Abstract
A variety of biological and pharmaceutical studies, such as for anti-cancer drugs, require the quantification of cell responses over long periods of time. This is performed with time-lapse video microscopy that gives a long sequence of frames. For this purpose, phase contrast imaging is commonly used since it is minimally invasive. The cell responses of interest in this study are the mitotic cell divisions. Their manual measurements are tedious, subjective, and restrictive. This study introduces an automated method for these measurements. The method starts with preprocessing for restoration and reconstruction of the phase contrast time-lapse sequences. The data are first restored from intensity non-uniformities. Subsequently, the circular symmetry of the contour of the mitotic cells in phase contrast images is used by applying a Circle Hough Transform (CHT) to reconstruct the entire cells. The CHT is also enhanced with the ability to “vote” exclusively towards the center of curvature. The CHT image sequence is then registered for misplacements between successive frames. The sequence is subsequently processed to detect cell centroids in individual frames and use them as starting points to form spatiotemporal trajectories of cells along the positive as well as along the negative time directions, that is, anti-causally. The connectivities of different trajectories enhanced by the symmetry of the trajectories of the daughter cells provide as topological by-products the events of cell divisions together with the corresponding entries into mitoses as well as exits from cytokineses. The experiments use several experimental video sequences from three different cell lines with many cells undergoing mitoses and divisions. The quantitative validations of the results of the processing demonstrate the high performance and efficiency of the method.
1. Introduction
Many scientific biological applications as well as preclinical pharmaceutical studies for drug development including for cancer treatment require the investigation of cell responses to a variety of stimuli. Some of these responses are cell migration, cell proliferation, as well as cell differentiation. This study, in particular, is about the detection of the spatiotemporal events of mitoses and of the accompanying cell divisions and cytokineses. It requires the observation of these events in samples for long periods of time to be able to draw the necessary experimental conclusions. An effective way to achieve this is with phase contrast microscopy.
1.1. Background on Phase Contrast Imaging for Cell Division
Phase contrast microscopy is the most widely used modality for long term imaging of cellular processes such as cell migration and cell division. Phase Contrast (PC) is a transmitted light based technique in which phase shifts created by the cells are translated into amplitude changes. Unlike fluorescence based methodologies, in which the cell is illuminated with very high intensity light to excite the fluorophores, phase contrast relies on the use of low intensity illumination. Thus, the serious issues of phototoxicity and photodamage elicited under fluorescence excitation are eliminated making phase contrast a far less invasive and gentle method for long term cellular imaging. In addition, phase contrast does not require labeling of cells and thus any cell type can easily be imaged while fluorescence based methodologies require the use of cells which are constitutively expressing fluorescent proteins, an important limitation. The above coupled with the higher cost of fluorescence microscopy systems as well as the higher cost of alternative transmitted light modalities such as Differential Interference Contrast (DIC) have established phase contrast the method of choice for live cell imaging.
The division of a cell starts with the division of the nucleus, namely, mitosis. It completes with the cleavage (division) of the cytoplasm into two daughter cells, namely, with cytokinesis. A cell rounds up starting with mitosis. In phase contrast imaging this results in a halo effect of increased contrast at the cell boundary. The contrast of the halos at the boundaries of the daughter cells remains high until cytokinesis completes.
The acquired phase contrast data sets of cell divisions are large and their manual analysis is tedious, subjective, and restrictive. Thus, this study proposes an automated method for their analysis. The method requires the reconstruction and restoration of phase contrast images of cells and the processing of large data sets of time-lapse movies. The processing is complicated by several factors. The first factor are the imaging artifacts. Other complicating factors are the varying sizes of the cells in different phases of mitosis and cytokinesis as well as the high cell density and the varying shapes of the surrounding cells. A complicating factor is also the motions of the cells that even result in some cells entering or leaving the field or plane of view.
1.2. Literature Review
An acquired image sequence is first preprocessed to remove imaging artifacts. This involves noise removal and restoration for intensity non-uniformity [1]. It also involves preprocessing for cell reconstruction [2,3,4,5]. This is followed by the analysis of the data to detect mitoses and cell divisions. The cell division detection methods can be classified into tracking-free, tracking-based, hybrid methods, as well as appearance-based with machine learning and neural networks [6,7]. The tracking-free methods involve the segmentation and detection of mitotic cells or of cell division events in individual frames. The tracking-based ones may still involve the segmentation or detection of cells from different frames of a sequence independently, for example from the first frame. However, tracking-based methods also involve a subsequent tracking of the cells throughout a sequence by updating the cells positions and detecting cellular cycle events.
The detection and even segmentation of cells in phase contrast microscopy has been done using a variety of low level features. It has been performed with morphological operations either directly [8,9,10,11] or in combination with texture features [12]. Other features that have been used are the intensity histogram [13] and gradient features. More involved features have been the Histogram of Oriented Gradients (HOG), GIST [14,15], and the Scale-Invariant Feature Transform (SIFT) [16]. More global shape characterizations and segmentation methods have been used. These include watersheds [17], symmetry [18], template matching [19], and statistical decomposition [20,21]. The segmentation has been performed with explicit modeling of the halo effect, which surrounds cells imaged with phase contrast microscopy, with active contours [9,11]. In some cases segmentation is semi-automatic [9,22]. Many of these techniques are tracking-free, independent of spatiotemporal trajectories, and hence allow causal online tracking [13].
The results of cell segmentation have been used as candidates to initialize automatic tracking of the motion of the cells in an image sequence to detect mitoses. The detection has been performed in a variety of ways. An approach estimates the cell division rate globally and implicitly from an image sequence based on cell area coverage without the explicit detection of individual cells or mitotic events [23,24]. Another method uses mean shift directly on an image sequence for cell event detection or motion tracking performed in forward or in reverse time direction and hence motion tracking may not be causal [8]. Spatial segmentation has also been addressed with active contours propagated in time to follow motion with linear prediction based on Kalman filtering [13,24,25,26,27,28]. Another representation for cell division has been with linear programming [22] or linear programming combined with level sets [29]. Some tracking-based approaches detect the lineage of each cell [30].
An alternative representation of tracking-based methods has been with global data associations between frames in a sequence. These have been established based on features of the detected or segmented cellular regions. Some methods in this context perform cell detection with the circle Hough transform for isolated cells [31] or its extension, the local circle Hough transform, as medial axis transform, to deal with cell detection in dense samples [1]. The temporal associations have been followed to form spatiotemporal trajectory segments that may then be linked together [1,32,33]. Such methods result in implicit detections of mitoses as a topological by-product [1,33]. Some of these tracking-based methods segment and follow cells using level sets-based active contours methods; however, they are sensitive to dense samples and have only been able to detect isolated dividing cells [25,31].
The distinction between the spatial and the temporal dimension is diminished using global flow networks [30,34]. In an approach resembling stochastic signal processing, a graph has been used for the detection of the sequence of the different phases of mitosis and of cell division explicitly with Markov Models (MM) and Hidden Markov Models (HMM) [35,36]. More elaborate Conditional Random Fields (CRFs) classifiers have also been used for mitosis detection [37,38]. Further elaborations of these types of classifiers that have been used are the Event Detection Conditional Random Field (EDCRF) that is able to not only detect mitosis events, but also to detect the completion of mitosis and the birth of the daughter cells [39]. Another elaboration has been the Multi-Grained Random Fields (MGRF) [40]. In addition, Max-Margin Hidden Conditional Random Field (MM-HCRF) with Max-Margin Semi-Markov Model (MM-SMM) classifiers have been used [41]. These classifiers require extensive training. They also involve at least a time-delay for the processing of a sequence. Stochastic Markov models have also been used at nano scale [42].
The methods that have been described thus far are geometric, graphical, or statistical. Beyond these methods that attempt to model the form of the cells and quantify the associated remaining error for the cellular events [31], many other techniques develop classifiers directly from the data. Some are based on machine learning methods and others are based on neural networks. In the context of machine learning, mitoses have been detected with the AdaBoost ensemble classifier [28]. As an extension to graph-based methods, graph neural networks have also been used [43]. Convolutional neural networks in 2D (2D CNN) provide the detection of mitoses in individual frames; however, the method is likely to lead to either repeated or missed mitoses detections depending on the temporal sampling points [44]. Features from a 3D Convolutional Neural Network (3D CNN) have been combined with a Support Vector Machine (SVM) classifier from the context of machine learning to detect the presence of a single mitosis, however without the ability to temporally resolve it [45]. A neural architecture that combines Fully Convolutional Networks (FCN) and 3D CNNs, namely, the F3D-CNN has also been developed [46]. The first step, FCN, detects areas with candidate divisions. A sequence of these candidates is formed, since a cell division can span multiple frames, and is then provided to a 3D CNN for cell division detection.
To enhance the temporal resolution of the detection, a 2D-U-Net [47] has been used to extract candidates in space that are then stacked and filtered in time with a Bidirectional Long Short-Term Memory (BLSTM) network to reject false positives [48]. To temporally resolve a mitosis in a spatiotemporal sequence, convolutional neural networks have been combined with a Two-Stream Bidirectional Long Short-Term Memory (TS-BLSTM) network [49,50,51]. In that method, candidate regions are first extracted and then processed with the TS-BLSTM using appearance and motion information to jointly detect the presence of a mitosis as well as to temporally localize it. In a similar vein, to improve temporal mitosis resolution, patch sequence candidates are extracted that are then processed by a convolutional neural network in space and subsequently with an LSTM (CNN-LSTM) along time. The combined results are used to build a 3D probability heat map that is processed to locate mitotic events. Its training is end-to-end and enables the joint detection of mitotic event in space and time [52,53]. These methods based on neural networks can only detect a single mitosis in a candidate spatiotemporal patch, which is a limitation particularly for dense samples. To address this limitation a method based on 3D CNN, namely, V-Net [54], computes a mitosis likelihood throughout a spatiotemporal sequence even for dense samples [55,56]. In another work, combining processing in space and time, a deep reinforcement learning-based Progressive Sequence Saliency Discovery (PSSD) network is used to train a Markov Decision Process (MDP) to discover salient frames and temporally locate mitoses [57].
Machine learning-based methods and neural networks-based methods have several characteristics in common. They both require extensive training that is computationally intensive. Machine learning-based methods are sometimes preferred, because they are often more efficient computationally [28]. Techniques belonging to both of these classes have been shown to be effective for samples from only a single cell line. This has been demonstrated particularly from the benchmark that has been developed for validation that only consists of myoblast cells and involves extensive manual annotations [7]. The neural networks detections are also sensitive to dense samples and do not provide explicit spatiotemporal cell information for a complete mitotic event cycle that leads to a cell division and the associated cytokinesis. This limits the applicability and sufficiency of neural networks based detections for biological studies of this phenomenon.
1.3. Overview of the Proposed Method and Contributions
An early version of the work on this method has been extended, made more robust, and validated in this study [1]. The proposed method starts with extensive preprocessing for restoration and reconstruction. It involves the detection of candidate mitotic cells or daughter cells centroids in individual frames that are tracked for cell displacements between frames. This is followed by the detection of candidate cell divisions in individual frames that are tested to potentially establish a cell division event. The detection and tracking of mitosis and of the accompanying cell division is the eventual objective of the proposed method.
The acquired video sequences are first preprocessed to correct for intensity non-uniformity artifacts due to non-uniform illumination. Subsequently, the halo effect surrounding mitotic cells is detected with Hessian-based image filtering. The principal components of the pixelwise Hessians and their eccentricity are used to perform a circle Hough transform that takes advantage of the circular symmetry of the cells and has several novelties as well [58,59,60]. The main novelty is that it identifies the pixel-based direction towards the center of curvature that is used to exclusively “vote” for the interior circle center using the expected radius of the cells. This makes the transform robust to a dense cell clutter in a sample. The emphasis of the circle Hough transform, that is also a novelty of the method, is to be particularly sensitive to the detection of the daughter cells that remain circular and with a high contrast for a short period of time during cytokinesis. The circle Hough transform images are then corrected for possible spatial misregistrations due to shifts among consecutive frames of a sequence.
A direct processing of the circle Hough transform images provides several cell centroids. The registered centroids are the starting points for cells trajectories extraction. This is based on pixel associations between pixels locations of successive frames in a sequence. The pixel associations are established and the cell trajectories are subsequently tracked in both the positive as well as in the negative time direction. That is, the trajectory extraction is also anti-causal. Candidates for cell division events are identified as a topological by-product of joining an already extracted trajectory of a parent cell together with one of its daughter cell with the trajectory corresponding to the second daughter cell. The trajectory of the second daughter cell is detected later along the progression of the method. The segment of the new trajectory joining the pre-existing one is extracted anti-causally. The candidate cell division events are further tested to establish the division. The method takes advantage of the symmetry between the daughter cells in cytokinesis and even of the symmetry of the cell cycle. The implementation of the proposed method emphasizes efficiency. The validation used fourteen time-lapse phase contrast movie sequences from real biological experiments of HeLa, Kyoto, and Fibonectin cell lines undergoing mitoses over long periods of time. The outputs of the method for these sequences were validated quantitatively that demonstrated the high performance and robustness of the method.
2. Data and Methods
The phase contrast cellular image sequences are processed in a pipeline. The first step is the restoration of the intensity uniformity along time. This is followed by intensity uniformity restoration in space. The images are then transformed with an extension of the circle Hough transform. Both the original and the circle Hough transform image sequences are used in subsequent processing steps. These are the extraction of cell trajectories as well as the detection of cell divisions. A summary of the preprocessing steps is given in Figure 1 and a summary of the steps for cell division detection is given in Figure 7.
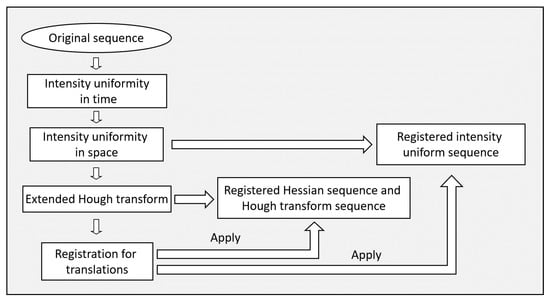
Figure 1.
A block diagram of the preprocessing steps of the method.
2.1. Data
Each image sequence corresponds to a different cell line. The sequences are from a total of three different cell lines, namely, of HeLa, Kyoto, and Fibronectin cells (the HeLa cell line was a kind gift from Dr. Katerina Strati (University of Cyprus), which acquired it from ATCC. The HeLa Kyoto cell line was kindly gifted to our lab by Dr. Niovi Santama (University of Cyprus). Regarding the Fibronectin null fibroblast cell line, it was a kind gift from Dr. Reinhard Fässler (Max Planck Institute of Biochemistry)). The cell divisions for the third type of sequences of Fibronectin cells in Table 1 were synchronized using the cell division inhibitor RO3306. The imaging was performed using a Carl Zeiss Axiovert 200M inverted microscope with a motorized stage, which was time sharing among several wells of different samples. Plan-Apochromatic lenses of magnification and were used, which gave total image magnification factors together with those of the ocular lens of and , respectively. The microscope was used for positive phase contrast microscopy. It was also linked to a Carl Zeiss AxioCam HRc digital camera. The resolution of the CCD and hence the size of the images is or approximately . Images were collected at intervals of 3–5 min for several hours to provide a time-lapse movie sequence.

Table 1.
Description of the acquisitions of the time-lapse sequences.
There are five sequences of the first cell line. They include two sequences that are from different positions (Fields of View (FOVs)) within the same sample that were imaged simultaneously during the experiment. Two more sequences were from different positions within another sample and were imaged simultaneously during the experiment. The last sequence was from a third sample. There are four sequences of the second cell line. They include two different positions within the same sample that were imaged simultaneously during the experiment. The other two sequences were acquired immediately afterwards from the same sample and they again correspond to different positions that were imaged simultaneously during the experiment. There are five sequences of the third cell line. All five of them show different positions within the same sample that were imaged simultaneously during the experiment.
The acquisition resulted in a total of time-lapse frames, . A phase-contrast image sequence is represented by , where spatial coordinates are and D is the domain of the images. The description of the acquisitions is in Table 1. The processing method also provides an option to decrease the spatial resolution of the input data to improve the efficiency of subsequent analysis.
2.2. Restoration of Intensity Uniformity along Time
The illumination of the lamp in phase contrast microscopy may vary along time. The lamp may also flicker. As a result the brightness of the images not only varies smoothly along time, but also some of the images may be much darker or brighter than the immediately adjacent ones in time both before and after.
It is necessary to restore the uniformity of the image intensities along time. This is achieved by making the average intensity along time uniform. That is, the mean intensity per frame is preserved along time. However, the total average sequence intensity is also preserved. To this end, the average intensity of each frame as well as the average intensity of the entire image sequence are calculated. The intensities of each pixel in a frame are multiplied by a correction factor, which is the ratio of the average intensity of the entire image sequence over the average intensity of the corresponding image. That is,
gives the sequence with time normalized intensity.
2.3. Restoration of Intensity Uniformity in Space
The intensity of the illumination is assumed to be and to remain uniform in space for all the frames of a sequence. However, in practice, the illumination in space across the sample is non-uniform. Hence, the acquired images suffer from spatial intensity non-uniformities. The non-uniformities are multiplicative and spatially smooth. Moreover, they cannot be computed directly. This is because they interfere with the inherent variation of the intensities primarily in the foreground of a sequence. In the analyzed sequences, the foreground is varying due to the dynamics and proliferation of the many cells present in a sample. In some frames, a spatial location may correspond to the background and in other frames the same spatial location may correspond to a cell.
The first objective is to remove the variability of the foreground to isolate a preliminary version of the background that is assumed constant along time. The background is considered equivalent to the intensity non-uniformity. The preliminary version of the background, , is estimated by projecting it along time. To this end the method considers a circle around every spatial location of radius approximately equal to that of a prespecified cell radius . This circle is the base of a cylinder covering all time in spatiotemporal space. The median intensity of this spatiotemporal volume becomes the value of .
The preliminary estimate of the background from the projection may still contain cells or their remnants from the foreground, particularly in regions occupied by stationary cells. Hence, is further processed to estimate a more accurate version of the background as its inverse that is the spatial intensity non-uniformity correction field. This is the objective of the remaining steps of the restoration performed with an adaptation of a Bayesian non-parametric method [61,62,63,64]. The histogram of the projection image is computed. The effect of the spatial intensity non-uniformity on the histogram is deconvolved to obtain a gain factor for each intensity in . The gain factors are back-projected in space to the preliminary background image to compute a spatial gain field. The gain field is spatially filtered with a Gaussian filter to give an incremental smooth correction image. This then multiplies a cumulative correction field , initialized to unity. The pixelwise multiplication of the correction field with each frame in the sequence gives the restored image sequence,
The cumulative restoration field is also scaled to ensure that it has a unit mean and that the intensity of the of the cumulative histogram of the spatiotemporal volume is preserved.
The spatial intensity restoration is iterative. The criterion to terminate the restoration uses the fact that the correction field has approximately unit mean and terminates the iterations when the standard deviation of the ratio between the correction fields of two consecutive iterations is very low, ≈. The restoration is also terminated at a maximum allowed number of iterations. This correction is the third step in the block diagram of the preprocessing in Figure 1. An overview of the steps of the spatial intensity restoration method is in Figure 2. An example of a frame from the intensity restoration of a sequence is shown in Figure 3.
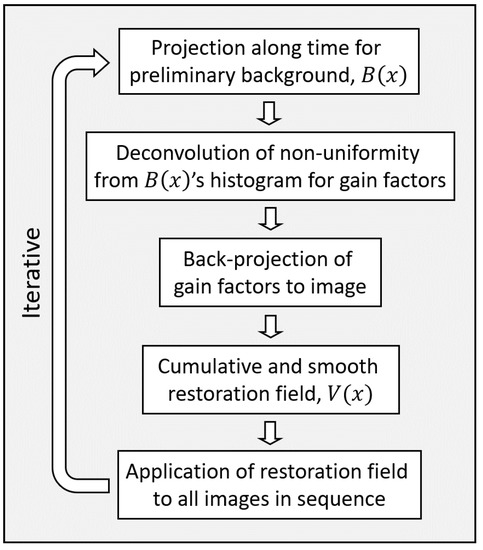
Figure 2.
A block diagram of the steps of the method for spatial intensity uniformity restoration.
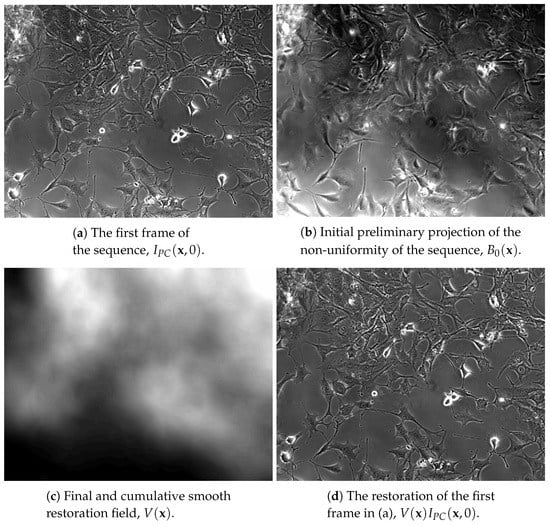
Figure 3.
Example restoration of a sequence of the third group in Table 1. In (a) is a frame from the original sequence. The projection of the non-uniformity along time for the first iteration is in (b). The cumulative restoration field of the entire sequence is in (c). In (d) is the intensity restoration of the frame in (a). Despite the non-uniformity of the original frame in (a), the restored frame in (d) is intensity uniform. All images in the figure have an enhanced dynamic range to better demonstrate the intensity non-uniformity effect and its correct removal.
2.4. Extended Circle Hough Transform of a Phase Contrast Sequence
The shape of the cells during mitosis and cytokinesis becomes rounded with a well formed boundary. The imaging of these cells with phase contrast microscopy creates an approximately circularly symmetric halo effect of high contrast around them. The pixels along the contour of the halo effect are detected with pixelwise Hessian-based filtering. This is a convolution of the images with second order Gaussian derivative filters for every entry of the pixelwise Hessian matrices. The principal components of the Hessian matrix of a pixel along a cell boundary gives an eigenvector, , normal to the boundary at that pixel with a corresponding eigenvalue of large magnitude, . The other eigenvector, , is tangent to the boundary at that pixel with corresponding eigenvalue of a lower magnitude, . The largest absolute eigenvalue corresponds to the direction normal to the boundary and satisfies .
In the phase contrast images in this study, the halo effect immediately surrounding the cells is bright as it is shown in the examples in Figure 5a,b. The bright cross-section of the contour along and considering the intensity as relief above the image plane is concave up and so it corresponds to an eigenvalue, , that is negative [65]. That is, along the cell contours the eigenvalue of largest magnitude is negative. Pixels where the eigenvalue of largest magnitude is positive are considered noisy and are pruned to zero. A summary of Hessian filtering over the entire sequence provides,
The entries of these images are used as modulating weights to extend the basic form of the circle Hough transform.
To further improve the efficiency of the circle Hough transform the set of pixels that participates in the transform is pruned even further. This is achieved using the statistics of the histogram of the sequence of the modulating weight images, , given in Equation (3). This histogram is unimodal with the mode corresponding to the dark background and the dark inner cytoplasm regions. The bright halo effects surrounding the mitotic cells correspond to the sidelobe of the higher intensity range. That sidelobe is detected as the intensity range above the intensity corresponding to the mode plus two times the full width half maximum of that mode. The pixels with intensities below that range are pruned and are not considered for the transform. This pruning only causes a minimal loss of accuracy to the transform and may even improve its accuracy by ignoring the contributions from noisy pixels.
The basic circle Hough transform [66] is extended to modulate the contribution of every pixel to the transform by a weight that is spatially localized with a two-dimensional Gaussian distribution, . The covariance of the Gaussian is aligned with the two eigenvectors of the Hessian. The direction along , across the contour for the centroid of the cell is assigned a standard deviation equal to . The direction along , tangent to the boundary smooths the transform and is assigned a standard deviation that is small and equal to a small fraction of the cell radius, but it still has a finite value, .
The transform considers the Gaussians only locally around their mean points relative to the radius parameter of the cells, . Thus, it contributes to only a subset of the image domain. It truncates the Gaussian normal to the boundary to and the Gaussian tangent to the boundary to . This not only expedites the transform, but also makes it more robust with respect to dense cells samples.
The proposed transform has two stages. The first stage for in Equation (5) transforms both along the positive as well as along the negative directions of the normal axes for the pixels assumed to be lying along cell contours. It is modulated with a Gaussian centered on the contour points at . The Hough transform of the image is:
is the Gaussian along and is the Gaussian along . In this resulting transform space, close to the contours points of the cells there is a higher density value towards the interiors rather than towards the exteriors of the cells. Sampling this transform space normal and close to the contours points identifies the direction from the contour points corresponding to higher transform densities. In the transform, “voting” is along lines normal to the contours. Hence, the higher densities are towards the centers of curvature [67]. The centers of curvature for the circular cells are in the directions towards the interiors of the cells. That is, the first stage provides the directions from the contour points towards the centers of curvature that are the directions towards the interiors of the cells.
The second stage that gives the final extended transform, , in Equation (6), is only towards the interiors of the cells. It involves exclusively the directions from the contour points towards the centers of curvature. Hence, it only uses the positive parts of the normal axes. The contributions along these semi-axes are also weighted with Gaussians centered at the estimated centers of the cells, which are at a positive distance from the points of the contours. The extended circle Hough transform of an image is given by:
is the Gaussian along and is the Gaussian along . The weighting Gaussian, , is shown schematically in the diagram in Figure 4.
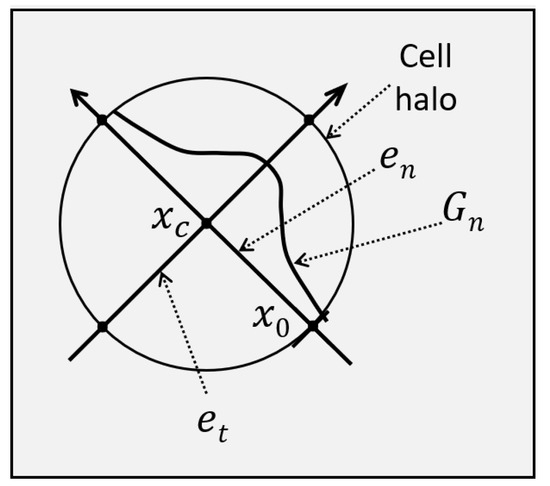
Figure 4.
A Gaussian is aligned at with the eigenvector direction corresponding to the largest Hessian eigenvalue magnitude. Its mean is the estimated center location of the cell, , and its standard deviation is larger than the cell radius, . The Gaussian contributes to the circle Hough transform.
The extended circle Hough transform is the fourth step in the block diagram of the preprocessing steps in Figure 1. Two example images that contain many rounded cells in mitosis and cytokinesis with a high contrast and the extended circle Hough transforms of these images are shown in Figure 5. It is recommended that the images in Figure 5 be viewed using the electronic/digital magnification functionality available. The extensions make the circle Hough transform images very intense and the cell centroids in these transform images to have a much higher contrast. After the cell division their contrast relaxes back to low levels.

Figure 5.
Two examples of phase contrast images transformed using the pixelwise Hessian-based filtering to the extended circle Hough transform images. The first example is from the second group of sequences in Table 1 and is shown in (a,c,e). The second example is from the third group of sequences in Table 1 and is shown in (b,d,f). In the first row, (a,b) are the original phase contrast images. In the second row (c,d) are the Hessian-based filtered images, . In the last row, (e,f) are the images transformed with the extension of the circle Hough transform, . It is recommended that the images be viewed magnified.
2.5. Registration for Translation Transformations
The time-lapse microscopy movies analyzed are acquired over long periods of time. During that time, there can be a mechanical shift of the sample and the microscope can be interleaving among several samples. Both of these result in a drift in the relative location between the sample and the camera that may be accumulating over time. In turn, this causes cumulative spatial misplacements between the frames of a sequence that are mainly translations.
This spatial artifact is addressed by registering for global image translations between consecutive frames of a sequence. The registration is based on the extended circle Hough transform images that contain a reconstructed and more explicit information about the sample with the cells being convex as opposed to the phase contrast images where the cytoplasm has the same intensity as that of the background. The translation of the zeroth time point frame, , is the identity . The registration computes the relative translation between two consecutive frames. The reference frame is that of the previous time point, , and the moving frame is the next one, . The objective is to spatially normalize the moving frame to the reference one. The sequence of registrations starts from the first frame and proceeds to the last frame. This gives the total frame registrations , . Several possible in-plane translations are considered with maximum extend . The candidate translations are examined using a regular spatial subsampling factor to improve computational efficiency. The optimal translation is selected.
The optimal translation is the one with minimal cost over the entire image. The basis of the cost of a candidate translation is the absolute value of the difference between a translated frame and the previous one that is the reference. This is the -norm of the difference. The -norm is further normalized with respect to the size of the common domain between the translated frame and the reference one. The trial translations are evaluated with exhaustive search to give the optimal one as:
The intersection of the domains of all the registered frames along time is the valid domain of the image sequence. That is, the valid domain is . The zeroth frame is not moved. The common domain of the sequence after registration with will be a subset of the valid domain of the zeroth image , .
The registration along a sequence starting from the first frame and finishing at the last one is repeated several times. This enables corrections for misregistrations that happen early in time to propagate to later time points. The repetitions of the registrations of the sequence follow a multiresolution approach. The search starts with a large displacement for the coarse resolution and completes with a small, local, displacement for the fine resolution. There are three levels of resolution and the extent of the maximum shift is for all of them. The initial value is pixels and the sampling is at . Both the extend of the maximum candidate shifts, 2h, and the number of sampling points for the displacement are refined to smaller values with the repetitions of the registration of the sequence. The decrease is linear and gradual to smaller values for finer resolutions with consecutive repetitions of the sequence registration. The sampling between candidate displacements is kept at for all resolutions.
The iterations over the spatiotemporal volume, , continue for a certain resolution up to the iteration at which the percentage of non-zero to zero optimal translations between frames over the entire sequence is very low, which implies that it is close to the optimal translations for a sequence or up to a maximum number of iterations, , for a specific resolution level. It then proceeds to the next finer resolution level until the finest resolution.
The transformations , computed with Equation (7), are applied to the corresponding frames of the phase contrast sequence , the Hessian filtered sequence , and the circle Hough transform sequence for the purposes of subsequent processing steps. The phase contrast image sequence becomes , where . The Hessian filtered image sequence becomes . Finally, the circle Hough transform sequence becomes . All three of them are defined in space only over the valid domain . The registration is the last step of the preprocessing as it is shown in the block diagram in Figure 1. A schematic diagram of the sequence registration is shown in Figure 6.
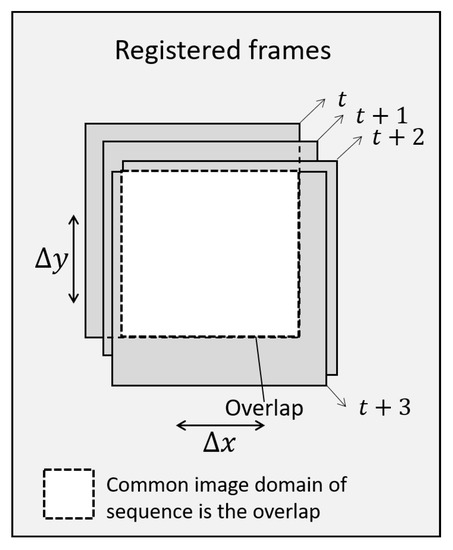
Figure 6.
A schematic diagram example of the registration for translations between successive frames in an image sequence. Only the overlap domain in white is considered for the entire sequence.
2.6. Extraction of Cell Centroids
The intensity restored and circle Hough transform registered sequences, as shown in Figure 7, are used for the extraction of the cell trajectories and the identification of the cell divisions. The extended circle Hough transform images provide the conditions to detect the cell centroids. The first condition is that a cell centroid be a local maximum within a circle of radius in a circle Hough transform image. This condition:
ensures a local maximum. The second condition uses the cumulative histogram of the 3D spatiotemporal volume of the framewise circle Hough transform sequence, . It requires that a cell centroid be of value that corresponds to at least 98% of . The condition
ensures that the candidate pixel has a value that is sufficiently high. Both of the two conditions above are necessary for a point to be classified as a cell centroid. These conditions in Equations (7) and (8) are drawn in Figure 8.
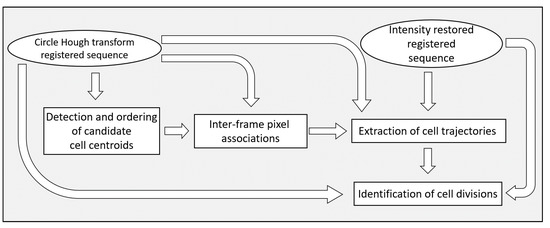
Figure 7.
A block diagram of the steps of the method for cell division detection.
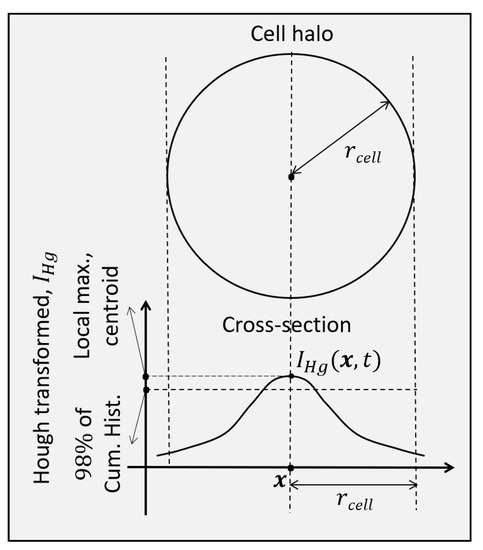
Figure 8.
Conditions for cell centroid detection in an extended circle Hough transform image.
A spatiotemporal volume is formed with non-zero values only at detected cell centroid pixels with actual values equal to those of the circle Hough transform images at those pixels:
This provides both the centroid pixels and their circle Hough transform values. The detection of cell centroids and their values is the second step of the processing pipeline in the block diagram shown in Figure 7.
2.7. Inter-Frame Associations between Pixels
The detected cell centroids are the starting points for the two trajectory segments. One proceeds in positive time direction and another in negative time direction. These establish the location of a cell centroid in previous and in subsequent time points, respectively. The tracking is performed not only along progressing time, but also anti-causally in the negative time direction. A cell division is detected as a join of two different trajectories at least one of which is almost always extracted in the negative time direction.
A cell motion trajectory is based on associations between pixels of successive frames. They are established by directing inter-frame pixel associations locally towards maxima of the circle Hough transform of the reference frame. Thus, the associations can be many to one and do form a function. Along the negative time direction the reference frame is the previous one and the association function is . In the positive time direction the reference frame is the next frame and the association function is .
An association of from a pixel at time t, , in reverse time direction to a pixel at time , , is established with a local search in frame . The search over candidate displacements is within a window of radius in the frame of the previous time point . The maximum with penalized displacement establishes an association from pixel in the frame at time t to pixel . The utility function for the association to pixel is:
where . The association is to the pixel which maximizes the utility in Equation (10) at time ,
and location . This maximization establishes to be the position of the association at the previous time point . That is, the selection is the pixel of maximum circle Hough transform intensity with penalized displacement. This search, within , is repeated three times. It creates a search path in that consists of a sequence of three local maxima and stops at the third maximum. The repetition contributes to representing not only the motion of a cell, but also possible remnants of misregistration between successive frames.
A reverse time association from is an entry in the function . The associations are computed in a lazy fashion for computational efficiency. They are computed only if starting from a certain detected or tracked cell centroid it is necessary to compute an association to continue with the extraction of a cell trajectory.
The same process as in Equations (10) and (11) is used to compute causal associations from a pixel at time t, , in forward time direction to a pixel at time , . Similarly to Equations (10) and (11), they are established by maximizing utility to give as the association at the next time point . These inter-frame pixel association of are also many to one and form a function. Thus, considering both functions and the pixel associations are many to many. They are mainly used to track cell centroids. The inter-frame pixel associations is the third step of the motion tracking part of the method summarized in Figure 7.
2.8. Extraction of Cell Trajectories and Detection of Cell Divisions
The temporal associations between pixels of adjacent time points are used to extract cell trajectories. The trajectories also establish cell centroids in frames where they might have been originally missed. Furthermore, joining two of these trajectories, extracted sequentially in the method, at a pixel in space and time gives a cell division candidate. That is, the joining of two trajectories identifies a candidate for cell division as a topological by-product. This happens when some of the extracted trajectories are of daughter cells during cytokineses. The cell division candidate, if verified, is actually immediately before the cytoplasm division to two daughter cells in cytokinesis and during the telophase of the mitotic cycle of the cell.
The extraction of the cell trajectories uses the inter-frame pixel associations from and , the circle Hough transform images , as well as the cell centroid images . The starting point, , for the extraction of a cell trajectory is the pixel of maximum intensity in the sequence of initially extracted centroids,
This point, , identified as starting point for a trajectory, has to also be at a distance of at least from all previously extracted trajectories.
The trajectory is extracted by progressing both reversely in time with as well as forwards in time with . This continues as long as the intensity of the trajectory at the circle Hough transform image , from Equation (6) and registered according to Equation (7), is greater that 20% of the reference Hough intensity that corresponds to the 95% of the cumulative histogram of the spatiotemporal volume of the circle Hough transform, .
In tracking reversely in time with , anti-causally, when the newly extracted trajectory approaches a previously extracted trajectory within a distance less than , it undergoes a test to examine if this corresponds to a trajectory joining event due to reverse cell division. That is, whether the two trajectories in that time point are within a single mitotic cell at telophase immediately before a cell division, in which case they are merged. The tests involve the Hessian filtered image, , as well as the circle Hough transform image, , intensities of the point in the pre-existing trajectory and the point in the newly tracked one at the same time t. The two points are considered as poles interconnected by a line segment. The first condition involves the Hessian filtered values, , and requires that any point along the line segment be less than that of the average of the two poles. The second condition involves the circle Hough transform values, , and requires that any point along the line segment be greater than that of the average of the two poles.
The third and last condition is topological for the trajectories and involves the extend of the pre-existing trajectory. It requires that the pre-existing trajectory exists for at least one time point both before and one time point after the time point for which the two points are investigated for merging due to cell division. This condition ensures that the newly extracted trajectory segment is not a simple continuation of the pre-existing trajectory.
The cell centroids in from Equation (9) in the region around a newly extracted trajectory are invalidated within a distance of so that no other trajectory is initiated from that region. The centroid pixel of maximum intensity from the remaining valid spatiotemporal centroid volume is identified. If it is non-zero, this centroid becomes the starting point for the extraction of a new trajectory. The sequential extraction of trajectories continues until the entire of volume becomes zero.
This is the last step of the method, namely, the step of cell trajectory extraction and cell division identification and is summarized in Figure 7. Figure 9 shows an example schematic of trajectories both in forward and in reverse time directions as well as a detected cell division. A flow chart of the method for the sequential extraction of cell trajectories and cell division detections is shown in Figure 10.
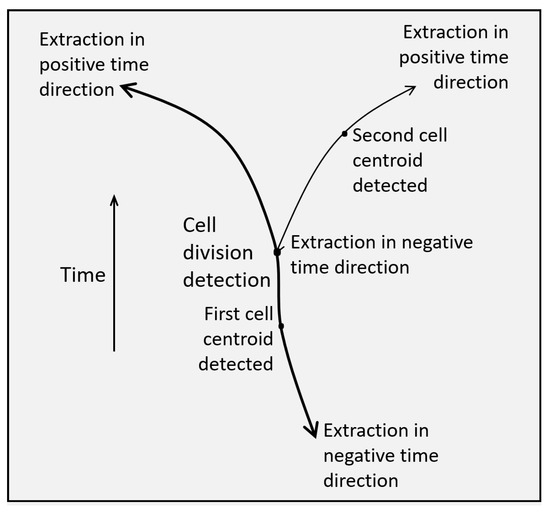
Figure 9.
Schematic of a cell division detected from the topological connectivity of two trajectories one of which corresponds to the trajectory segment of one of the two daughter cells that is extracted in the negative time direction.
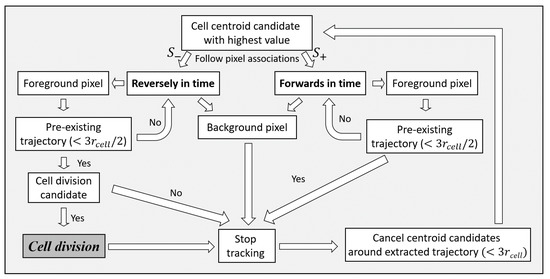
Figure 10.
A flow chart for the sequential extraction of cell trajectories and the detection of cell divisions. A cell centroid is the starting point for a cell trajectory. The extraction proceeds in both positive and negative time directions using inter-frame pixel associations. The termination of the extraction depends on the circle Hough transform value of a tracked centroid in a frame. The extraction also terminates if it approaches close enough or meets an already extracted trajectory. In the case of approaching or meeting an already extracted trajectory in negative time direction, anti-causally, a test examines if it corresponds to a cell division.
3. Experiments
The method has been applied and validated with multiple image sequences of several cell lines that were acquired over many hours and hence have a large temporal size as described in Table 1. These sequences are from real biological experiments. The analysis of the data has multiple processing steps that normally have considerable computational requirements. To maintain a reasonable running time for the method, despite the extensive processing required, the implementation of the various steps of the method has been made efficient. The method is implemented in the C++ programming language. The spatial resolution of the imaging sequence used is over each spatial axis or over the whole spatiotemporal volume. The experiments were performed on a laptop with an Intel Core i7 processor of GHz and GB of RAM.
3.1. Validation Measures for Cell Division Detection
The objective of the method is to detect mitoses and accompanying cell divisions. These events are considered for the evaluation as long as they happen in the spatiotemporal interval imaged to obtain a sequence. In particular, a cell division is considered if the image sequence contains at least the late phases of mitosis, namely, anaphase and telophase, as well as the early part of cytokinesis including the cleavage of the cytoplasm into two daughter cells. The reference quantities considered for the performance evaluation of the cell division detection method for a sequence are the True Positive (), False Positive (), False Negative (), and True Negative () detections. The number of cells is the number of cells in the spatiotemporal sequence not undergoing mitosis.
The performance of the system is quantified in terms of the and , the and , as well as the . The recall is equivalent to the sensitivity. The is the fraction of the detected cell divisions that are actually cell divisions and is given by:
The is the fraction of the cell divisions that are actually detected out of all the cell divisions. This is equivalent to the and are given by:
The is the proportion of the cells not undergoing cell division that are not detected and is given by:
The is the harmonic mean of the and the , that is, it is their average rate:
These quantities are computed for every sequence independently.
3.2. Experimental Performance of the Image Sequences
The data for the validation of the method were fourteen spatiotemporal sequences corresponding to three different cell lines. Each cell line belongs to a different group. The description of the first, second, and third groups are in the three columns of Table 1, respectively. The output of the method for a microscopy sequence is a new movie with background the original movie with preprocessing and foreground annotations for the automatically tracked cells that undergo cell division for the duration of the sequence. The annotation starts from mitosis during metaphase, anaphase, or telophase. The annotations continue for the tracking of the daughter cells during cytokinesis immediately following the division of the cytoplasm. A parent during mitosis and prior to cell division is annotated with a mark of a specific shape, color, and length. The same mark annotates the two resulting daughter cells. The actual point in space and time before the parent cell divides is indicated with a thick cross of the same color as that of the cell mark.
Some examples of representative frames from the output sequences from each of the first, second, and third groups of sequences are shown in Figure 11, Figure 12 and Figure 13. These frames show multiple cells in mitosis as well as in cytokinesis both before and after the division of the cytoplasm. It is recommended that the images in Figure 11, Figure 12 and Figure 13 be viewed in their original color and using the electronic/digital magnification functionality available. The sequence in Figure 11 is of the cell line of the first group of Table 1. It lasted for 3 h 15 min and gave 65 frames. The sequence in Figure 12 is of the cell line of the second group of Table 1. It lasted for 5 h and gave 100 frames. The sequence in Figure 13 is of the cell line of the third group of Table 1. It lasted for 5 h and gave 60 frames.
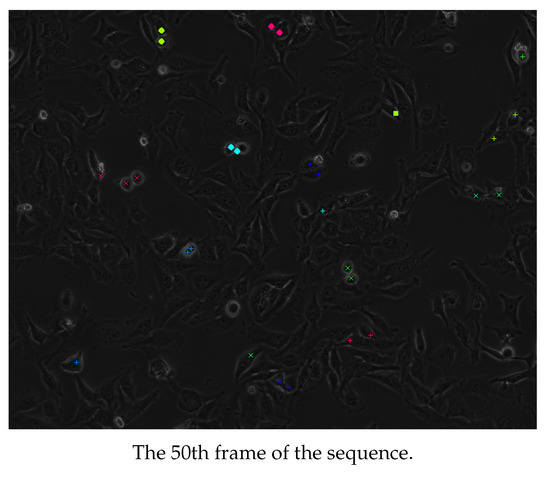
Figure 11.
Cells detected as dividing from an image sequence of the first group of Table 1. Adjacent cells with annotations of the same color and shape are siblings. It is recommended that the image be viewed in color and magnified.
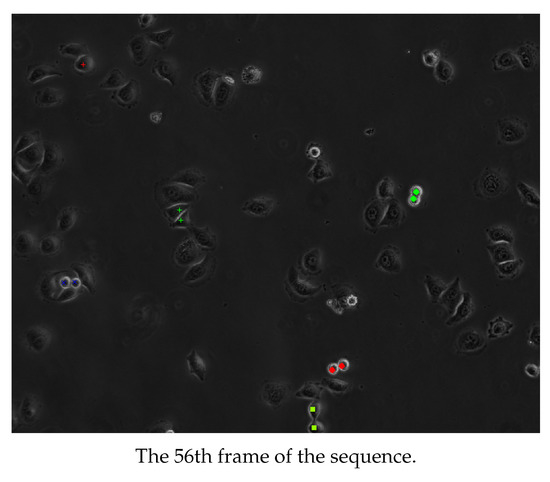
Figure 12.
Cells detected as dividing from an image sequence of the second group of Table 1. Adjacent cells with annotations of the same color and shape are siblings. It is recommended that the image be viewed in color and magnified.
Figure 13.
Cells detected as dividing from an image sequence of the third group of Table 1. Adjacent cells with annotations of the same color and shape are siblings. It is recommended that the image be viewed in color and magnified.
The results of the method for the fourteen sequences were evaluated by a scientist, MD, with specialty in radiology as well as a PhD in neuroscience and expertise in microscopy and biomedical quantitative image analysis. The evaluator annotated and counted the s, s, and s for every movie. These provided the , and the , which is equivalent to the , as well as the . The results of the evaluation of the method with these sequences are in Table 2. The evaluations for the sequences described in the columns of Table 1 are given in the corresponding groups of rows of Table 2. The results show that the overall performance of the method is very good. This is shown particularly by the as well as the .

Table 2.
Evaluation of the performance of the detections of cell divisions.
The cell divisions that happen before the beginning of the acquisition of the sequence are not considered even if apparent sibling cells are adjacent in the movies. Similarly, mitotic cells are ignored if the cleavage of their cytoplasms does not occur before the end of the sequence. Some were identified in the outer field of the movies close to the borders of the images. The cells close to the boundaries of the images may be blurry due to artifacts such as spherical aberrations of the lenses present in the outer field as well as due to artifacts of the outer field specific to phase contrast imaging. The method may also not detect cell divisions that occur partly beyond the borders of the frames of the registered sequence, in which case the s are ignored. A limited number of s were identified from planes originating from below or above the one imaged due to cells moving in and out of the imaged plane.
In many sequences, particularly those with dense samples, the contrast of many of the cells not undergoing mitosis is low and the cells may even be unnoticeable. In these cases it is also difficult to discriminate between adjacent cells. Thus, the presence of was only assessed qualitatively. The number of cells was still found to be significantly higher than the number of cells, that is, . Thus, the was also very high.
The average time to process the sequences in the first group in Table 1 was ≈16 min, the average time to process the sequences of the second group in Table 1 was ≈26 min, and the average time to process the sequences of the third group in the same table was ≈17 min. This short running time for all the steps of preprocessing and processing of the whole method demonstrates its efficient implementation.
4. Discussion and Future Work
This study is in the context of interpreting dynamic cellular data from phase contrast microscopy to follow and quantify cell motility and proliferation. In particular, the objectives of the developed method have been to detect mitoses associated with cell divisions as spatiotemporal events from phase contrast cellular video data. The method takes advantage of the circular symmetry of mitotic cells, the symmetry of cell division and cytokinesis, as well as the symmetry of the entire cell cycle.
4.1. Summary
The method starts with a necessary preprocessing of the image sequences. Individual frames in a time sequence can be subject to different lighting due to flickering of the microscopy lamp. Thus, some frames can be brighter or darker with jumps in contrast. The method accounts for this with normalization of the mean intensities of the frames of the sequences along time. This is followed by restoration for the spatial distribution of the intensity non-uniformity of the microscope lamp that is assumed constant along time in a sequence. The intensity restored images are processed with a circle Hough transform enhanced with the ability to “vote” only towards the centers of curvature. That is, “vote” only around the cell centroid in the cell interior. This enhancement improves the robustness of the transform for a dense cell sample and the discriminability of the cell centroids.
A sufficient number of cell centroids of parents as well as necessarily of daughter cells are then identified and serve as starting points for the extraction of cell trajectories. Their extraction proceeds both towards the positive as well as towards the negative time directions. Therefore, the extraction of a cell trajectory is non-causal. The extraction of the trajectory of a daughter cell in negative time direction enables the identification of a candidate cell division as being a connection to an already extracted trajectory consisting of the motion of its sibling daughter cell together with that of their parent cell. That is, candidate cell divisions are detected as a by-product of topological connectivity of trajectories at least one of which is that of a daughter cell extracted in the negative time direction. Thus, the cell division detection is anti-causal. The candidate cell divisions are also verified by testing within the corresponding frames that they indeed belong to the same cell. In cancer cell samples, with abnormalities in the cell cycle, the cell divisions can lead to more than two daughter cells. Such abnormal cell divisions are also detected by the proposed method. This is made possible by the more than two to one inter-frame pixel associations that are possible since they preserve the function property of the associations. These associations are estimated and followed on a per need basis for trajectory extraction. The method can detect cell trajectories around the cell division time point as well as a limited cell ancestry from a grandparent.
A parameter of the method for a sequence is the expected radius of the cells, . The value of the radius is set by considering the approximate size of the cells in the sample as well as of the actual image magnifications both optical and digital. The prior knowledge of the value of tunes the circle Hough transform to improve its discriminability for the daughter cells as well as its efficiency. The method has been found robust to the value of . The circle Hough transform is robust to cells which have a convex boundary as well. The value of is also used to restrict the motion displacement estimates, since mitotic and dividing cells have limited mobility. The second parameter, set to in Equation (8) for the circle Hough transform image sequence, affects the cut-off value for consideration of the cell centroids. The implementation of all of the preprocessing and processing steps of the method make efficiency considerations. The sequences are spatially subsampled, the pixels used for the circle Hough transform are pruned, the optimization for the registration uses spatial subsampling, and the inter-pixel associations are computed in a lazy fashion. The experiments used fourteen sequences of three different cell lines. The sequences were processed with the developed cell division detection method. The outputs were validated quantitatively in terms of , (), and even in terms of . The validation demonstrated the high performance of the method.
4.2. Discussion
A cell that rounds up and increases its contrast may be mitotic, but may fail to undergo cytokinesis. However, not all cells that round up and increase their contrast are mitotic. Thus the method requires that a detected rounded cell also undergoes cytokinesis that leads to two detected daughter cells to classify the event as a mitotic cell division. This may miss some divisions, that is, it may create some . However, this is much less than the that would result without the requirement for the presence of two daughter cells.
The method does not consider adjacent cells as being in cytokineses even if associated cell divisions appear to have happened immediately before the beginning of the sequence. It also does not consider mitotic cells even if they divide immediately after the end of the sequence. Cell divisions close to the image boundary may not be detected and lead to misses of the method for several reasons. Some cells, particularly close to the image boundaries, are blurry due to lens distortions and phase contrast imaging limitations. The boundary effects also limit Hessian filtering. Finally, due to compensation for misregistration shifts, the domain of some of the frames close to the image boundaries may not be part of the common domain of the sequence. In the latter two cases, the misses are ignored.
The cell division detection methods can be classified into tracking-based approaches, tracking-free approaches, and appearance-based approaches [6,31]. There exist trade-offs between these different types of approaches. Tracking-based methods can better represent the progression of mitoses and cell divisions over time. However, the performance of tracking-based methods can be lower for dense samples or in the presence of excessive cellular motion. They are also non-causal and involve delays in sequence processing. To address these issues, tracking-free approaches are also used. Tracking-free approaches suffer from limitations as well, specifically since mitoses and cell divisions are a temporal sequence of events that cannot be completely analyzed in each frame in time independently. On the other hand, appearance-based approaches with machine learning and neural networks, can detect cell divisions events, even spatiotemporally, robustly to their variations in appearance. However, appearance-based methods have several limitations. They require extensive training, they have not been shown able to generalize among cell lines to the authors knowledge, they are sensitive to dense samples, and do not characterize the complete mitoses and cytokineses events. In addition, in the last two types of approaches, namely, tracking-free and appearance-based, when treating spatiotemporal frame sequences as samples, the precise temporal resolutions for the cell divisions becomes weak.
The proposed method benefits from the spatial shapes and symmetries of the cells and the cell divisions rather than simply treating the sequences as signals or samples. It is also able to take advantage of the cell trajectories in space and time to robustly and uniquely detect cell divisions even in dense samples.
4.3. Future Work
The proposed method can be further improved and generalized in a variety of ways. The method was developed and validated with real experimental movies to demonstrate its applicability to the practice of a biological laboratory. A benchmark with movies of not only a single cell line [7,68], but of multiple cell lines, specifically acquired and annotated for method development can contribute to methodology improvements.
The imaging of the sequences analyzed was performed with positive phase contrast. This has been considered to interpret the Hessian-based filtering and to adjust the circle Hough transform as well as several other processing steps. In negative phase contrast, the appearance of the cells and of the halo effect around them is different. It requires a generalization in the interpretation of Hessian-based filtering in Equation (3) and of other subsequent processing steps. In low spatial resolution sequences, the thickness of the Halo effect at the boundary of the cells is larger and Hessian filtering must be adjusted accordingly.
The spatial intensity correction method computes the non-uniformity retrospectively from the entire sequence. To achieve real time intensity correction, the processing could be made causal by only considering preceding frames of the sequence. Further, the intensity uniformity restoration over time and over space could be performed jointly to better preserve contrast in the restored sequence.
The cell division detection with circle Hough transform based on shape symmetry can be made more specific by detecting circles of only a prespecified radius. That would avoid false positives from which conventional circle Hough transform in this context suffers due to “votes” into rings of smaller or larger radii as well as due to cells in interphase that may be elongated. A parent cell prior to its division has a larger radius compared to its daughter cells immediately following the division. Abnormal cell divisions that lead to three daughter cells have even smaller radii. Thus, it will be productive to extend the cell detection with multiresolution to consider prespecified cell radii for both before and after the cell division. Concerning the cell radii, another novelty in the extended circle Hough transform method would be to not only detect the direction towards the center of curvature, but also to detect the center of curvature itself. That would obviate the need to specify a cell radius parameter that is present explicitly or implicitly in most relevant methods in the literature. The cells in the sequences processed are convex. However, nerve cells can be concave and can have shapes that resemble some forms of starlike superquadrics [69]. The detection of such cells would require the development of a novel Hough transform methodology applicable to such superquadrics.
The circle Hough transform sequences are used to establish inter-frame pixel associations. For sequences with extensive cell motility, the associations can be established with the contribution of a stochastic motion tracking method as well [42]. The extraction of the trajectories of the daughter cells that drift apart can also use a stochastic motion tracking method [42]. The spatiotemporal angle between the trajectories of two daughter cells after a cell division is symmetric and is a “fingerprint” of a particular cell line. Abnormalities in the spatiotemporal division angle can also be a “fingerprint” of specific pathologies such as a wider neuronal precursor division angle for the Miller Diecker Lissencephaly syndrome that is causing retardation and epilepsy [70]. The spatiotemporal division angle can be considered as an additional condition in the methodology to determine whether to join two trajectories to form a cell division to improve the accuracy of the detection.
An abnormal cell division can lead to cell death, namely, apoptosis. The detection of such events would make the method more representative and further improve its performance. It would also be informative for high throughput screening applications to compute overall statistics for a movie, such as the number of cells dividing per minute, the duration of the cell cycle, the planar orientations of the mitotic axes, and the number of apoptoses. In a pharmaceutical context, the method can be used to quantify and study the response of treating cells with compounds whose objective is to affect the cell cycle by inhibiting mitotic progression or targeting abnormal cell divisions [42]. An example use would be for the development and screening of anti-cancer drugs.
Author Contributions
Conceptualization, S.H. and P.S.; data curation, R.H. and A.C.; methodology, S.H.; project administration, I.P. (Ioanna Panayiotou) and P.S.; resources, R.H. and A.C.; supervision, I.P. (Ioanna Panayiotou) and P.S.; validation, I.P. (Ismini Papageorgiou). All authors have read and agreed to the published version of the manuscript.
Funding
This research was co-funded by the European Regional Development Fund and the Republic of Cyprus through the Research and Innovation Foundation of Cyprus [Grant: INFRASTRUCTURES/1216/0060 to P.S.].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available on request.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| True Positive | |
| False Positive | |
| True Negative | |
| False Negative |
References
- Hadjidemetriou, S.; Gabrielli, B.; Pike, T.; Stevens, F.; Mele, K.; Vallotton, P. Detection and tracking of cell divisions in phase contrast video microscopy. In Proceedings of the Third International Workshop on Microscopic Image Analysis with Applications in Biology (MIAAB)-in Conjunction with MICCAI, New York, NY, USA, 6 September 2008. [Google Scholar]
- Yin, Z.; Kanade, T.; Chen, M. Understanding the phase contrast optics to restore artifact-free microscopy images for segmentation. Med. Image Anal. 2012, 16, 1047–1062. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yin, Z.; Kanade, T.; Huh, S. Phase contrast image restoration via dictionary representation of diffraction patterns. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Nice, France, 1–5 October 2012; Volume 7512, pp. 615–622. [Google Scholar]
- Huh, S.; Su, H.; Chen, M.; Kanade, T. Efficient phase contrast microscopy restoration applied for muscle myotube detection. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Nagoya, Japan, 22–26 September 2013; Volume 8149, pp. 420–427. [Google Scholar]
- Su, H.; Yin, Z.; Huh, S.; Kanade, T. Cell segmentation via spectral analysis on phase retardation features. In Proceedings of the IEEE 10th International Symposium on Biomedical Imaging (ISBI), San Francisco, CA, USA, 7–11 April 2013; pp. 1477–1483. [Google Scholar]
- Liu, A.; Lu, Y.; Chen, M.; Su, Y. Mitosis detection in phase contrast microscopy image sequences of stem cell populations: A critical review. IEEE Trans. Big Data 2017, 3, 443–457. [Google Scholar] [CrossRef]
- Su, Y.T.; Lu, Y.; Liu, J.; Chen, M.; Liu, A.A. Spatio-temporal mitosis detection in time-lapse phase-contrast microscopy image sequences: A benchmark. IEEE Trans. Med. Imaging 2021, 40, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Debeir, O.; Ham, P.; Kiss, R.; Decaestecker, C. Tracking of migrating cells under phase-contrast video microscopy with combined mean-shift processes. IEEE Trans. Med. Imaging 2005, 24, 697–711. [Google Scholar] [CrossRef]
- Altman, M.; Wang, S.; Whitlock, J.; Roeske, J. Cell detection in phase-contrast images used for alpha-particle track-etch dosimetry: A semi-automated approach. Phys. Med. Biol. 2005, 50, 305–318. [Google Scholar] [CrossRef]
- Kofahi, O.; Radke, R.; Roysam, B.; Banker, G. Automated semantic analysis of changes in image sequences of neurons in culture. IEEE Trans. Biomed. Eng. 2006, 53, 1109–1123. [Google Scholar] [CrossRef]
- Tscherepanow, M.; Zollner, F.; Kummert, F. Automatic segmentation of unstained living cells in bright-field microscope images. In Proceedings of the Industrial Conference on Data Mining-Workshops, Leipzig, Germany, 14–15 July 2006; pp. 86–95. [Google Scholar]
- Colin, F.; Cisneros, M.; Cervantes, J.; Martinez, J.; Debeir, O. Detection of biological cells in phase-contrast microscopy images. In Proceedings of the Fifth IEEE Mexican International Conference on Artificial Intelligence, Washington, DC, USA, 13–17 November 2006. [Google Scholar]
- Li, K.; Miller, E.; Weiss, L.; Campbell, P.; Kanade, T. Online tracking of migrating and proliferating cells imaged with phase-contrast microscopy. In Proceedings of the Conference on Computer Vision and Pattern Recognition Workshop (CVPRW’06), New York, NY, USA, 17–22 June 2006. [Google Scholar]
- Oliva, A.; Torralba, A. modeling the shape of the scene: A holistic representation of the spatial envelope. Int. J. Comput. Vis. 2001, 42, 145–175. [Google Scholar] [CrossRef]
- Liu, A.; Li, K.; Kanade, T. Mitosis sequence detection using hidden conditional random fields. In Proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI), Rotterdam, The Netherlands, 14–17 April 2010. [Google Scholar]
- Nie, W.; Cheng, H.; Su, Y. Modeling temporal information of mitotic for mitotic event detection. IEEE Trans. Big Data 2017, 3, 458–469. [Google Scholar] [CrossRef]
- Jayalakshmi, N. Cell lineage construction of neural progenitor cells. Int. J. Comput. Appl. 2014, 90, 40–47. [Google Scholar] [CrossRef]
- Gilad, T.; Bray, M.; Carpenter, A.; Raviv, T. Symmetry-based mitosis detection in time-lapse microscopy. In Proceedings of the IEEE 12th International Symposium on Biomedical Imaging (ISBI), Bridge, NY, USA, 16–19 April 2015; pp. 164–167. [Google Scholar]
- Miroslaw, L.; Chorazyczewski, A.; Buchholz, F.; Kittler, R. Correlation-based method for automatic mitotic cell detection in phase contrast microscopy. Comput. Recognit. Systems. Adv. Soft Comput. 2005, 30, 627–634. [Google Scholar]
- Liu, A.; Li, K.; Kanade, T. Spatiotemporal mitosis event detection in time-lapse phase contrast microscopy image sequences. In Proceedings of the IEEE International Conference on Multimedia and Expo, Singapore, 19–23 July 2010; pp. 161–166. [Google Scholar]
- Liu, A.; Hao, T.; Gao, Z.; Su, Y.; Yang, Z. Nonnegative mixed-norm convex optimization for mitotic cell detection in phase contrast microscopy. Comput. Math. Methods Med. 2013, 2013, 176272. [Google Scholar] [CrossRef] [PubMed]
- Kofahi, O.; Radke, R.; Goderie, S.; Shen, Q.; Temple, S.; Roysam, B. Automated cell lineage construction: A rapid method to analyze clonal development established with murine neural progenitor cells. Cell Cycle 2006, 5, 327–335. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wang, X.; Metaxas, D.; Mathew, R.; White, E. Cell segmentation for division rate estimation in computerized video time-lapse microscopy. In Proceedings of the Microscopy Image Analysis with Applications in Biology (MIAAB)–in Conjunction with MICCAI, San Jose, CA, USA, 20 January 2007; Available online: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/6431/643109/Cell-segmentation-for-division-rate-estimation-in-computerized-video-time/10.1117/12.717590.short (accessed on 13 July 2022).
- Wang, X.; He, W.; Metaxas, D.; Mathew, R.; White, E. Cell segmentation and tracking using texture adaptive snakes. In Proceedings of the IEEE International Symposium of Biomedical Imaging (ISBI), Washington, DC, USA, 12–15 April 2007. [Google Scholar]
- Yang, F.; MacKey, M.; Ianzini, F.; Gallardo, G.; Sonka, M. Cell segmentation, tracking, and mitosis detection using temporal context. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Palm Springs, CA, USA, 26–29 October 2005; pp. 302–309. [Google Scholar]
- Li, K.; Chen, M.; Kanade, T. Cell population tracking and lineage construction with spatiotemporal context. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Brisbane, Australia, 29 October–2 November 2007; pp. 295–302. [Google Scholar]
- Li, K.; Kanade, T. Cell population tracking and lineage construction using multiple-model dynamics filters and spatiotemporal optimization. In Proceedings of the International Workshop on Microscopic Image Analysis with Applications in Biology (MIAAB)-in Conjunction with MICCAI, Piscataway, NY, USA, 21 September 2007; Available online: https://www.ri.cmu.edu/publications/cell-population-tracking-and-lineage-construction-using-multiple-model-dynamics-filters-and-spatiotemporal-optimization/ (accessed on 13 July 2022).
- Li, K.; Miller, E.; Chen, M.; Kanade, T.; Weiss, L.; Campbell, P. Computer vision tracking of stemness. In Proceedings of the IEEE International Symposium of Biomedical Imaging (ISBI), Paris, France, 14–17 May 2008; pp. 847–850. [Google Scholar]
- Padfield, D.; Rittscher, J.; Roysam, B. Spatio-temporal cell segmentation and tracking for automated screening. In Proceedings of the IEEE International Symposium of Biomedical Imaging (ISBI), Paris, France, 14–17 May 2008; pp. 376–379. [Google Scholar]
- Becker, T.; Rapoport, D.H.; Mamlouk, A.M. From time lapse-data to genealogic trees: Using different contrast mechanisms to improve cell tracking. In Proceedings of the 9th IEEE International Symposium on Biomedical Imaging (ISBI), Barcelona, Spain, 2–5 May 2012; pp. 386–389. [Google Scholar]
- Grah, J.; Harrington, J.; Koh, S.; Pike, J.; Schreiner, A.; Burger, M.; Schoenlieb, C.; Reichelt, S. Mathematical imaging methods for mitosis analysis in live-cell phase contrast microscopy. Methods 2017, 115, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Miller, E.; Chen, M.; Kanade, T.; Weiss, L.; Campbell, P. Cell population tracking and lineage construction with spatiotemporal context. Med. Image Anal. 2008, 12, 546–566. [Google Scholar] [CrossRef]
- Bise, R.; Yin, Z.; Kanade, T. Reliable cell tracking by global data association. In Proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI), Chicago, IL, USA, 30 March–2 April 2011. [Google Scholar]
- A. Massoudi, D.S.; Sowmya, A. Cell tracking and mitosis detection using splitting flow networks in phase-contrast imaging. In Proceedings of the 34th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), San Diego, CA, USA, 28 August–1 September 2012; pp. 5310–5313. [Google Scholar]
- Zhou, X.; Li, F.; Yan, J.; Wong, S.T.C. A novel cell segmentation method and cell phase identification using Markov model. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, G.M.; Yang, F.; Ianzini, F.; Mackey, M.; Sonka, M. Mitotic cell recognition with hidden Markov models. In Medical Imaging 2004: Visualization, Image-Guided Procedures, and Display; Galloway, R.L., Jr., Ed.; International Society for Optics and Photonics, SPIE: San Diego, CA, USA, 2004; Volume 5367, pp. 661–668. [Google Scholar] [CrossRef]
- Liang, L.; Zhou, X.; Li, F.; Wong, S.; Huckins, J.; King, R. Mitosis cell identification with conditional random fields. In Proceedings of the IEEE/NIH Life Science Systems and Applications Workshop, Bethesda, MD, USA, 8–9 November 2007; pp. 9–12. [Google Scholar]
- Liu, A.; Li, K.; Hao, T. A hierarchical framework for mitosis detection in time-lapse phase contrast microscopy image sequences of stem cell populations. In Medical Imaging; InTech: Vienna, Austria, 2011; pp. 355–374. [Google Scholar]
- Huh, S.; Ker, D.F.E.; Bise, R.; Chen, M.; Kanade, T. Automated mitosis detection of stem cell populations in phase-contrast microscopy images. IEEE Trans. Med. Imaging 2011, 30, 586–596. [Google Scholar]
- Liu, A.; Tang, J.; Nie, W.; Su, Y. Multi-grained random fields for mitosis identification in time-lapse phase contrast microscopy image sequences. IEEE Trans. Med. Imaging 2017, 36, 1699–1710. [Google Scholar] [CrossRef]
- Liu, A.; Li, K.; Kanade, T. A semi-Markov model for mitosis segmentation in time-lapse phase contrast microscopy image sequences of stem cell populations. IEEE Trans. Med. Imaging 2012, 31, 359–369. [Google Scholar]
- Sherin, L.; Farwa, S.; Sohail, A.; Li, Z.; Bég, O. Cancer drug therapy and stochastic modeling of “nano-motors”. Int. J. Nanomed. 2018, 13, 6429–6440. [Google Scholar] [CrossRef]
- Ben-Haim, T.; Riklin-Raviv, T. Graph neural network for cell tracking in microscopy videos. arXiv 2022, arXiv:2202.04731. [Google Scholar]
- Shkolyar, A.; Gefen, A.; Benayahu, D.; Greenspan, H. Automatic detection of cell divisions (mitosis) in live-imaging microscopy images using convolutional neural networks. In Proceedings of the 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 743–746. [Google Scholar]
- Nie, W.; Li, W.; Liu, A.; Hao, T.; Su, Y. 3D convolutional networks-based mitotic event detection in time-lapse phase contrast microscopy image sequences of stem cell populations. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Las Vegas, NV, USA, 1–26 July 2016; pp. 1359–1366. [Google Scholar]
- Zhou, Y.; Mao, H.; Yi, Z. Cell mitosis detection using deep neural networks. Knowl. Based Syst. 2017, 137, 19–28. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Verma, E.; Singh, V.; Safwan, M. Mitosis detection in phase contrast microscopy image sequences using spatial segmentation and spatio-temporal localization refinement. In Proceedings of the IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1112–1116. [Google Scholar] [CrossRef]
- Mao, Y.; Yin, Z. A hierarchical convolutional neural network for mitosis detection in phase-contrast microscopy images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Athens, Greece, 17–21 October 2016; Volume 9901. [Google Scholar]
- Mao, Y.; Yin, Z. Two-stream bidirectional long short-term memory for mitosis event detection and stage localization in phase-contrast microscopy images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Quebec City, QC, Canada, 10–14 September 2017; Volume 10434, pp. 56–64. [Google Scholar]
- Mao, Y.; Han, L.; Yin, Z. Cell mitosis event analysis in phase contrast microscopy images using deep learning. Med. Image Anal. 2019, 57, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Lu, Y.; Chen, M.; Liu, A. Spatiotemporal joint mitosis detection using CNN-LSTM network in time-lapse phase contrast microscopy images. IEEE Access 2017, 5, 18033–18041. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, A.A.; Chen, M.; Nie, W.Z.; Su, Y.T. Sequential saliency guided deep neural network for joint mitosis identification and localization in time-lapse phase contrast microscopy images. IEEE J. Biomed. Health Inform. 2020, 24, 1367–1378. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. arXiv 2016, arXiv:1606.04797. [Google Scholar]
- Nishimura, K.; Bise, R. Spatial-temporal mitosis detection in phase-contrast microscopy via likelihood map estimation by 3DCNN. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1811–1815. [Google Scholar] [CrossRef]
- Hayashida, J.; Nishimura, K.; Bise, R. Consistent cell tracking in multi-frames with spatio-temporal context by object-level warping loss. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 4–8 January 2022; pp. 1759–1768. [Google Scholar] [CrossRef]
- Su, Y.T.; Lu, Y.; Chen, M.; Liu, A.A. Deep reinforcement learning-based progressive sequence saliency discovery network for mitosis detection in time-lapse phase-contrast microscopy images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Hough, P. Method and Means for Recognizing Complex Patterns. U.S. Patent No. 3,069,654, 18 December 1962. [Google Scholar]
- Rosenfeld, A. Picture Processing by Computer. ACM Comput. Surv. 1969, 1, 147–176. [Google Scholar] [CrossRef]
- Kimme, C.; Ballard, D.; Sklansky, J. Finding circles by an array of accumulators. Commun. ACM 1975, 18, 120–122. [Google Scholar] [CrossRef]
- Sled, J.; Zijdenbos, A.; Evans, A. A nonparametric method for automatic correction of intensity nonuniformity in MRI Data. IEEE Trans. Med. Imaging 1998, 17, 87–97. [Google Scholar] [CrossRef]
- Likar, B.; Maintz, J.; Viergever, M.; Pernus, F. Retrospective shading correction based on entropy minimization. J. Microsc. 1999, 197, 285–295. [Google Scholar] [CrossRef]
- Hadjidemetriou, S.; Studholme, C.; Mueller, S.; Weiner, M.; Schuff, N. Restoration of MRI data for intensity non-uniformities using local high order intensity statistics. Med. Image Anal. 2009, 13, 36–48. [Google Scholar] [CrossRef]
- Tustison, N.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 2010, 29, 1310–1320. [Google Scholar] [CrossRef] [PubMed]
- Marsden, J.; Tromba, A. Vector Calculus, 6th ed.; W.H. Freeman: New York, NY, USA, 2011. [Google Scholar]
- Gonzalez, R.; Woods, R. Digital Image Processing; Prentice Hall: Hoboken, NJ, USA, 1993. [Google Scholar]
- Borovik, A.; Katz, M.G. Who gave you the Cauchy–Weierstrass tale? The dual history of rigorous calculus. Found. Sci. 2011, 17, 245–276. [Google Scholar] [CrossRef][Green Version]
- Ker, D.; Eom, S.; Sanami, S.; Bise, R.; Pascale, C.; Yin, Z.; Huh, S.; Osuna-Highley, E.; Junkers, S.N.; Helfrich, C.J.; et al. Phase contrast time-lapse microscopy datasets with automated and manual cell tracking annotations. Sci. Data 2018, 5, 1–12. [Google Scholar] [CrossRef]
- Barr, A.H. Superquadrics and angle-preserving transformations. IEEE Comput. Graph. Appl. 1981, 1, 11–23. [Google Scholar] [CrossRef]
- Bershteyn, M.; Nowakowski, T.; Pollen, A.; Lullo, E.; Nene, A.; Wynshaw-Boris, A.; Kriegstein, A. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 2017, 20, 435–449.e4. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).