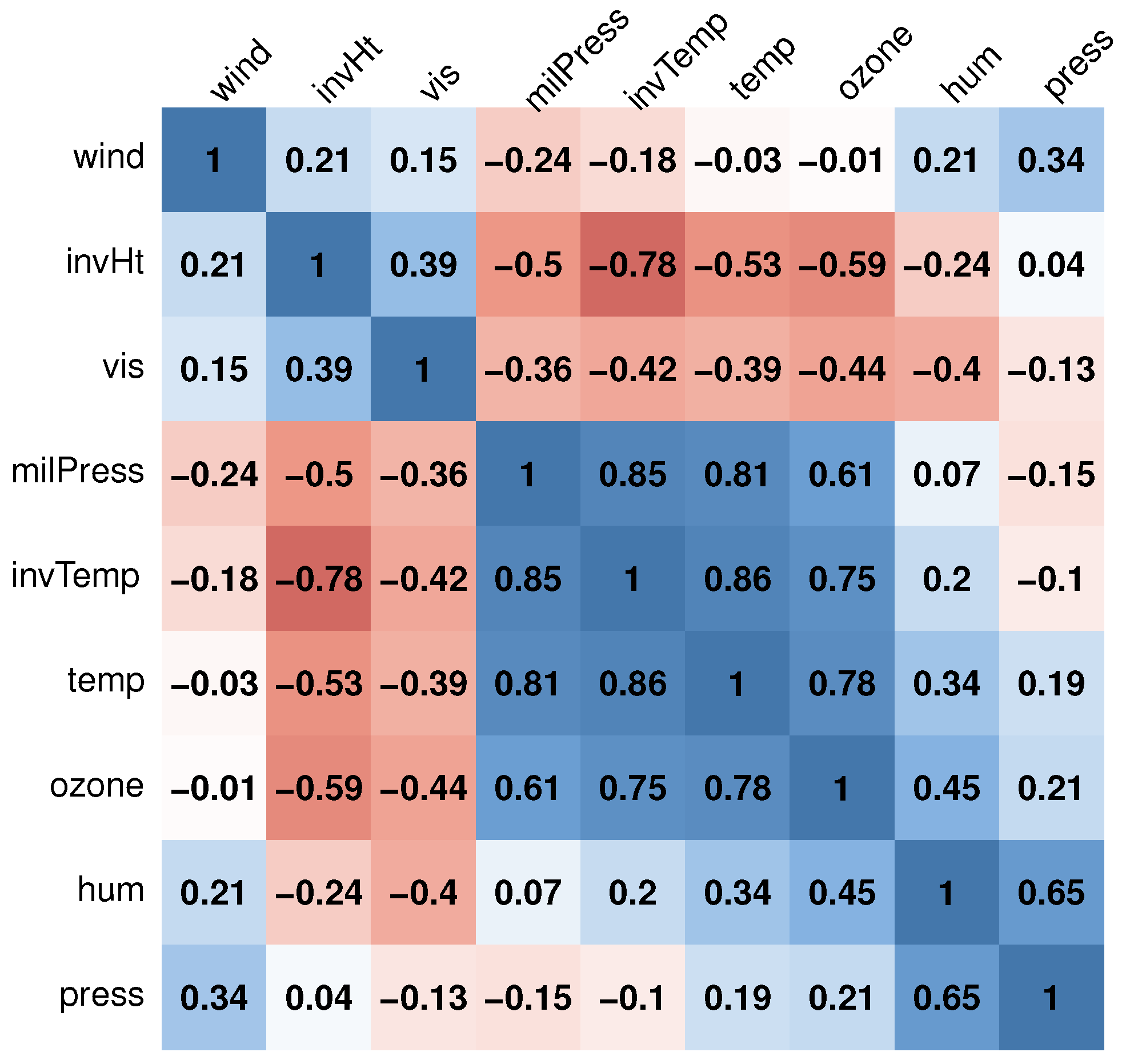
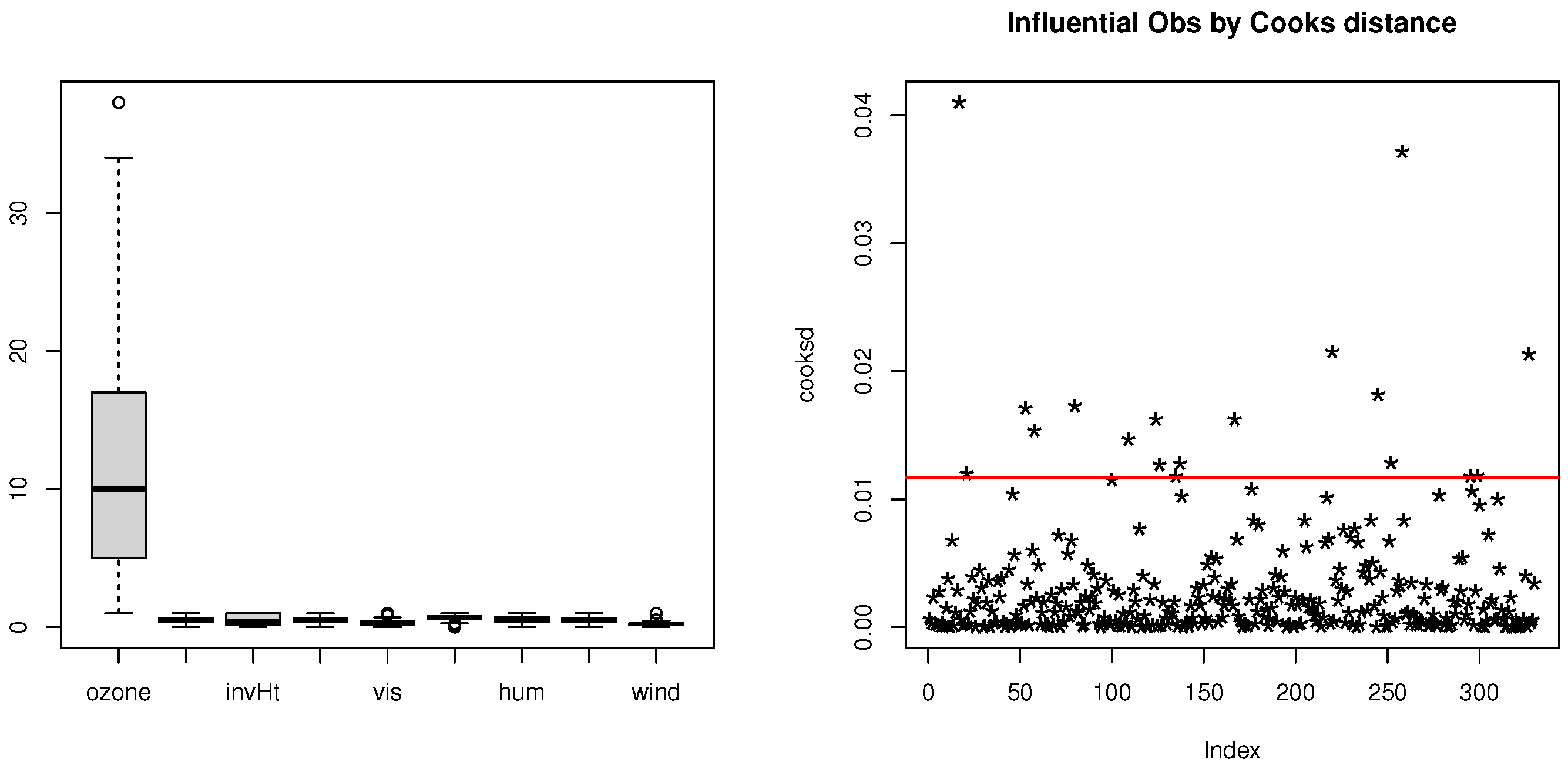
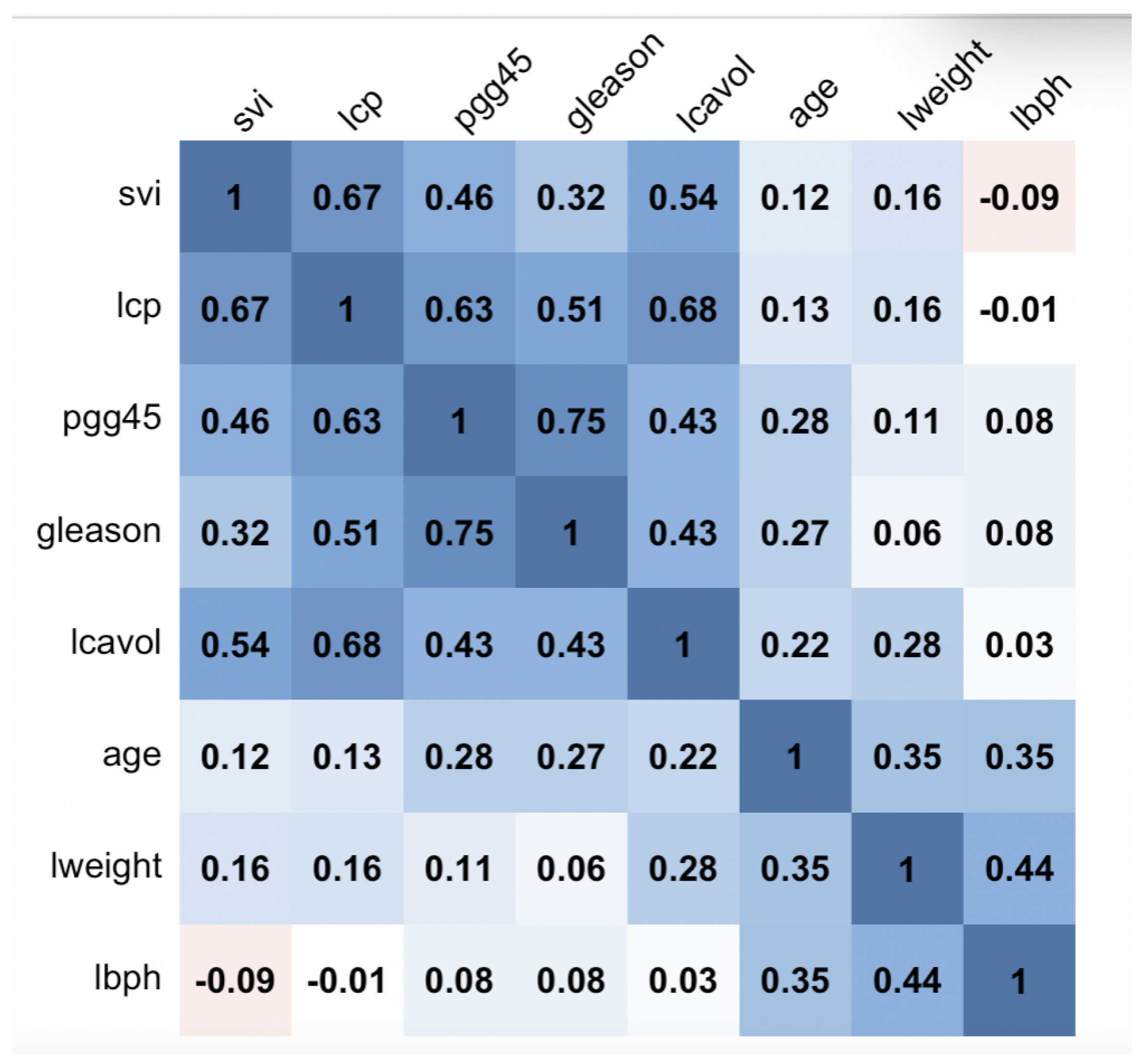
1. Introduction
The usual estimators are inapplicable in cases where the matrix is singular. In practice, when estimating () or pursuing variable selection, high empirical correlations between two or a few other covariates (multicollinarity) lead to unstable outcomes. This study uses the least absolute shrinkage and selection operator (Lasso) estimation method in order to circumvent this issue.
Lasso estimation is a type of variable selection. It was first described by Tibshirani [
1] and was further studied by Fan and Li [
2]. They explored a class of penalized likelihood approaches to address these types of problems, including the Lasso problem.
In contrast, when outliers are present in the sample, classical least squares and maximum likelihood estimation methods often fail to produce reliable results. In such situations, there is a need for an estimation method that can effectively handle multicollinearity and is robust in the presence of outliers.
Zou [
3] introduced the concept of assigning adaptive weights to penalize coefficients with different degrees of skewness. Due to the convex nature of this penalty, it typically leads to convex optimization problems, ensuring that the estimators do not suffer from local minima issues. These adaptive weights in the penalty term allow for oracle properties.
To create a robust Lasso estimator, the authors of [
4] proposed combining the least absolute deviation (LAD) loss with an adaptive Lasso penalty (LAD-Lasso). This approach results in an estimator that is robust against outliers and proficient at variable selection. Nevertheless, it is important to note that the LAD loss is not designed for handling small errors; it penalizes small residuals severely. Consequently, this estimator may be less accurate than the classic Lasso when the error distribution lacks heavy tails or outliers.
In a different approach, Lambert-Lacroix, S. and Zwald, L. [
5] introduced a novel estimator by combining Huber’s criterion with an adaptive Lasso penalty. This estimator demonstrates resilience to heavy-tailed errors and outliers in the response variable.
Additionally, ref. [
6] proposed the Sparse-LTS estimator, which is a least-trimmed-squares estimator with an
penalty. The study by Alfons demonstrates that the Sparse-LTS estimator exhibits robustness to contamination in both response and predictor variables.
Furthermore, ref. [
7] combined MM-estimators with an adaptive
penalty, yielding lower bounds on the breakdown points of MM-Lasso and adaptive MM-Lasso estimators.
Recently, ref. [
8] introduced c-lasso, a Python tool, while [
9] proposed robust multivariate Lasso regression with covariance estimation.
While recent years have seen a significant focus on outlier detection using direct approaches, a substantial portion of this research has centered on the utilization of single-case diagnostics (as seen in [
10,
11,
12,
13]). For high-dimensional data, ref. [
12] introduced the identification of numerous influential observations within linear regression models.
This article introduces a novel Lasso estimator named D-Lasso. D-Lasso is grounded in diagnostic techniques and involves the creation of a clean subset of data, free from outliers, before calculating Lasso estimates for the clean samples. We anticipate that these modified Lasso estimates will exhibit greater robustness against the presence of outliers. Moreover, they are expected to yield minimized sums of squares of residuals and possess a breakdown point of 50%. This is achieved through the elimination of outlier influence, as well as addressing multicollinearity and variable selection via Lasso regression.
The paper’s structure is as follows:
Section 2 provides a review of both classical and robust Lasso-type techniques.
Section 3 introduces the diagnostic-Lasso estimator. Regression diagnostic measures are presented and discussed in
Section 4.
Section 5 offers a comparison of the proposed method’s performance against existing approaches, while
Section 6 presents an analysis of the Los Angeles ozone data as an illustrative example. Finally, a concluding remark is presented in
Section 7.
5. Simulation Study
In this part, a simulation study that compared the proposed D-Lasso estimator’s performance to those of some other Lasso estimators is described. There are six estimators in the study: (i) D-Lasso, (ii) MM-Lasso, (iii) Sparse-LTS, (iv) Huber-LTS, (v) LAD-Lasso, and (vi) classical Lasso. Consider three different simulation scenarios:
Simulation 1: In the first simulation, multiple linear regression is taken into account with a sample size of 50 (n = 50) and 25 variables (p = 25), where each variable is selected from a joint Gaussian marginal distribution with a correlation structure of = 0.5.
The true regression parameters
are set to be
. The distribution of random errors
e is generated from the following contamination model:
where
is the contamination ratio,
is the signal to noise, which is chosen to be 3,
is the standard normal distribution, and
H is the Cauchy distribution to create a heavy-tailed distribution. The response variables are then calculated as follows:
The percentage of zero coefficients (Z.coef) equals 80%, and the percentage of non-zero coefficients (N.Z coef.) equals 20%.
Simulation 2: The second simulation process is similar to the first, except that the
p and
n values are different (
p = 50,
n = 150), and the response variables are calculated as follows:
The percentage of true zero coefficients (Z.coef) equals 90% and the percentage of true non-zero coefficients (N.Z.coef) equals 10%.
Simulation 3: The third simulation is similar to the second, but
n is increased to 500 and
=
The response variables are then calculated as follows:
The percentage of true zero coefficients (Z.coef) equals 90% and the percentage of true non-zero coefficients (N.Z.coef) equals 10%.
The following data are looked at to see how well the approaches stand up to outliers and leverage points: (a) uncontaminated data; (b) vertical contamination (outliers on the response variables); (c) bad leverage points (outliers on the covariates).
The response variables and covariates are contaminated by certain ratios (
= 0.05, 0.10, 0.15, and 0.20) of vertical and high leverage points; these are created by randomly replacing some original observations with large values equal to 15 [
24].
The simulations were performed in statistical software
R. D-Lasso, MM-Lasso, Sparse-LTS, Huber-Lasso, LAD-Lasso, and classical Lasso. Using the
measure suggested by [
12], D-Lasso was assessed in order to determine which observations were influential, and
was chosen as described in
Section 4.3. For MM-Lasso we used the functions available in the github repository
https://github.com/esmucler/mmlasso (accesses on 25 October 2017) [
7].
The estimator was calculated using the sparseLTS() function from the robustHD package in R, and
was chosen using a
criterion as advocated by references [
6]. Huber-Lasso used the package
and the LAD-Lasso estimator was calculated using the package
. The Lasso estimator was calculated using the lars() function from the lars package ([
25]), where
was chosen based on 5-fold cross-validation.
, and
,
were chosen by applying the classical
.
In each simulation run, there were 1000 replications. Four criteria are considered to evaluate the performances of the six methods, namely: (1) the percentage of zero coefficients (Z.coef), (2) the percentage of non-zero coefficients (N.Z.coef), (3) the average of mean squares of errors (), and (4) the median of the mean squares of errors Med(mse). A good method for Simulation 1 is the one that possesses the percentage of Z.coef closed to 80% and the percentage of N.Z.coef closed to 20%. However, a good method for Simulations 2 and 3 is the one that possesses the percentage of Z.coef and N.Z.coef reasonably close to 90% and 10%, respectively, with a good method having the least () and Med(mse) values.
The results clearly show the merit of D-Lasso. It can be observed from
Table 1,
Table 2 and
Table 3 that the D-Lasso has the smallest values of
and Med(mse) compared to the other methods.
In the case of no contamination,
Table 1 shows that both classical and D-Lasso methods perform well in model selection ability. For example, in the scenario of Simulation 1, the classical Lasso successfully selected 80% of Z.coef and 20% N.Z.coef, followed by D-Lasso, which selected 77.6% and 22.4% for Z.coef and N.Z.coef, respectively.
However, the performance of other robust lasso methods is good, but it has a larger and Med(mse) than the other two methods. Furthermore, none of the methods suffer from false selection variables.
In the case of vertical outliers and leverage points, the classical Lasso is clearly influenced by the outliers, as reflected in the much higher and Med(mse). Furthermore, it tended to select more variables in the final model (overfitting) when the percentage of contamination increased to 20%.
On the other hand, in the case of vertical outliers, the robust Lasso methods (MM-Lasso, Sparse, Huber-Lasso, and LAD-Lasso) clearly maintain their excellent behavior. Sparse-LTS has a considerable tendency toward false selection when the percentage of contamination increases to 20%.
Table 2 shows that the robust Lasso methods (Sparse, Huber-Lasso, and LAD-Lasso) were affected by the presence of leverage points in the data. The effect was worse with a higher percentage of bad leverage points in the data.
The results of D-Lasso and MM-Lasso are consistent for all percentages of contamination, but MM-Lasso has a larger than D-Lasso, which indicates that the performance of D-Lasso is more efficient than the other methods. For further illustrative purposes, the forthcoming section analyzes some real data sets.
7. Conclusions
The classical Lasso technique is often utilized for creating regression models, but it can be influenced by the presence of vertical and high leverage points, leading to potentially misleading results. A robust version of the Lasso estimator is commonly derived by replacing the ordinary squared residuals () function with a robust alternative.
This article aims to introduce robust Lasso methods that utilize regression diagnostic tools to detect suspected outliers and high leverage points. Subsequently, the D-Lasso is computed following diagnostic checks.
To assess the effectiveness of our newly proposed approaches, we conducted comparisons with the classical Lasso and existing robust Lasso methods based on LAD, Huber, Sparse-LTS, and MM estimators using both simulations and real datasets.
In this article, D-Lasso regression serves as the primary variable selection technique. Future endeavors may delve into exploring the asymptotic theoretical aspects and establishing the oracle properties of D-Lasso.