A Novel Autoencoder-Based Feature Selection Method for Drug-Target Interaction Prediction with Human-Interpretable Feature Weights
Abstract
1. Introduction
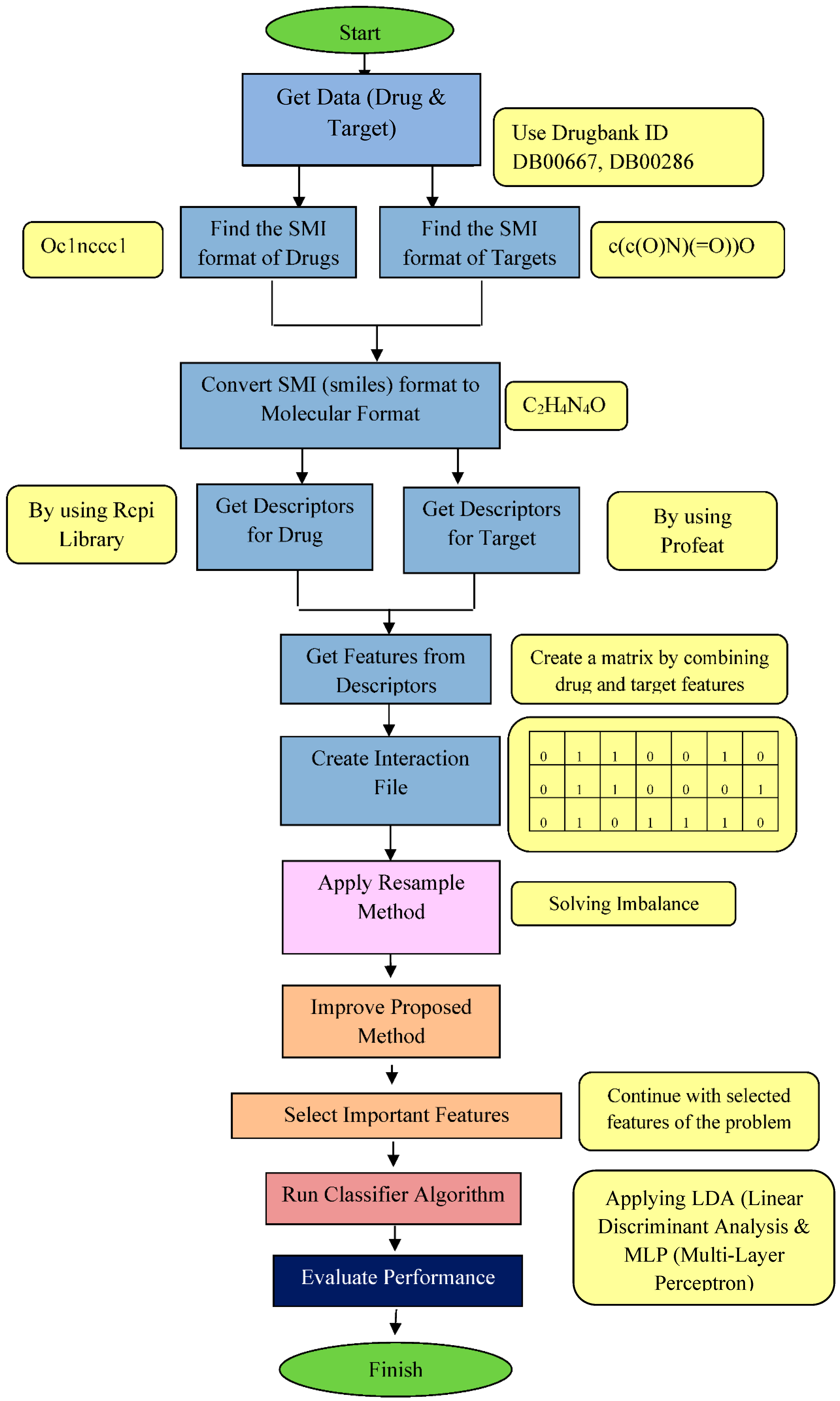
2. The Proposed Method
- First, we explain how we constructed drug-target feature vectors and how we extracted and transformed relevant data from the available databases.
- Then, we explain how we used these vectors in training an autoencoder that allows us to reduce the dimensionality of the feature vectors.
- Next, we train a classification network for predicting whether a given drug and a target are likely to interact.
- Lastly, we experiment with different network structures and discuss the outcomes of these experiments.
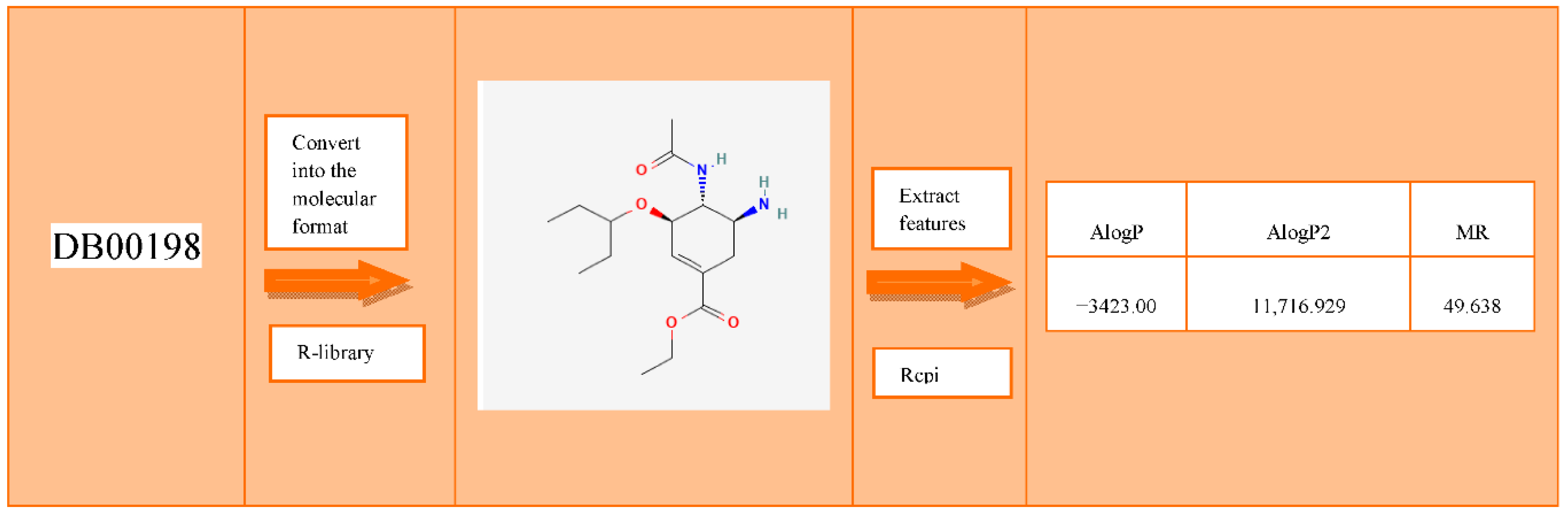
2.1. Construction of Drug-Target Interaction Vectors
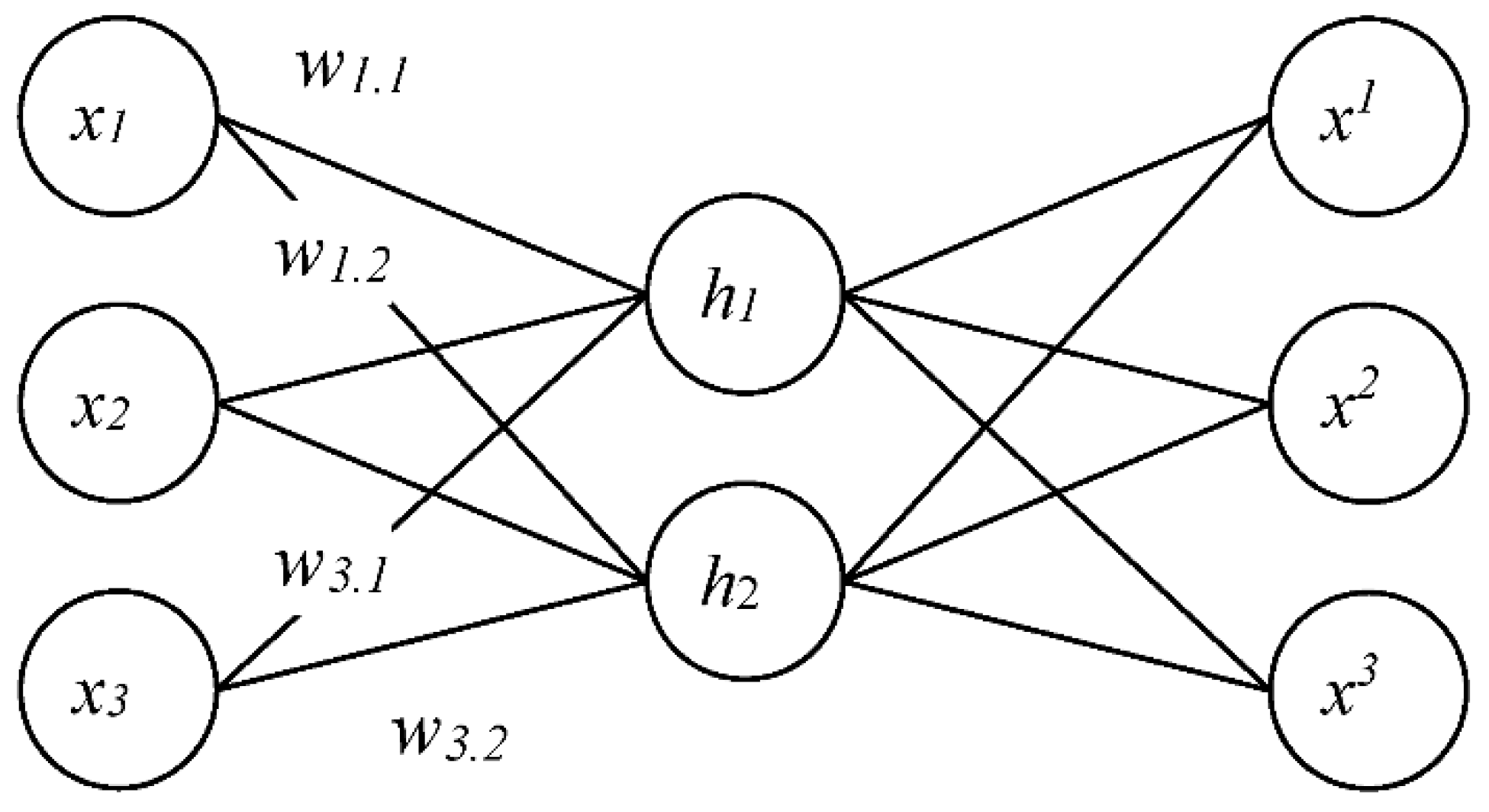
2.2. The Modified Autoencoder
2.3. Ablation Study for the Classifier
3. Performance Evaluation
3.1. The Performance Metric
3.2. Feature Selection Approaches
test = SelectKBest(score_func=chi2, k=n_feature)
fit = test.fit(X, y)
# summarize scores
np.set_printoptions(precision=3)
#print(fit.scores_)
features = fit.transform(X)
selector = VarianceThreshold()
fit=selector.fit(X,y)
features=fit.transform(X)
1,k=features.shape
n_feature=k
features_index = fit.get_support(indices=True)
clf = ExtraTreesClassifier(n_estimators=50)
clf = clf.fit(X, y)
clf.feature_importances_
model = SelectFromModel(clf, prefit=True)
features = model.transform(X)
l,n_feature=features.shape
features_index=model.get_support(indices=True)
4. Results and Discussion
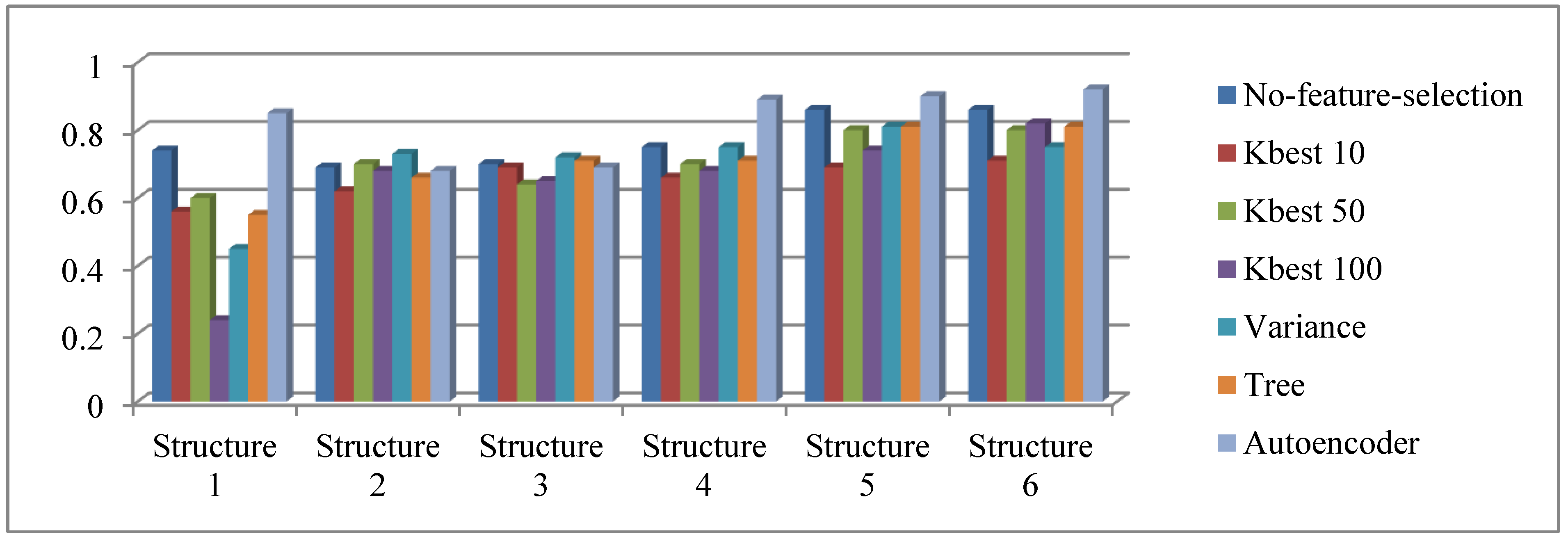
- All methods delivered consistently better results as the MLP structures become larger in width and depth. The proposed approach was no exception.
- When we used all 715 features without elimination, the delivered performance was competitive (second best on four out of six MLP structures). However, the proposed approach performed better with only 350 features, both in terms of accuracy and F-Scores.
- The proposed method delivers the best accuracy values with 350 features out of 715, and with larger sizes of MLP. We surmise that the capacity of the autoencoder was not sufficient for proper encoding on smaller networks.
- The input data is heavily imbalanced, with approximately less than 13,000 interacting cases vs. 27 million non-interacting cases. Although we used oversampling to alleviate this problem, the adverse effects of class imbalance persist, as indicated by very low F-Scores. In this regard, AE 360 delivered the best F-Scores (albeit still small, 0.1), outranking other methods by a large margin (about 0.003 on average).
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Live Science. Available online: https://www.livescience.com/37247-dna.html (accessed on 10 November 2021).
- Padmanabhan, S. Handbook of Pharmacogenomics and Stratified Medicines; Academic Press: Cambridge, MA, USA, 2014; Chapter 18. [Google Scholar]
- Sachdev, K.; Gupta, M.K. A comprehensive review of feature-based methods for drug target interaction prediction. J. Biomed. Inform. 2019, 93, 103159. [Google Scholar] [CrossRef]
- Chen, R.; Liu, X.; Jin, S.; Lin, J.; Liu, J. Machine Learning for Drug-Target Interaction Prediction. Molecules 2018, 23, 2208. [Google Scholar] [CrossRef]
- Lindsay, M.A. Target discovery. Nat. Rev. Drug Discov. 2003, 2, 831–838. [Google Scholar] [CrossRef]
- Yang, Y.; Adelstein, S.J.; Kassis, A.I. Target discovery from data mining approaches. Drug Discov. Today 2009, 14, 147–154. [Google Scholar] [CrossRef]
- Mousavian, Z.; Masoudi-Nejad, A. Drug–target interaction prediction via chemogenomic space: Learning-based methods. Expert Opin. Drug Metab. Toxicol. 2014, 10, 1273–1287. [Google Scholar] [CrossRef]
- Johnson, M.A.; Maggiora, G.M. Concepts and Applications of Molecular Similarity; Wiley: Hoboken, NJ, USA, 1991. [Google Scholar]
- He, H.; Garcia, E.A. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar] [CrossRef]
- Ezzat, A.; Wu, M.; Li, X.; Kwoh, C. Drug-target interaction prediction via class imbalance-aware ensemble learning. In Proceedings of the 2016, 15th International Conference On Bioinformatics (INCOB 2016), Queenstown, Singapore, 21–23 September 2016. [Google Scholar]
- Zhang, Y.; Jiang, Z.; Chen, C.; Wei, Q.; Gu, H.; Yu, B. DeepStack-DTIs: Predicting drug-target interactions using LightGBM feature selection and deep-stacked ensemble classifier. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 311–330. [Google Scholar] [CrossRef]
- Hasan Mahmud, S.M.; Chen, W.; Jahan, H.; Dai, B.; Din, S.U.; Dzisoo, A.M. DeepACTION: A deep learning-based method for predicting novel drug-target interactions. Anal. Biochem. 2020, 610, 11. [Google Scholar] [CrossRef]
- You, J.; McLeod, R.D.; Hu, P. Predicting drug-target interaction network using deep learning model. Comput. Biol. Chem. 2019, 80, 90–101. [Google Scholar] [CrossRef]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef]
- Wang, L.; You, Z.; Chen, X.; Xia, S.-X.; Liu, F.; Yan, X.; Zhou, Y.; Song, K.-J. A computational-based method for predicting drug-target interactions by using a stacked autoencoder deep neural network. J. Comput. Biol. 2018, 25, 361–373. [Google Scholar] [CrossRef]
- Monteiro, N.R.; Ribeiro, B.; Arrais, J. Drug-target interaction prediction: End-to-end deep learning approach. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 2364–2374. [Google Scholar] [CrossRef]
- Redkar, S.; Mondal, S.; Joseph, A.; Hareesha, K.S. A machine learning approach for drug-target interaction prediction using wrapper feature selection and class balancing. Mol. Inform. 2020, 39, 1900062. [Google Scholar] [CrossRef]
- Eslami Manoochehri, H.; Nourani, M. Drug-target interaction prediction using semi-bipartite graph model and deep learning. BMC Bioinform. 2020, 21, 248. [Google Scholar] [CrossRef]
- Peng, J.; Li, J.; Shang, X. A learning-based method for drug-target interaction prediction based on feature representation learning and deep neural network. BMC Bioinform. 2020, 21 (Suppl. S13), 394. [Google Scholar] [CrossRef]
- Wang, Y.-B.; You, Z.-H.; Yang, S.; Yi, H.-C.; Chen, Z.-H.; Zheng, K. A deep learning-based method for drug-target interaction prediction based on long short-term memory neural network. BMC Med. Informatics Decis. Mak. 2020, 20 (Suppl. S2), 49. [Google Scholar] [CrossRef]
- Meena, T.; Roy, S. Bone fracture detection using deep supervised learning from radiological images: A paradigm shift. Diagnostics 2022, 12, 2420. [Google Scholar] [CrossRef] [PubMed]
- Pal, D.; Reddy, P.B.; Roy, S. Attention UW-Net: A fully connected model for automatic segmentation and annotation from Chest X-ray. Comput. Biol. Med. 2022, 150, 106083. [Google Scholar] [CrossRef]
- Xu, X.; Gu, H.; Wang, Y.; Wang, J.; Qin, P. Autoencoder-based feature selection method for classification of anticancer drug response. Front. Genet. 2019, 10, 233. [Google Scholar] [CrossRef]
- Abid, A.; Balin, M.F.; Zou, J. Concrete autoencoders for differentiable feature selection and reconstruction. arXiv 2019, arXiv:1901.09346. [Google Scholar]
- DrugBank. DrugBank Fall 2019 Feature Release. Available online: https://go.drugbank.com/ (accessed on 4 March 2021).
- O’Boyle, N.M. Towards a Universal SMILES representation—A standard method to generate canonical SMILES based on the InChI. J. Cheminform. 2012, 4, 22. [Google Scholar] [CrossRef]
- PackageRcpi. Available online: https://www.rdocumentation.org/packages/Rcpi/versions/1.8.0 (accessed on 4 March 2021).
- Yu, H.; Chen, J.; Xu, X.; Li, Y.; Zhao, H.; Fang, Y.; Li, X.; Zhou, W.; Wang, W.; Wang, Y. A systematic prediction of multiple drug-target interactions from chemical, genomic, and pharmacological data. PLoS ONE 2012, 7, e37608. [Google Scholar] [CrossRef]
- Xia, Z.; Xia, Z.; Wu, L.Y.; Zhou, X.; Wong, S. Semi-Supervised Drug-Protein Interaction Prediction from Heterogeneous Biological Spaces. BMC Syst. Biol. 2010, 4, S6. [Google Scholar] [CrossRef]
- Ballesteros, J.; Palczewski, K. G protein-coupled receptor drug discovery: Implications from the crystal structure of rhodopsin. Curr. Opin. Drug Discov. Dev. 2001, 4, 561–574. [Google Scholar]
- Imrie, C.E.; Durucan, S. A River flow prediction using artificial neural networks: Generalization beyond the calibration range. J. Hydrol. 2000, 233, 138–153. [Google Scholar] [CrossRef]
- Fletcher, R.H.; Suzanne, W. Clinical Epidemiology: The Essentials, 4th ed.; Lippincott Williams & Wilkins: Baltimore, MD, USA, 2005; p. 45.126. ISBN 0-7817-5215-9. [Google Scholar]
- Raschka, S.; Mirjalili, V. Python Machine Learning: MAchine Learning and Deep Learning with Python, Scikit-Learn, and TensorFlow, 2nd ed.; Packt: Birmingham, UK, 2017. [Google Scholar]
- Scikit Learn. Available online: https://scikit-learn.org/stable/modules/feature_selection.html (accessed on 4 March 2021).
- GeÌron, A. Hands-on Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems, 2nd ed.; O’Reilly: Sebastopol, CA, USA, 2019. [Google Scholar]

| No | Author | Imbalance Solving | Dimension Reduction | Method | Description | Performance |
|---|---|---|---|---|---|---|
| The studies that use deep learning methods for DTI prediction problem | ||||||
| 1 | Sachdev and Gupta, 2019 [3] | No | PCA | SVM | Proposes a novel technique by using ensemble classifiers to increase the prediction of drug-target interaction. | AUC 0.96 |
| 2 | Ezzat et al., 2016 [10] | Yes | No | SVM, RF, DT | Proposes an ensemble learning method predict DTI. | AUC 0.90 |
| 3 | Zhang et al., 2022 [11] | Yes | LightGBM | DeepStack-DTI | Proposes a novel method, DeepStack-DTI, for predicting DTI using deep learning. | Accuracy 97.54 |
| 4 | Hasan Mahmut et al., 2020 [12] | Yes/MMIB | LASSO (Least Absolute Shrinkage and Selection) | DeepAction | Proposes a deep-learning method to predict potential or unknown DTIs. | AUC 0.98 |
| 5 | You et al., 2019 [13] | No | LASSO | LASSO-DNN | Shows that the efficient representations of drug and target features are key for building learning models for predicting DTIs. | AUC 0.89 Accuracy 0.81 |
| 6 | Beck et al., 2020 [14] | No | No | MT-DTI (Molecule Transformer-Drug Target Interaction) | Uses a pre-trained deep-learning-based drug-target interaction model called MT-DTI to identify commercially available drugs that could act on viral proteins of SARS-CoV-2. | Generally similar results compared to the conventional 3D structure-based prediction model. |
| 7 | Wang et al., 2018 [15] | Yes | - | Stacked AutoEncoder-RF | Presents a new computational method for predicting DTIs from drug molecular structure and protein sequence by using the stacked autoencoder of deep learning. | Accuracy 0.94 |
| 8 | Monteiro et al., 2021 [16] | Yes | - | Deep Neural Network Architecture | Presents a deep-learning architecture model, which exploits the particular ability of CNNs to obtain 1D representations from protein sequences and SMILES strings. | Accuracy 0.91 Sensitivity 0.82 F1-score 0.87 |
| 9 | Redkar et al., 2019 [17] | Yes | Synthetic minority oversampling technique (SMOTE) | RF, SVM | Addresses challenges faced by datasets with the class imbalance and high dimensionality to develop a predictive model for DTI prediction. | Accuracy 95.9 |
| 10 | Manoochehri and Nourani, 2020 [18] | Yes | - | Deep Neural Network | Models DTI prediction problems as link prediction in a semi-bipartite graph and uses deep learning as a learning tool. | The model improves the performance 14% AUROC |
| 11 | Peng et al., 2020 [19] | Denoising Autoencoder | Depp CNN | Implements a learning-based method for DTI prediction based on feature representation learning and deep neural networks. | AUROC 0.94 AUPR 0.94 | |
| 12 | Wang et al., 2020 [20] | SPCA | Deep Long Short-Term Memory | Develops a deep-learning-based model for DTIs prediction. | AUC 0.99 | |
| The studies that use deep learning methods in other medical areas | ||||||
| 13 | Meena and Roy, 2022 [21] | - | - | Deep Learning based methods | Reviews some fracture detection and classification approaches and claims that CNN-based models performed very well. | - |
| 14 | Pal et al., 2022 [22] | Yes | - | UW-Net | To reduce the use of tedious and prone-to-error manual annotations from chest X-rays, the author gave a probabilistic map for automatic annotation from a small dataset. | F1-Score 95.7% |
| The studies that use autoencoders for feature selection in DTI prediction problem | ||||||
| 15 | Xu et. al., 2019 [23] | Yes | AutoEncoder& Boruta | SVM-RFE, KNN, Autohidden, Naive Bayes | Determines a small set of features for the random forest to predict drug response by using the Boruta algorithm. | AUC 0.70 |
| 16 | Abid et al., 2019 [24] | No | Concrete Autoencoder | Proposes a new method for differentiable, end-to-end feature selection via backpropagation. | Concrete autoencoders show good performance. | |
| 17 | Our Proposal | Resampling Method | Autoencoder, DT, Kbest, Variance Threshold | MLP | Presents a new autoencoder-based feature selection approach for DTI prediction problem. | Accuracy 0.92 |
| FD1 | FD2 | … | FD41 | |
|---|---|---|---|---|
| D1 | 1 | 2 | … | 3 |
| D2 | 4 | 5 | … | 9 |
| … | … | … | … | … |
| D5771 | 6 | 7 | 8 |
| FT1 | FT2 | … | FT674 | |
|---|---|---|---|---|
| T1 | A | B | … | C |
| T2 | D | E | … | F |
| … | … | … | … | … |
| T4845 | X | Y | … | Z |
| FD1 (f1) | FD2 (f2) | … | FD41 | FT1 | FT2 | … | FT674 (f715) | |
|---|---|---|---|---|---|---|---|---|
| x1 | 1 | 2 | … | 3 | A | B | … | C |
| x2 | 1 | 2 | … | 3 | D | E | … | F |
| … | … | … | … | … | … | … | … | … |
| X27,960,495 | 6 | 7 | … | 8 | X | Y | … | Z |
| Parameter | Value |
|---|---|
| Number of neurons in the hidden layer | 13 |
| Input Dimension Size | 715 |
| Activation Functions | (ReLu, Sigmoid) |
| Loss Function | Binary Cross Entropy |
| Batch Size | (25, 100) |
| Structure | Designation | Structure |
|---|---|---|
| S1 | MLP-5-2 | Input + 5 hidden neurons + 2 hidden neurons + 2 output neurons |
| S2 | MLP-5-3 | Input + 5 hidden neurons + 3 hidden neurons + 2 output neurons |
| S3 | MLP-10-10 | Input + 10 hidden neurons + 10 hidden neurons + 2 output neurons |
| S4 | MLP-20-10 | Input + 20 hidden neurons + 10 hidden neurons + 2 output neurons |
| S5 | MLP-50-20 | Input + 50 hidden neurons + 20 hidden neurons + 2 output neurons |
| S6 | MLP-150-50 | Input + 150 hidden neurons + 50 hidden neurons + 2 output neurons |
| Parameter | Value |
|---|---|
| Number of neurons in the hidden layer | [2–20] |
| Learning rate | (0–1) |
| Momentum | (0–1) |
| Maximum iteration | 10,000 |
| Number of epochs | 500 |
| S1 | S2 | S3 | S4 | S5 | S6 | |
|---|---|---|---|---|---|---|
| Kbest10 | 0.56 | 0.62 | 0.69 | 0.66 | 0.69 | 0.71 |
| Kbest 50 | 0.6 | 0.7 | 0.64 | 0.7 | 0.8 | 0.8 |
| Kbest 100 | 0.24 | 0.68 | 0.65 | 0.68 | 0.74 | 0.82 |
| Kbest 160 | 0.22 | 0.56 | 0.62 | 0.6 | 0.69 | 0.71 |
| Kbest 200 | 0.24 | 0.63 | 0.67 | 0.54 | 0.6 | 0.56 |
| Kbest 240 | 0.2 | 0.45 | 0.53 | 0.25 | 0.67 | 0.61 |
| Kbest 300 | 0.32 | 0.3 | 0.32 | 0.2 | 0.43 | 0.56 |
| Kbest 350 | 0.3 | 0.54 | 0.65 | 0.54 | 0.43 | 0.52 |
| Structure 1 | Structure 2 | Structure 3 | Structure 4 | Structure 5 | Structure 6 | |
|---|---|---|---|---|---|---|
| Autoencoder 10 | 0.12 | 0.23 | 0.12 | 0.14 | 0.13 | 0.18 |
| Autoencoder 50 | 0.15 | 0.41 | 0.16 | 0.23 | 0.26 | 0.28 |
| Autoencoder 100 | 0.38 | 0.32 | 0.17 | 0.28 | 0.35 | 0.39 |
| Autoencoder 160 | 0.42 | 0.27 | 0.25 | 0.38 | 0.47 | 0.46 |
| Autoencoder 200 | 0.35 | 0.15 | 0.24 | 0.34 | 0.43 | 0.45 |
| Autoencoder 240 | 0.38 | 0.26 | 0.37 | 0.48 | 0.73 | 0.75 |
| Autoencoder 300 | 0.43 | 0.39 | 0.45 | 0.56 | 0.73 | 0.78 |
| Autoencoder 350 | 0.85 | 0.68 | 0.69 | 0.89 | 0.9 | 0.92 |
| Accuracy | F-Score | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Approach | S1 | S2 | S3 | S4 | S5 | S6 | S1 | S2 | S3 | S4 | S5 | S6 |
| No FS | 0.74 | 0.69 | 0.7 | 0.75 | 0.86 | 0.86 | 0.0024 | 0.001 | 0.002 | 0.0024 | 0.003 | 0.003 |
| Kbest 10 | 0.56 | 0.62 | 0.69 | 0.66 | 0.69 | 0.71 | 0.0014 | 0.0028 | 0.0021 | 0.0022 | 0.0026 | 0.003 |
| Kbest 50 | 0.6 | 0.7 | 0.64 | 0.7 | 0.8 | 0.8 | 0.0017 | 0.003 | 0.0022 | 0.0021 | 0.0026 | 0.0025 |
| Kbest 100 | 0.24 | 0.68 | 0.65 | 0.68 | 0.74 | 0.82 | 0.0012 | 0.0026 | 0.0025 | 0.0026 | 0.0034 | 0.004 |
| Variance | 0.45 | 0.73 | 0.72 | 0.75 | 0.81 | 0.75 | 0.0011 | 0.002 | 0.002 | 0.023 | 0.074 | 0.01 |
| Tree | 0.55 | 0.66 | 0.71 | 0.71 | 0.81 | 0.81 | 0.0017 | 0.0025 | 0.0023 | 0.0023 | 0.0035 | 0.0032 |
| AE 350 | 0.85 | 0.68 | 0.69 | 0.89 | 0.9 | 0.92 | 0.0031 | 0.002 | 0.002 | 0.005 | 0.08 | 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozsert Yigit, G.; Baransel, C. A Novel Autoencoder-Based Feature Selection Method for Drug-Target Interaction Prediction with Human-Interpretable Feature Weights. Symmetry 2023, 15, 192. https://doi.org/10.3390/sym15010192
Ozsert Yigit G, Baransel C. A Novel Autoencoder-Based Feature Selection Method for Drug-Target Interaction Prediction with Human-Interpretable Feature Weights. Symmetry. 2023; 15(1):192. https://doi.org/10.3390/sym15010192
Chicago/Turabian StyleOzsert Yigit, Gozde, and Cesur Baransel. 2023. "A Novel Autoencoder-Based Feature Selection Method for Drug-Target Interaction Prediction with Human-Interpretable Feature Weights" Symmetry 15, no. 1: 192. https://doi.org/10.3390/sym15010192
APA StyleOzsert Yigit, G., & Baransel, C. (2023). A Novel Autoencoder-Based Feature Selection Method for Drug-Target Interaction Prediction with Human-Interpretable Feature Weights. Symmetry, 15(1), 192. https://doi.org/10.3390/sym15010192





