Abstract
In recent years, much research has focused on using machine learning (ML) for disease prediction based on gene expression (GE) data. However, many diseases have received considerable attention, whereas some, including Alzheimer’s disease (AD), have not, perhaps due to data shortage. The present work is intended to fill this gap by introducing a symmetric framework to predict AD from GE data, with the aim to produce the most accurate prediction using the smallest number of genes. The framework works in four stages after it receives a training dataset: pre-processing, gene selection (GS), classification, and AD prediction. The symmetry of the model is manifested in all of its stages. In the pre-processing stage gene columns in the training dataset are pre-processed identically. In the GS stage, the same user-defined filter metrics are invoked on every gene individually, and so are the same user-defined wrapper metrics. In the classification stage, a number of user-defined ML models are applied identically using the minimal set of genes selected in the preceding stage. The core of the proposed framework is a meticulous GS algorithm which we have designed to nominate eight subsets of the original set of genes provided in the training dataset. Exploring the eight subsets, the algorithm selects the best one to describe AD, and also the best ML model to predict the disease using this subset. For credible results, the framework calculates performance metrics using repeated stratified k-fold cross validation. To evaluate the framework, we used an AD dataset of 1157 cases and 39,280 genes, obtained by combining a number of smaller public datasets. The cases were split in two partitions, 1000 for training/testing, using 10-fold CV repeated 30 times, and 157 for validation. From the testing/training phase, the framework identified only 1058 genes to be the most relevant and the support vector machine (SVM) model to be the most accurate with these genes. In the final validation, we used the 157 cases that were never seen by the SVM classifier. For credible performance evaluation, we evaluated the classifier via six metrics, for which we obtained impressive values. Specifically, we obtained 0.97, 0.97, 0.98, 0.945, 0.972, and 0.975 for the sensitivity (recall), specificity, precision, kappa index, AUC, and accuracy, respectively.
1. Introduction
Alzheimer’s disease (AD) is the most common cause of dementia and memory loss, in addition to being a major cause of death. It is a chronic neurodegenerative disease that starts silently and worsens gradually over time [1]. In 2015, 47 million people worldwide were suffering from AD, costing more than USD 818 billion. Both figures are expected to rise as time goes by [2]. It is also anticipated that 1 out of every 85 people will have AD by 2050 [3].
There are many known symptoms of AD, the most common being the difficulty to remember recent events. As the disease advances, symptoms can include problems with orientation, language, self motivation, mood, memory, self-care, and behavior [4]. As the condition of AD patients deteriorates, they begin to withdraw from family and society. Gradually, body functions are lost, eventually leading to death. Although the speed of progression can vary, the typical life expectancy after AD becomes visible is 3 to 9 years [5]. Thus, early diagnosis of AD can even save lives, and that is where the present work comes in.
Traditionally, AD diagnosis has been primarily carried out via brain magnetic resonance imaging (MRI) and neuropsychological testing [6]. Recognizing the molecular-level of AD is lacking due to the difficulty of sampling posterior brains of normal and AD patients. Thankfully, recent trails have produced large-scale omics data for various brain areas. Using these data, it is easy nowadays to develop prediction methods, such as those in this article, whereby machine learning (ML) models are leveraged to diagnose AD as early as possible [7]. Such methods can also be advantageous to the patient in that they are convenient and inexpensive. It has been even shown that they can better predict AD than clinicians in certain circumstances [8]. This fact has led to much research focusing on ML application to AD diagnosis by using medical data in different forms, such as MRI.
MRI scans can be used with support vector machine (SVM), or variants thereof, to detect AD, as in [9], where an approach that leverages recursive feature elimination to select the features is introduced. The approach demonstrates higher accuracy in classifying mild cognitive impairment (MCI), control normal (CN), and AD cases (subjects or instances). A related SVM work is given in [10], where the authors incorporate universum to develop a twin SVM model. First, a universum hyperplane is constructed, then the classifying hyperplane is constructed by minimizing the angle with the universum hyperplane. The model is applied to AD detection and high-accuracy is reported. Another related SVM work appears in [11], where three variants of SVM classifiers are employed to detect AD using 30 features, selected from an original total of 420 features.
MRI scans can also be used with other ML models, especially neural networks and their derivatives, to detect AD, as in [12], where a convolutional neural network (CNN) is used for feature extraction, and k-means clustering is used to classify AD, MCI, and normal cognition (NC). The proposed method is reported to achieve high accuracy. CNN is also used in [13] to classify MCI and AD cases from normal (N) cases. They study the impact of incorporating data from MRI and diffusion tensor imaging (DTI). Their techniques achieve high-accuracy, specificity, and AUC values. CNN is used as well in [2] to recognize the patterns that identify each AD stage. To this end a time series is processed for each patient. In [14], partial least squares is used for dimensionality reduction, ANOVA is used for feature selection, and a random forest (RF) classifier is used for classification of AD. The authors in [15] use resting-state functional MRI and a deep learning (DL) technique to classify AD. They use an expanded network architecture to apply transfer learning with and without fine-tuning. In [16], the authors propose also a DL model for all level feature extraction and fuzzy hyperplane based least square twin support vector machine (FLS-TWSVM) for the detection of AD. Furthermore, in [17], the authors propose an ensemble of deep neural networks for the classification of AD. The proposed ensemble leverages the diversity introduced by many different locally optimal solutions reached by individual networks through the randomization of hyper-parameters. In [18], the authors propose an improved twin support vector regression model for brain age estimation, which can be helpful in mental health issues in general.
The diagnosis of various diseases is nowadays possible thanks to gene expression, which is the basis of the present work. Such data are obtained through the powerful technology of DNA microarrays [19]. It provides expression levels of thousands of genes [20]. The level of gene expression signifies the combination of different messenger molecule ribonucleic acid (mRNA) in the cell. By using this level, it is possible not only to detect diseases, but also to select the best treatment and discover mutations in other processes [21].
For example, the authors in [22] use a blood-derived gene expression biomarkers to distinguish AD cases from other sick and healthy cases. They use XGBoost classification models and succeed in detecting AD in a heterogeneous aging population by adding related mental and elderly health disorders. Nevertheless, improving the sensitivity of the model is still required to define a more specific blood signature to AD. In [23], three independent datasets, AddNeuroMed1 (ANM1), ANM2 and Alzheimer’s Disease Neuroimaging Initiative (ADNI), are used to distinguish AD from CN. Different gene selection (GS) methods, such as variational autoencoder, transcription factor, hub genes, and convergent functional genomics (CFG) are used to select the most informative genes. Five models, SVM, RF, logistic regression (LR), L1-regularized LR (L1-LR), and DNN, are employed for classification. The AUC values obtained are , , and for ANM1, ANM2, and ADNI, respectively. The authors also analyze the biological functions of the blood genes related to AD and compare the blood bio-signature with the brain bio-signature. They employ 1291 brain genes extracted from a gene expression dataset with 2021 blood genes extracted from the other three datasets, given that there are 140 common genes between the two. In [24], a study is presented to identify expression genes from a blood dataset, and to explore the correlation between the blood and brain genes of an AD patient. They identify 789 deferentially expressed genes common in both blood and the brain. Least absolute shrinkage and selection operator (LASSO) regression is used as a GS method. Logistic ridge regression (RR), SVM, and RF models are used for classification. They succeed in discriminating AD cases from control cases with accuracy. In [25], multiple brain regions are used to identify prospective diagnostic biomarkers of AD. Gene expression data from six brain regions are employed to determine AD biomarkers. A t-test is used to select the most informative genes. Significance tests are used to check those biomarkers and evaluate their potential for clinical diagnosis. The authors of [26] integrate gene expression and DNA methylation datasets, forming a multi-omics dataset, to predict AD using on a deep neural network (DNN). Principal component analysis (PCA) and t-stochastic nearest neighbor techniques are used to select and the most informative features. In [27], the authors use blood gene expression data obtained from the ANM and dementia case registry (DCR) cohorts. They employed recursive feature elimination for GS and used RF for classifying AD cases. They used ANM1 for training the classifier and integrated both ANM2 and DCR to use them for testing. They obtained for AUC and for ACC.
In Table 1, we present a summary of eminent studies to diagnose AD and to identify genes that qualify to be its biomarkers. The Table shows the original number of genes in each research work and the number of genes used after the GS step. We deduce that the number of selected genes has no obvious pattern or rule, and is largely dataset and model dependent. In other words, each diagnosis experiment can select a different subset of relevant genes and end up with a different accuracy value based on the ML model used. The table is also a testimony of the main obstacle facing the analysis of gene expression data—the small number of cases and the large number of genes.

Table 1.
Summary of some recent studies on the prediction of AD using gene expression data, employing different GS methods, and ML models.
In the present article, we propose a symmetric framework to predict AD, made of steps. First, we use a number of statistical metrics to evaluate the relevance of the genes of a dataset to AD prediction. We apply each metric individually, then average for each gene the values obtained from all applied metrics. Next, we select the genes that have the highest such averages, where highest is assessed with respect to some user defined threshold. Finally, we feed these genes into a number of ML models and monitor the classification performance. The model with the highest performance is considered for future use of the AD prediction system which is our ultimate outcome.
To validate the strategy, we use for gene evaluation the chi-squared (), analysis of variance (ANOVA), and mutual information (MI) metrics. They end up selecting the genes that are most relevant for AD detection. For classification, we use four different ML models, SVM, RF, LR, and AdaBoost. We keep on varying the number of informative genes to test which are essential for AD prediction. The AD prediction system obtained this way shows excellent results on four omics datasets.
The contribution of this article is multifaceted as follows.
- A comprehensive framework to diagnose AD from GE data;
- A novel GS methodology based on hybrid filter/wrapper selection methods;
- The use of 6 different performance metrics to evaluate the proposed framework;
- High-performance exceeding, as demonstrated by experimental results, state of the art GE-based AD prediction frameworks;
- An enrichment to the literature on AD prediction based GE data, which is admittedly poor compared to the literature on other diseases.
The article is organized as follows. In Section 2, we introduce the foundations and elements of the strategy used in our study. In Section 3, we validate the strategy by applying four specific ML models to classify AD cases with the datasets and display the results. In Section 4 we present our concluding remarks.
2. Materials and Methods
In this section, we introduce the proposed approach for GS and for AD classification, which is illustrated in Figure 1. The approach consists of four stages: integration of the datasets, preprocessing, GS, and classification. The details of the proposed approach are presented below.
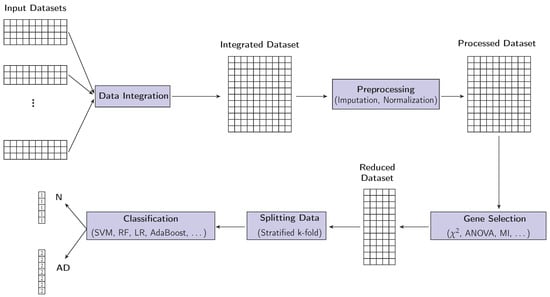
Figure 1.
Proposed symmetrical AD prediction framework. It takes as input a GE dataset, possibly by integrating a number of smaller datasets, as well as a set of classifiers. In a multistage operation, it selects a minimal set of genes to represent the data, identifies the best classifier, then finally gives as output the correct AD classification of an unseen case: positive/negative.
2.1. Integration of Datasets
A typical problem with gene expression data in general, including that of AD, is that the number of genes is huge (usually in the thousands) whereas the number of cases is small (usually in the tens). This imbalance makes classification a difficult problem. A possible solution is to concatenate two or more datasets, provided that they have the same set of genes, which we have done in the present study.
2.2. Preprocessing
We start this step by first normalizing the gene values in order to avoid heavy variations among the different genes. We use for normalization the min–max method which re-scales the range of values for each gene to the interval . In particular, given a set C of cases described by a set G of genes, the normalized gene value , and , of some gene value is given by
where and are the minimum and maximum values, respectively, of gene across all cases .
After normalization, we handle the problem of missing values, which is quite persistent in almost all experimental datasets. There are many approaches in the literature to handle missing values, and we have selected from them imputation, due to its simplicity and efficiency [28]. In particular, we compute the mean of the existing values for each gene to fill in the missing values of that gene. Let be the set of cases that have values for gene , and let be the complementary set of cases without a value for that gene. Then, for each gene , we assign to each case that does not have a value for gene the value
where denotes the cardinality of set x.
2.3. Gene Selection (GS)
Selecting genes relevant to AD prediction from the raw gene expression dataset is crucial. Simply, relevant genes are class, model, and dataset dependent. Meanwhile, inclusion of inconsequential and redundant genes can negatively affect the classification accuracy significantly. Thus, in our work, we pay special attention to gene selection. We, first, inspect the significance of each gene with respect to AD prediction. For that we use three filter-based metrics. Then, we evaluate the significance of each gene with respect to each of four ML models that we have used. At the end of these two stages, we can identify the most relevant genes and the most accurate model for predicting AD.
GS is particularly challenging because the number of genes is typically very large and the number of cases is typically very small. This imbalance can be noticed in Table 2. To overcome this problem, we introduce in the present work a novel scheme for GS. The scheme uses three symmetric filter-based techniques to rank the genes with respect to their ability to predict AD, , ANOVA (F statistic), and MI. Filter-based gene evaluation techniques are preferred due to their computational feasibility.

Table 2.
Summary of the four datasets integrated in the present study into one dataset of 1157 cases, each described by 39,280 genes.
- Chi squared ():is a well-known statistical metric used to examine the dependence between two random variables, in our situation a gene and the target output, which is the case diagnosis, AD or N. In order to calculate we first build a contingency table, having r rows, where r denotes the number of distinct gene values, and c columns, where c denotes the number of distinct classes of the target output, in our situation 2. At the () entry of the table, we place both the observed value and expected value for gene value i of class value j. The observed value is the number of times value i appears associated with class j, whereas the expected value is the fraction of times value i appears as a value for the gene, multiplied by the number of cases having class j. With this table at hand, thenThe higher the value, the more dependent the two variables, hence the more important the gene under consideration for predicting AD. Conversely, the smaller the value the more independent the two variables, and, hence, the more irrelevant the gene for predicting AD.
- Analysis of variance (ANOVA-Fstatistic):Analysis of variance (ANOVA) is a powerful family of techniques to test the significance of the difference between the means of two random variables. In our situation, the two variables are a gene and the target output, which is the case diagnosis, AD or N. The F statistic is one metric of the ANOVA family. For a given dataset with two classes, 1 and 2, the F statistic of a certain gene and the class variable is calculated, after first determining the sum of squares and degrees of freedom, as [14]where represents the number of cases with class 1, the number of cases with class 2, the mean of all values of the gene, the mean of the values of the gene with class 1, the mean of the values of the gene with class 2, the kth value, with class 1, of the gene, and the kth value, with class 2, of the gene. A larger F statistic value means that the gene is important for determining the class, AD or N, and vice versa.
- Mutual Information (MI):Let us first introduce entropy, which is a well-known metric in information theory. It is used as a measure of uncertainty in random variables. In particular, given a discrete random variable X, let be the probability that , where is the domain set of X. The entropy of X, denoted by , is given byHaving introduced entropy, we are in a position to introduce the mutual information which measures the shared information between two random variables X and Y. In our situation, the two variables are a gene and the target output, which is the diagnosis, AD or N. The MI is given bywhereis the conditional entropy of Y given X, with the joint distribution of X and Y and the domain of Y.
2.4. Classification
After identifying the genes most relevant to AD prediction, in the GS stage, the next stage in our framework is classification. In general, we can use any ML model for AD prediction, but we will focus in our experiments below on the four that proved most powerful for the task, as per the recent studies surveyed in Section 1, namely: SVM, RF, LR, and Adaboost [29]. The classification in the present work is binary, using the one-versus-all concept. That is, even if we provide our framework with a dataset of multiple classes, AD one of them, we consider AD one class and everything else the other class.
- 1.
- SVM:SVM is a famous supervised ML model that classifies data by first mapping, in a nonlinear way, the data to high-dimensional gene spaces. Then, it finds a linear optimal hyperplane, a decision boundary, to separate the points of one class from that of the other. SVM aims to maximize the distances (called functional margin) between the hyperplane and closest training data points of any type. The hyperplane, which is basically the SVM classifier, is expressed aswhere is a weight vector, b some bias and a nonlinear mapping. The optimal hyperplane is defined by and b that minimize the functionwhere the are some slack variables, n the number of cases, and A some factor.
- 2.
- RF:RF is a popular ensemble ML model, which means it combines predictions from multiple ML algorithms together to improve accuracy. In particular, it is a collection of decision trees, comprising a forest, trained with the bagging method. Prediction is made for a new case by a majority vote according to these steps. First, Given a set X of cases for training, , with labels , each node chooses a random case with g genes. Second, split the g genes and calculate the D node using the best split point, where D refers to next node. Third, continue splitting the tree until just one leaf node remains and the tree is complete. At this point, the algorithm is trained on each case individually. Finally, The prediction data from the n trained trees are collected by voting, and the highest votes are used to make the RF decision.
- 3.
- LR:LR is usually used to estimate or predict the probability of categorical variables, especially in binary classification. The logistic regression Sigmoid activation is defined asThe probability of the categorical dependent variable X equalswhere is the regression coefficient, determined by minimizing the cost function of logistic regression.
- 4.
- AdaBoostWith AdaBoost, predictions are made iteratively by computing the weighted average of the weak classifiers. The whole process can be summarized as follows. First, all cases in the training set are given the same weight. Second, a weak classifier is used to classify the cases, and the classification error rate is calculated, and used to update the weight of each case and to calculate the weight of the weak classifier in the next iteration. The classification error rate of the weak classifier for the training set is given bywhere , denotes input case i, denotes the labels of the classes, t is the current iteration number, is the prediction result of the weak classifier, is the true label, I is an indicator function that returns 1 for a correctly classified case and 0 for a misclassified case, and is the weight of the current weak classifier. The weights of the weak classifiers areBy combining the weak classifiers and optimizing their weights, the following strong classifier is obtainedwhere T is the total number of iterations and is the prediction result of weak classifier .
3. Experimental Work
In this section we report the findings of the extensive experimentation we carried out to validate the proposed framework and its GS Algorithm 1. The experimental work was carried out using a composite dataset made up of four multi-tissue GE profiles of the human brain for DNA microarray data. The profiles come from three different parts in the brains of AD patients, prefrontal cortex (PFC), visual cortex (VC), and cerebellum (CR). Downloaded from the National Center for Biotechnology Information-Gene Expression Omnibus (NCBI-GEO) database [30], the four datasets have the access numbers GSE33000, GSE44770, GSE44771, and GSE44768 [30], with GSE33000, GSE44770 [26] focusing exclusively on the PFC, GSE44771 on the VC, and GSE44768 on the CR [31]. We integrated the four gene-expression datasets carefully as the integration is known to be error prone [32]. Specifically, we integrated those datasets that were generated from the same platform (GPL4372), with normal (non-demented, healthy) present for control. The integrated dataset, summarized in Table 2, consists of 1157 cases, 697 AD and 460 Normal, each described by 39,280 genes.
At the outset, preprocessing and GS were performed on the integrated dataset as per Algorithm 1, which was coded in Python version 3.7.3 with the Scikit-learn packages. For reprehensibility and the common good, we have uploaded the code to the GitHub repository at the URL provided at the end of the article. The code was run on an Intel (R) Core (TM) i7-8550U CPU, 8 GB RAM, and 64-bit OS Win 10 configuration. The algorithm was used principally to select the most relevant and informative genes for AD and remove the remaining genes which would produce poor results if they remained. It was also used to identify the best classifier, out of four classifiers used, to work with those genes.
Once the dataset was pre-processed, the genes were then evaluated individually for their relevance in predicting AD, using the three filter metrics mentioned above. Part of the result of this evaluation is shown in Figure 2, which shows the 30 genes with the highest average of the three metrics. One can look at these 30 genes as the most relevant for predicting AD.
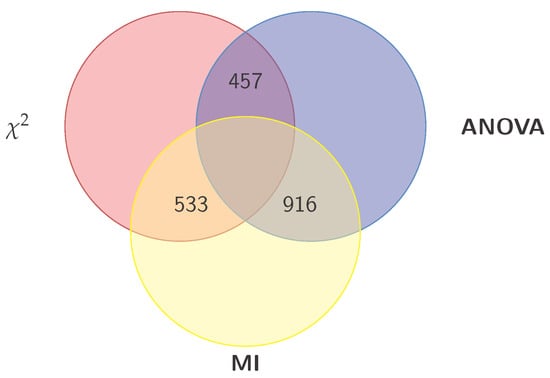
Figure 2.
Venn diagram of the three gene subsets obtained from , ANOVA, and MI. Five other subsets are generated (using the union and intersection operations) from these three, for a total of eight subsets, that are then explored by different classifiers. The exploration results in both the best subset to represent the data and the best classifier to predict the disease.
The crux of the present work is its unique GS methodology. The methodology depends basically on the three gene subsets , and , as a start. From these three subsets, we proceed as follows.
- Construct from the above three sets, the following four intersection sets:
- Construct from the last three sets, the following set:
We then train and test, using a repeated stratified k fold cross validation approach, every classifier on each of the above 8 sets, calculating the six performance metrics (sensitivity (recall), specificity, precision, kappa index, AUC, and accuracy) in the process. Simply, the best classifier and best gene subset (out of these 8 subsets) will be the ones that produce the highest values for the metrics (or the majority of them).
Proceeding with Algorithm 1, the final step was to apply the four ML models to the integrated dataset, with increasing numbers of genes, at an increment size , and calculate the accuracy. For this exercise, we first partitioned the input dataset into two sets of cases: 1000 cases for training/testing and 157 cases for final validation. The 1000 cases were used for the selection of the best classifier, and the 157 cases were isolated to test that classifier with. The objective of this isolation is to ensure the credibility of the performance of the classifiers, since it would classify cases it never saw before. For further credibility, we used repeated stratified k-fold cross validation, with and the repetition number being 30. That is, in each fold of the cases was used for training and for testing, and this was repeated 30 times, for a total of 300 tests. In other words, for each model, on each fold, six performance metrics were evaluated. This process is repeated 30 times, for a total of 300 times. The results of the 10 folds are averaged, providing at the end of the test only 30 values for each metric, one value per repetition.
| Algorithm 1: GS and best classifier identification, using enhanced filter-based methods and multiple hand-crafted gene subsets. |
Input : // AD gene expression dataset for training/testing // AD gene expression dataset for validation //Set of genes //Ranking increment Output: //8 gene subsets //Pre-processing:  |
We found out that the highest performance was consistently that of the SVM model, when used with the 700 genes with the highest value, 1000 genes with the highest ANOVA (F statistic) value, and 1700 genes with the highest MI vlaue. Having identified these three sets of genes, we began to get their pairwise intersections, as per the proposed Algorithm, which are depicted visually in Figure 2. As can be seen, the intersection between the and ANOVA sets contains 457 genes, between ANOVA and MI contains 916 genes, and finally between , and MI contains 533 genes. Having obtained the pairwise intersections, we then obtained their union which contains 1058 genes. As can be seen from the bar charts, the 1058 genes of the subset represent the most relevant (producing the highest performance) for predicting AD by the SVM model.
Incidentally, We compared the genes selected by Algorithm 1 with the list of genes reported in the well-known database AlzGene [33], which represents 695 influential AD genes obtained from 1395 studies, and found that the 30 genes of Table 3 are actually in that database.

Table 3.
Listing of the 30 genes having the highest averages of the three metrics: , ANOVA, and MI, and overlap with the AlzGene database of the most influential AD genes.
Figure 3, Figure 4, Figure 5, Figure 6, Figure 7 and Figure 8 show the results of the training/test phase for six metrics, sensitivity (recall), specificity, precision, kappa index, AUC and accuracy, whose equations can be found in any ML reference, see e.g., [29]. Each figure displays the values of this metric for for four classifiers, SVM, RF, LR and Adaboost, and eight gene subsets. As mentioned earlier, the training/test phase was carried out on only 1000 cases of the original dataset of 1157 cases. Further, in this phase, the testing was done using a repeated stratified k-fold approach, with and the number of repetitions equal 30, to ensure credible results. It is evident from the Figures that best performing classifier, the one with the highest values of the metrics, is the SVM classifier. It is also evident that the gene subset associated with this high performance is the subset, which contains 1058 genes out of an original number of 39,280 genes. For the box plot of Figure 8, we plotted the 30 accuracy values obtained from the 30 repetitions. The 10 results of the 10-fold in each repetition were first averaged, producing only one value, which was then considered the value of one repetition.
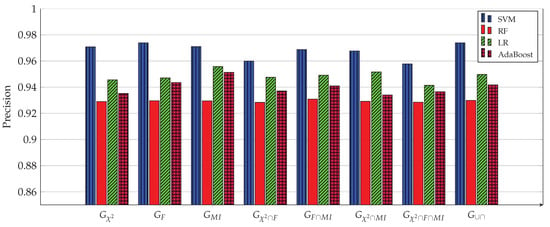
Figure 3.
Precision of four ML models for 8 gene subsets. The SVM model achieves the highest precision (0.9739) when used with the 1058 genes of the subset.
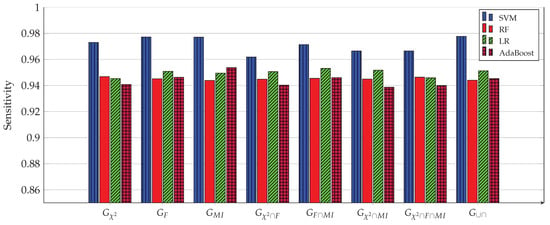
Figure 4.
Sensitivity (recall) of all four ML models for 8 gene subsets. The SVM model achieves the highest sensitivity (0.9777) when used with the 1058 genes of the subset.
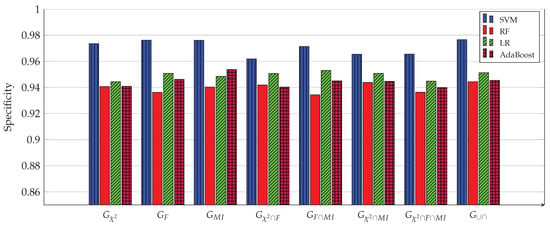
Figure 5.
Specificity of all four ML models for 8 gene subsets. The SVM model achieves the highest specificity (0.9766) when used with the 1058 genes of the subset.
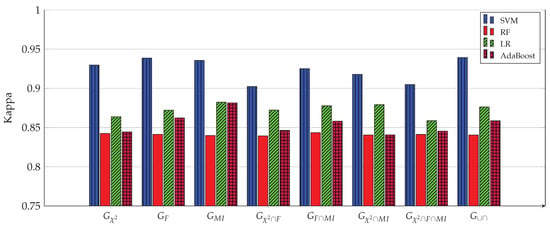
Figure 6.
Kappa index of all four ML models for 8 gene subsets. The SVM model achieves the highest kappa (0.9393) when used with the 1058 genes of the subset.
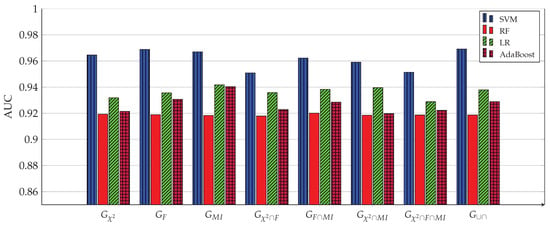
Figure 7.
AUC of all four ML models for 8 gene subsets. The SVM model achieves the highest AUC (0.9692) when used with the 1058 genes of the subset.
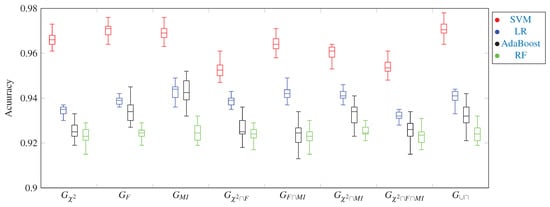
Figure 8.
A box plot of the accuracy metric of four ML models for 8 gene subsets. Once again in this plot, we can see that the SVM model achieves the highest accuracy when used with the 1058 genes of the subset.
After finishing the training/test phase, we moved on to the validation phase, where the best classifier, SVM, was evaluated on the remaining 157 cases, using the minimal gene subset, . As was the situation in the training/testing phase, the validation was done using a repeated stratified k-fold approach, with and the number of repetitions equal 30, to ensure credible results. Table 4 shows the confusion matrix resulting from the validation phase. This matrix was used to calculate the six performance metrics of the SVM when the gene subset is used. Table 5 shows the impressive values of 0.97, 0.97, 0.98, 0.945, 0.972, and 0.975 for the sensitivity (recall), specificity, precision, kappa index, AUC and accuracy, respectively. Compared with the results of the state of the art shown in Table 1, these values are much better. In fact, the proposed framework could achieve the same results of Table 1 using much fewer genes, telling how powerful our framework is, and how selective our GS algorithm can be.

Table 4.
Confusion matrix of the final validation experiment on the best classifier—SVM. The experiment was carried out on 157 cases never seen by the classifier in any training/testing, using 1058 genes selected by the proposed algorithm out of an original total of 39,280 genes.

Table 5.
Validation results of the SVM classifier obtained from 157 cases that were kept for final testing of the best classifier, using the the 1058 genes of the () subset.
Code of the experimental work is available at:https://github.com/aliaa2007/AD_Classification (accessed on 22 February 2022).
4. Conclusions
In this article, we have presented a framework for the prediction of a disease that has not found enough attention in the literature—Alzheimer’s disease (AD), using GE data. The framework has been shown to predict AD from GE data accurately and with a minimal number of genes, compared with recently published competitive frameworks. We have developed an efficient algorithm for GS and used it to identify the most relevant genes for AD prediction. The algorithm produces eight sets of genes, which are then explored by a number of ML models. The best model is the one that achieves the highest performance with the smallest number of genes. In our experiments on an integrated dataset of 39,280 genes, this model has turned out to be SVM. It reached an accuracy of 97.5% using the 1058 hand-crafted genes, obtained by intersections and unions of genes with high filter values. The framework is characterized by its openness and symmetry. It can deal any number of ML models, any number of filter metrics and any number of genes. It can be generalized for other diseases as well. It demonstrated experimentally that it outperforms the state of the art in that it either achieves the same performance with fewer genes or higher performance with the same number of genes.
The present work, though reliable and meticulous, has one limitation as far as we can see. Specifically, it predicts an input case for being only AD or normal, meaning it is based on a binary classification scheme. As such, it cannot predict the various stages of AD, for example, which require a multi-classification scheme. We intend to address this limitation in a future work, producing a more powerful framework that is capable of predicting two or more stages of AD.
Author Contributions
Conceptualization, A.E.-G.; methodology, A.E.-G., M.A.M., B.S.T. and H.N.; software, A.E.-G., M.A.M., B.S.T. and H.N.; validation, A.E.-G., M.A.M., B.S.T. and H.N.; formal analysis, A.E.-G., M.A.M. and H.N.; investigation, A.E.-G., M.A.M., B.S.T. and H.N.; resources, A.E.-G., M.A.M., B.S.T. and H.N.; data curation, A.E.-G., M.A.M. and H.N.; writing–original draft preparation, A.E.-G.; writing–review and editing, A.E.-G. and H.N.; visualization, A.E.-G., M.A.M., B.S.T. and H.N.; supervision, H.N.; project administration, A.E.-G., M.A.M., B.S.T. and H.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tanveer, M.; Richhariya, B.; Khan, R.; Rashid, A.; Khanna, P.; Prasad, M.; Lin, C. Machine learning techniques for the diagnosis of Alzheimer’s disease: A review. TOMM 2020, 16, 1–35. [Google Scholar] [CrossRef]
- Bringas, S.; Salomón, S.; Duque, R.; Lage, C.; Montaña, J.L. Alzheimer’s disease stage identification using deeplearning models. J. Biomed. Inform. 2020, 109, 103514. [Google Scholar] [CrossRef]
- Wang, S.H.; Phillips, P.; Sui, Y.; Liu, B.; Yang, M.; Cheng, H. Classification of alzheimer’s disease based on eightlayer convolutional neural network with leaky rectified linear unit and max pooling. J. Med. Syst. 2018, 42, 85. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; He, Y.; Ji, J.; Shi, Y. A machine learning method for identifying critical interactions between gene pairs in alzheimer’s disease prediction. Front. Neurol. 2019, 10, 1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhao, Y.; Chen, X.; Xiao, Y.; Qin, Y. Detecting alzheimer’s disease on small dataset: A knowledge transfer Perspective. IEEE J Biomed Health Inform. 2018, 23, 1234–1242. [Google Scholar] [CrossRef]
- Bryan, R.N. Machine learning applied to Alzheimer disease. Radiology 2016, 281, 665–668. [Google Scholar] [CrossRef] [Green Version]
- Neelaveni, J.; Devasana, M.G. Alzheimer disease prediction using machine learning algorithms. In Proceedings of the 6th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 6–7 March 2020. [Google Scholar]
- Alam, S.; Kwon, G.R. Alzheimer disease classification using KPCA, LDA, and multi-kernel learning SVM. Int. J. Imaging Syst. Technol. 2017, 27, 133–143. [Google Scholar] [CrossRef]
- Richhariya, B.; Tanveer, M.; Rashid, A.H. Diagnosis of Alzheimer’s disease using universum support vector machine based recursive feature elimination (USVM-RFE). Biomed Signal Process Control 2020, 59, 101903. [Google Scholar] [CrossRef]
- Richhariya, B.; Tanveer, M. An efficient angle-based universum least squares twin support vector machine for classification. ACM Trans. Internet Technol. 2021, 21, 1–24. [Google Scholar] [CrossRef]
- Khan, R.U.; Tanveer, M.; Pachori, R.B. A novel method for the classification of Alzheimer’s disease from normal controls using magnetic resonance imaging. Expert Systems 2021, 38, e12566. [Google Scholar] [CrossRef]
- Bi, X.; Li, S.; Xiao, B.; Li, Y.; Wang, G.; Ma, X. Computer aided alzheimer’s disease diagnosis by an unsupervised deep learning technology. Neurocomputing 2020, 392, 296–304. [Google Scholar] [CrossRef]
- Marzban, E.N.; Eldeib, A.M.; Yassine, I.A.; Kadah, Y.M. Alzheimer’s disease diagnosis from diffusion tensor images using convolutional neural networks. PLoS ONE 2020, 15, e0230409. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, J.; Górriz, J.M.; Ortiz, A.; Martínez-Murcia, F.J.; Segovia, F.; Salas-Gonzalez, D.; Castillo-Barnes, D.; Illán, I.A.; Puntonet, C.G. Ensemble of random forests One vs. Rest classifiers for MCI and AD prediction using ANOVA cortical and subcortical feature selection and partial least squares. J. Neurosci. Methods 2018, 302, 47–57. [Google Scholar] [CrossRef]
- Ramzan, F.; Khan, M.U.G.; Rehmat, A.; Iqbal, S.; Saba, T.; Rehman, A.; Mehmood, Z. A deep learning approach for automated diagnosis and multi-class classification of alzheimer’s disease stages using resting-state fmri and residual neural networks. J. Med. Syst. 2020, 44, 37. [Google Scholar] [CrossRef]
- Sharma, R.; Goel, T.; Tanveer, M.; Murugan, R. FDN-ADNet: Fuzzy LS-TWSVM based deep learning network for prognosis of the Alzheimer’s disease using the sagittal plane of MRI scans. Appl. Soft Comput. 2022, 115, 108099. [Google Scholar] [CrossRef]
- Tanveer, M.; Rashid, A.H.; Ganaie, M.A.; Reza, M.; Razzak, I.; Hua, K.L. Classification of Alzheimer’s disease using ensemble of deep neural networks trained through transfer learning. IEEE J Biomed Health Inform. 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ganaie, M.A.; Tanveer, M.; Beheshti, I. Brain age prediction using improved twin SVR. Neural. Comput. Appl. 2022, 1–11. [Google Scholar] [CrossRef]
- Ayyad, S.M.; Saleh, A.I.; Labib, L.M. Gene expression cancer classification using modified K-Nearest Neighbors technique. Biosystems 2019, 176, 41–51. [Google Scholar] [CrossRef]
- Vanitha, C.D.A.; Devaraj, D.; Venkatesulu, M. Gene expression data classification using support vector machine and mutual information-based gene selection. Procedia Comput. Sci. 2015, 47, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Ayyad, S.M.; Saleh, A.I.; Labib, L.M. A new distributed feature selection technique for classifying gene expression data. Int. J. Biomath. 2019, 12, 1950039. [Google Scholar] [CrossRef]
- Patel, H.; Iniesta, R.; Stahl, D.; Dobson, R.J.; Newhouse, S.J. Working Towards a Blood-Derived Gene Expression Biomarker Specific for Alzheimer’s Disease. J. Alzheimer’s Dis. 2022, 74, 545–561. [Google Scholar] [CrossRef]
- Lee, T.; Lee, H. Prediction of Alzheimer’s disease using blood gene expression data. Sci. Rep. 2020, 10, 3485. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, H.; Long, J.; Pan, G.; He, T.; Anichtchik, O.; Belshaw, R.; Albani, D.; Edison, P.; Green, E.K.; et al. Systematic analysis and biomarker study for Alzheimer’s disease. Sci. Rep. 2018, 8, 17394. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Z.P. Detecting diagnostic biomarkers of Alzheimer’s disease by integrating gene expression data in six brain regions. Front. Genet. 2019, 10, 157. [Google Scholar] [CrossRef]
- Park, C.; Ha, J.; Park, S. Prediction of Alzheimer’s disease based on deep neural network by integrating gene expression and DNA methylation dataset. Expert Syst. Appl. 2020, 140, 112873. [Google Scholar] [CrossRef]
- Voyle, N.; Keohane, A.; Newhouse, S.; Lunnon, K.; Johnston, C.; Soininen, H.; Kloszewska, I.; Mecocci, P.; Tsolaki, M.; Vellas, B.; et al. A pathway based classification method for analyzing gene expression for Alzheimer’s disease diagnosis. J. Alzheimer’s Dis. 2016, 49, 659–669. [Google Scholar] [CrossRef] [Green Version]
- De Souto, M.C.; Jaskowiak, P.A.; Costa, I.G. Impact of missing data imputation methods on gene expression clustering and classification. BMC Bioinform. 2015, 16, 64. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, C.C. Machine Learning for Text; Springer: New York, NY, USA, 2018. [Google Scholar]
- Gene Expression Omnibus. Available online: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 22 January 2022).
- Zhang, B.; Gaiteri, C.; Bodea, L.G.; Wang, Z.; McElwee, J.; Podtelezhnikov, A.A.; Zhang, C.; Xie, T.; Tran, L.; Dobrin, R.; et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 2013, 153, 707–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fajarda, O.; Duarte-Pereira, S.; Silva, R.M.; Oliveira, J.L. Merging microarray studies to identify a common gene expression signature to several structural heart diseases. BioData Min. 2020, 13, 8. [Google Scholar] [CrossRef]
- AlzGene. Available online: http://www.alzgene.org/ (accessed on 22 January 2022).
- Amidfar, M.; de Oliveira, J.; Kucharska, E.; Budni, J.; Kim, Y.K. The role of CREB and BDNF in neurobiology and treatment of alzheimer’s disease. Life Sci. 2020, 257, 118020. [Google Scholar] [CrossRef]
- Bobińska, K.; Gałecka, E.; Szemraj, J.; Gałecki, P.; Talarowska, M. Is there a link between tnf gene expression and cognitive deficits in depression? Acta Biochim. Pol. 2017, 64, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Paudel, Y.N.; Angelopoulou, E.; Piperi, C.; Othman, I.; Aamir, K.; Shaikh, M. Impact of HMGB1, RAGE, and TLR4 in Alzheimer’s disease (AD): From risk factors to therapeutic targeting. Cells 2020, 9, 383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.R.; Smith, R.G.; Pishva, E.; Hannon, E.; Roubroeks, J.A.; Burrage, J.; Troakes, C.; Al-Sarraj, S.; Sloan, C.; Mill, J.; et al. Parallel profiling of DNA methylation and hydroxymethylation highlights neuropathology-associated epigenetic variation in Alzheimer’s disease. Clin. Epigenetics 2019, 11, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).