Outlier Based Skimpy Regularization Fuzzy Clustering Algorithm for Diabetic Retinopathy Image Segmentation
Abstract
1. Introduction
2. Related Works
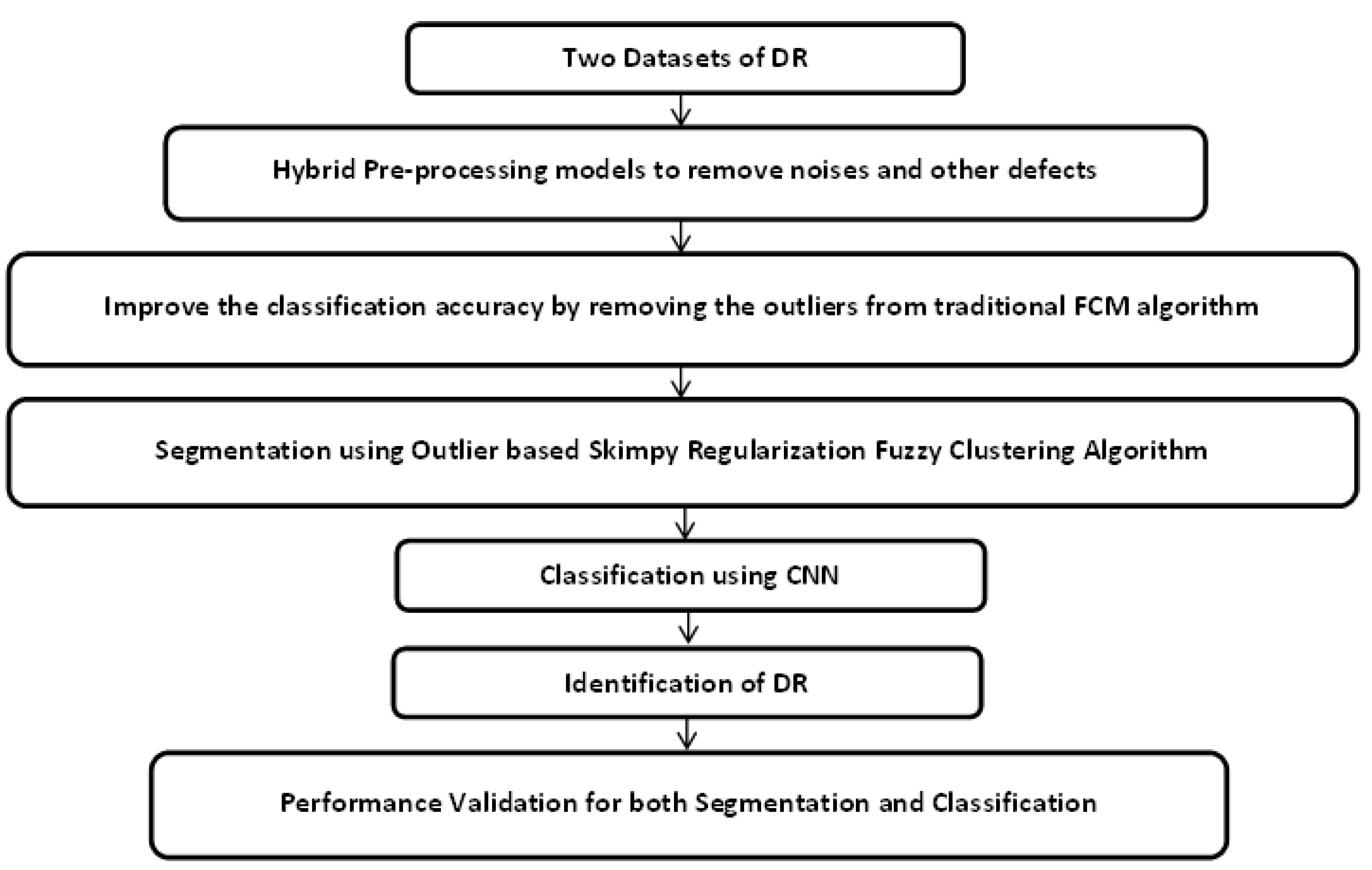
3. Proposed Methodology
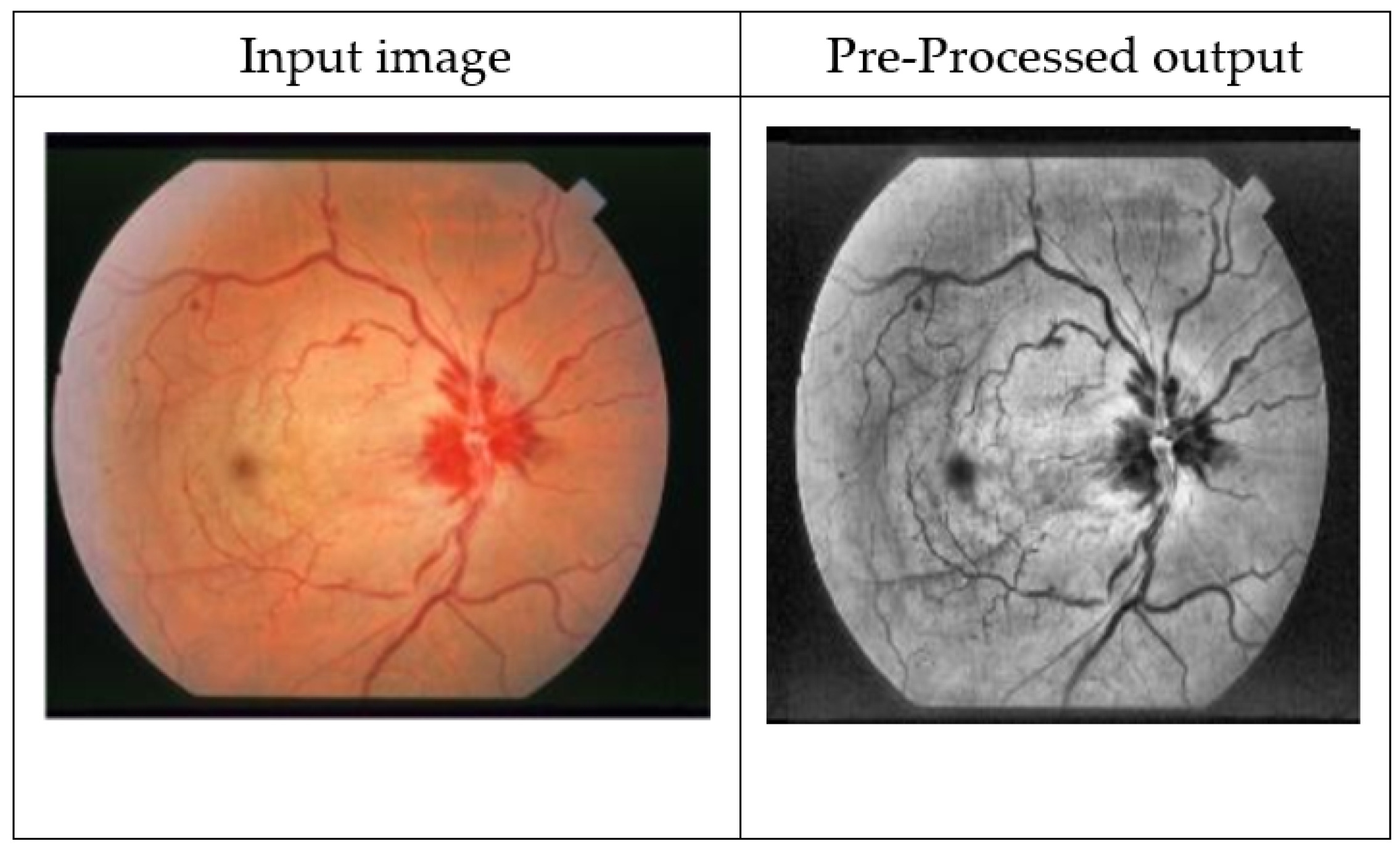
3.1. Pre-Processing
3.2. Segmentation
Brief Description of OSR-FCA
- (1)
- Set the number of clusters c, regularization parameter , convergence threshold , and maximum iteration number T.
- (2)
- Initialize the membership , the clustering centres , and the covariance matrix using the FCM algorithm.
- (3)
- Set the loop counter .
- (4)
- (5)
- Update the objective function using Equation (6).
- (6)
- If max stop; otherwise, update and go to step 4.
3.3. Classification Using Deep Learning Network
- (1)
- Local Connectivity: For example, the first layer of units receives data solely from the pixels in their receptive field (RF), which is a narrow rectangle of picture pixels (for the subsequent layers). Units in a layer are normally spaced apart by a stride. The layer’s dimensions are determined by the combined effects of image size, RF size, and stride. Because the image is 5 × 5 monochromatic (a single-channel image), just 9 units are needed to cover the entire area of the image with a layer of 3 × 3 units with one-pixel strides. Smaller layers result from greater strides and larger RFs. Comparing fully-connected traditional networks to those with local connectivity, the number of weights is drastically reduced. The spatial nature of visual information is also consistent, and several elements of natural visual systems are mimicked by this method [27].
- (2)
- Parameter Sharing: In which weights are shared across units in the same tier. It is possible to create a feature map when the units in a given layer all have the same vector of weights, but each calculates a separate local feature from the image. As a result, the derived features are equivariant, significantly reducing the number of parameters. For example, regardless of the number of units, a layer of units with three three RFs coupled to single-channel image requires just 10 parameters.
- (3)
- Pooling: Convolution is not the only way to combine the outputs of many units, but it is the most common one. Most commonly, max-pooling aggregates data so that each aggregating unit can return its RF’s full potential. Translational invariance is provided through pooling, which degrades resolution in relation to the prior layer.
- (4)
- Slide RFs across an input image by the number of pixels defined in stride makes subsequent layers to be smaller, therefore the final grid sent into the fully-connected is frequently considerably smaller than the initial image. It is common to see multiple feature maps running in tandem, each extracting a different feature. Several dozens of feature maps may be required for large networks [28]. If an image has more than one channel, such as RGB, then distinct feature maps are used to connect the various channels of information. It is possible to mix data from various maps in the previous layer in the succeeding layers. If a unit has numerous RFs with different weight vectors, the composed constitute the excitation of that unit.
3.4. Network Training
4. Results and Discussion
4.1. Dataset Description
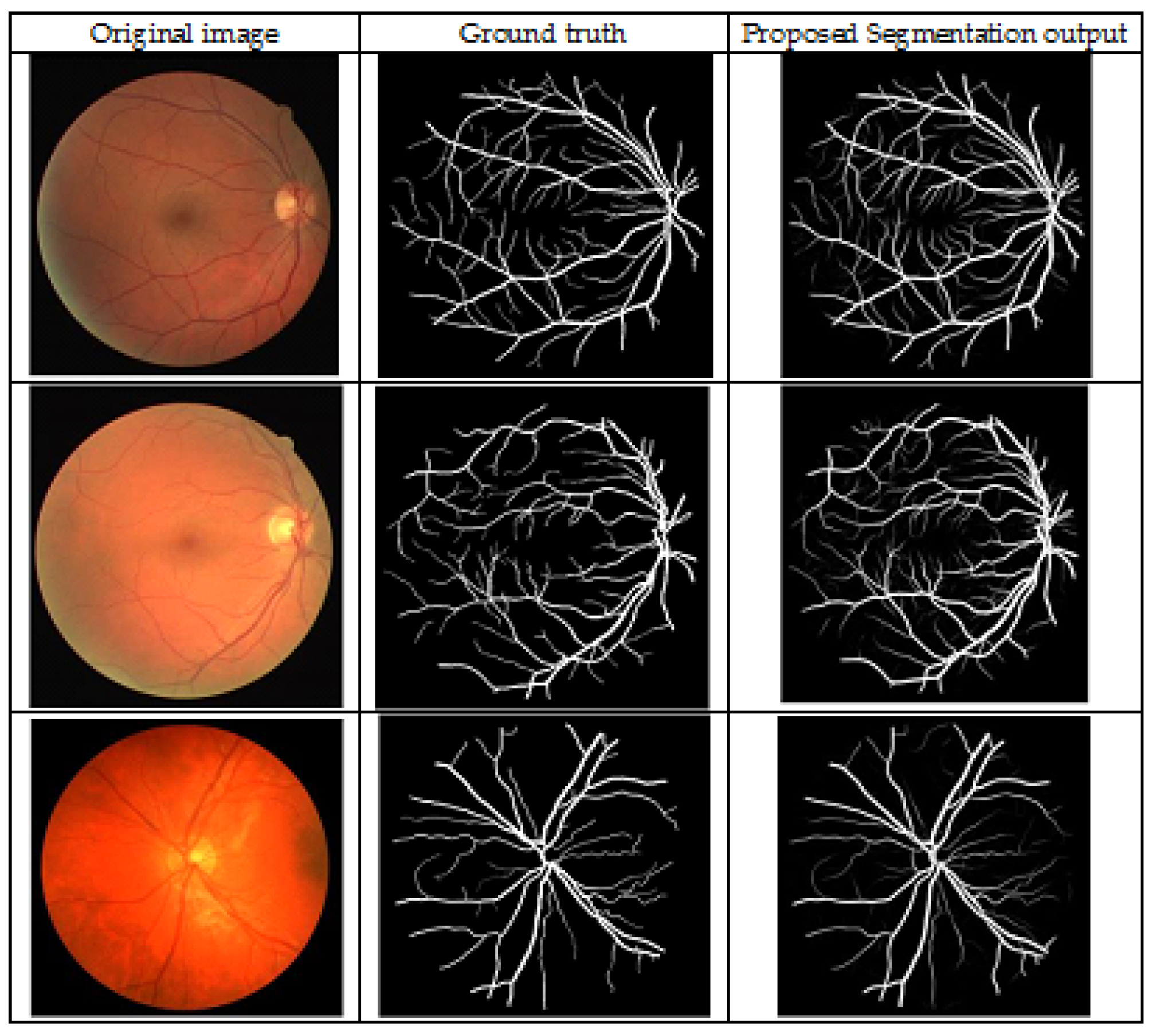
4.2. Segmentation Analysis
4.2.1. Evaluation Metrics
4.2.2. Discussion
4.3. Classification Analysis
Performance Measure
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Aswini, S.; Suresh, A.; Priya, S.; Santhosh Krishna, B.V. Retinal vessel segmentation using morphological top hat approach on diabetic retinopathy images. In Proceedings of the 2018 Fourth International Conference on Advances in Electrical, Electronics, Information, Communication and Bio-Informatics (AEEICB), Chennai, India, 27–28 February 2018. [Google Scholar]
- Xue, J.; Yan, S.; Qu, J.; Qi, F.; Qiu, C.; Zhang, H.; Chen, M.; Liu, T.; Li, D.; Liu, X. Deep membrane systems for multitask segmentation in diabetic retinopathy. Knowl. Based Syst. 2019, 183, 104887. [Google Scholar] [CrossRef]
- Li, Q.; Fan, S.; Chen, C. An intelligent segmentation and diagnosis method for diabetic retinopathy based on improved U-NET network. J. Med. Syst. 2019, 43, 304. [Google Scholar] [CrossRef] [PubMed]
- Jebaseeli, T.J.; Deva Durai, C.A.; Peter, J.D. Retinal blood vessel segmentation from diabetic retinopathy images using tandem PCNN model and deep learning based SVM. Optik 2019, 199, 163328. [Google Scholar] [CrossRef]
- Jebaseeli, T.J.; Durai, C.A.D.; Peter, J.D. Segmentation of retinal blood vessels from ophthalmologic Diabetic Retinopathy images. Comput. Electr. Eng. 2019, 73, 245–258. [Google Scholar] [CrossRef]
- Gupta, A.; Chhikara, R. Diabetic retinopathy: Present and past. Procedia Comput. Sci. 2018, 132, 1432–1440. [Google Scholar] [CrossRef]
- Furtado, P.; Travassos, C.; Monteiro, R.; Oliveira, S.; Baptista, C.; Carrilho, F. Segmentation of Eye Fundus Images by density clustering in diabetic retinopathy. In Proceedings of the 2017 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Orlando, FL, USA, 16–19 February 2017. [Google Scholar]
- Aujih, A.B.; Izhar, L.I.; Mériaudeau, F.; Shapiai, M.I. Analysis of retinal vessel segmentation with deep learning and its effect on diabetic retinopathy classification. In Proceedings of the 2018 International Conference on Intelligent and Advanced System (ICIAS), Kuala Lumpur, Malaysia, 13–14 August 2018. [Google Scholar]
- Cui, Y.; Zhu, Y.; Wang, J.C.; Lu, Y.; Zeng, R.; Katz, R.; Wu, D.M.; Vavvas, D.G.; Husain, D.; Miller, J.W.; et al. Imaging artifacts and segmentation errors with wide-field swept-source optical coherence tomography angiography in diabetic retinopathy. Transl. Vis. Sci. Technol. 2019, 8, 18. [Google Scholar] [CrossRef]
- Mann, K.S.; Kaur, S. Segmentation of retinal blood vessels using artificial neural networks for early detection of diabetic retinopathy. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2017; Volume 1836, p. 020026. [Google Scholar]
- Burewar, S.; Gonde, A.B.; Vipparthi, S.K. Diabetic Retinopathy Detection by Retinal segmentation with Region merging using CNN. In Proceedings of the 2018 IEEE 13th International Conference on Industrial and Information Systems (ICIIS), Rupnagar, India, 1–2 December 2018. [Google Scholar]
- Nur, N.; Tjandrasa, H. Exudate segmentation in retinal images of diabetic retinopathy using saliency method based on region. In Journal of Physics: Conference Series; IOP Publishing: Britol, UK, 2018; Volume 1108, p. 012110. [Google Scholar]
- Arsalan, M.; Owais, M.; Mahmood, T.; Cho, S.W.; Park, K.R. Aiding the diagnosis of diabetic and hypertensive retinopathy using artificial intelligence-based semantic segmentation. J. Clin. Med. 2019, 8, 1446. [Google Scholar] [CrossRef]
- He, Y.; Jiao, W.; Shi, Y.; Lian, J.; Zhao, B.; Zou, W.; Zhu, Y.; Zheng, Y. Segmenting diabetic retinopathy lesions in multispectral images using low-dimensional spatial-spectral matrix representation. IEEE J. Biomed. Health Inform. 2020, 24, 493–502. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Sengupta, D.; Terzopoulos, D. End-to-end trainable deep active contour models for automated image segmentation: Delineating buildings in aerial imagery. In Computer Vision—ECCV 2020; Springer International Publishing: Cham, Switzerland, 2020; pp. 730–746. [Google Scholar]
- Jin, Q.; Meng, Z.; Pham, T.D.; Chen, Q.; Wei, L.; Su, R. DUNet: A deformable network for retinal vessel segmentation. Knowl. Based Syst. 2019, 178, 149–162. [Google Scholar] [CrossRef]
- Hasan, M.K.; Alam, M.A.; Elahi, M.T.E.; Roy, S.; Martí, R. DRNet: Segmentation and localization of optic disc and Fovea from diabetic retinopathy image. Artif. Intell. Med. 2021, 111, 102001. [Google Scholar] [CrossRef]
- Sambyal, N.; Saini, P.; Syal, R.; Gupta, V. Modified U-Net architecture for semantic segmentation of diabetic retinopathy images. Biocybern. Biomed. Eng. 2020, 40, 1094–1109. [Google Scholar] [CrossRef]
- Ali, A.; Qadri, S.; Khan Mashwani, W.; Kumam, W.; Kumam, P.; Naeem, S.; Goktas, A.; Jamal, F.; Chesneau, C.; Anam, S.; et al. Machine learning based automated segmentation and hybrid feature analysis for diabetic retinopathy classification using fundus image. Entropy 2020, 22, 567. [Google Scholar] [CrossRef]
- Wan, C.; Chen, Y.; Li, H.; Zheng, B.; Chen, N.; Yang, W.; Wang, C.; Li, Y. EAD-net: A novel lesion segmentation method in diabetic retinopathy using neural networks. Dis. Markers 2021, 2021, 6482665. [Google Scholar] [CrossRef] [PubMed]
- Kandhasamy, J.P.; Balamurali, S.; Kadry, S.; Ramasamy, L.K. Diagnosis of diabetic retinopathy using multi level set segmentation algorithm with feature extraction using SVM with selective features. Multimed. Tools Appl. 2020, 79, 10581–10596. [Google Scholar] [CrossRef]
- Tang, Y.; Pan, Z.; Pedrycz, W.; Ren, F.; Song, X. Viewpoint-based kernel fuzzy clustering with weight information granules. IEEE Trans. Emerg. Top. Comput. Intell. 2022, 1–15. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, X.; Fan, R.; Wang, Z. Deviation-sparse fuzzy C-means with neighbor information constraint. IEEE Trans. Fuzzy Syst. 2019, 27, 185–199. [Google Scholar] [CrossRef]
- Guo, L.; Chen, L.; Lu, X.; Chen, C.L.P. Membership affinity lasso for fuzzy clustering. IEEE Trans. Fuzzy Syst. 2020, 28, 294–307. [Google Scholar] [CrossRef]
- Mukaidono, S.M.A. Fuzzy C-means as a regularization and maximum entropy approach. In Proceedings of the 7th International Fuzzy Systems Association World Congress (IFSA’97), Prague, Czech Republic, 25–29 June 1997; pp. 86–92. [Google Scholar]
- Huang, J.; Nie, F.; Huang, H. A New Simplex Sparse Learning Model to Measure Data Similarity for Clustering. Available online: https://www.ijcai.org/Proceedings/15/Papers/502.pdf (accessed on 20 October 2022).
- Fukushima, K. ImageNet Neocognition: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position Biol. Cybernet 1980, 36, 193–202. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving Neural Networks by Preventing Co-Adaptation of Feature Detectors. arXiv 2012, arXiv:1207.0580. [Google Scholar]
- Patry, G.; Gauthier, G.; Bruno, L.A.Y.; Roger, J.; Elie, D.; Foltete, M.; Donjon, A.; Maffre, H. Messidor. 2019. Available online: http://www.adcis.net/en/third-party/messidor/ (accessed on 20 October 2022).
- Porwal, P.; Pachade, S.; Kamble, R.; Kokare, M.; Deshmukh, G.; Sahasrabuddhe, V.; Meriaudeau, F. Indian diabetic retinopathy image dataset (IDRiD): A database for diabetic retinopathy screening research. Data 2018, 3, 25. [Google Scholar] [CrossRef]


| Model Name | Sensitivity | Speficity | F1 | Accuracy | AUC |
|---|---|---|---|---|---|
| Multi-scale, multi-path FCM | 0.8259 | 0.9841 | 0.8295 | 0.9703 | 0.9870 |
| Multi-scale, multi-output fusion FCM | 0.8063 | 0.9866 | 0.8286 | 0.9708 | 0.9871 |
| Basic FCM | 0.8196 | 0.9848 | 0.8286 | 0.9703 | 0.9870 |
| Multi-scale FCM | 0.8115 | 0.9860 | 0.8290 | 0.9707 | 0.9873 |
| Multi-path FCM | 0.8118 | 0.9858 | 0.8287 | 0.9706 | 0.9871 |
| Multi-output fusion FCM | 0.8192 | 0.9850 | 0.8293 | 0.9705 | 0.9870 |
| Multi-path, multi-output fusion FCM | 0.8320 | 0.9828 | 0.8304 | 0.9701 | 0.9873 |
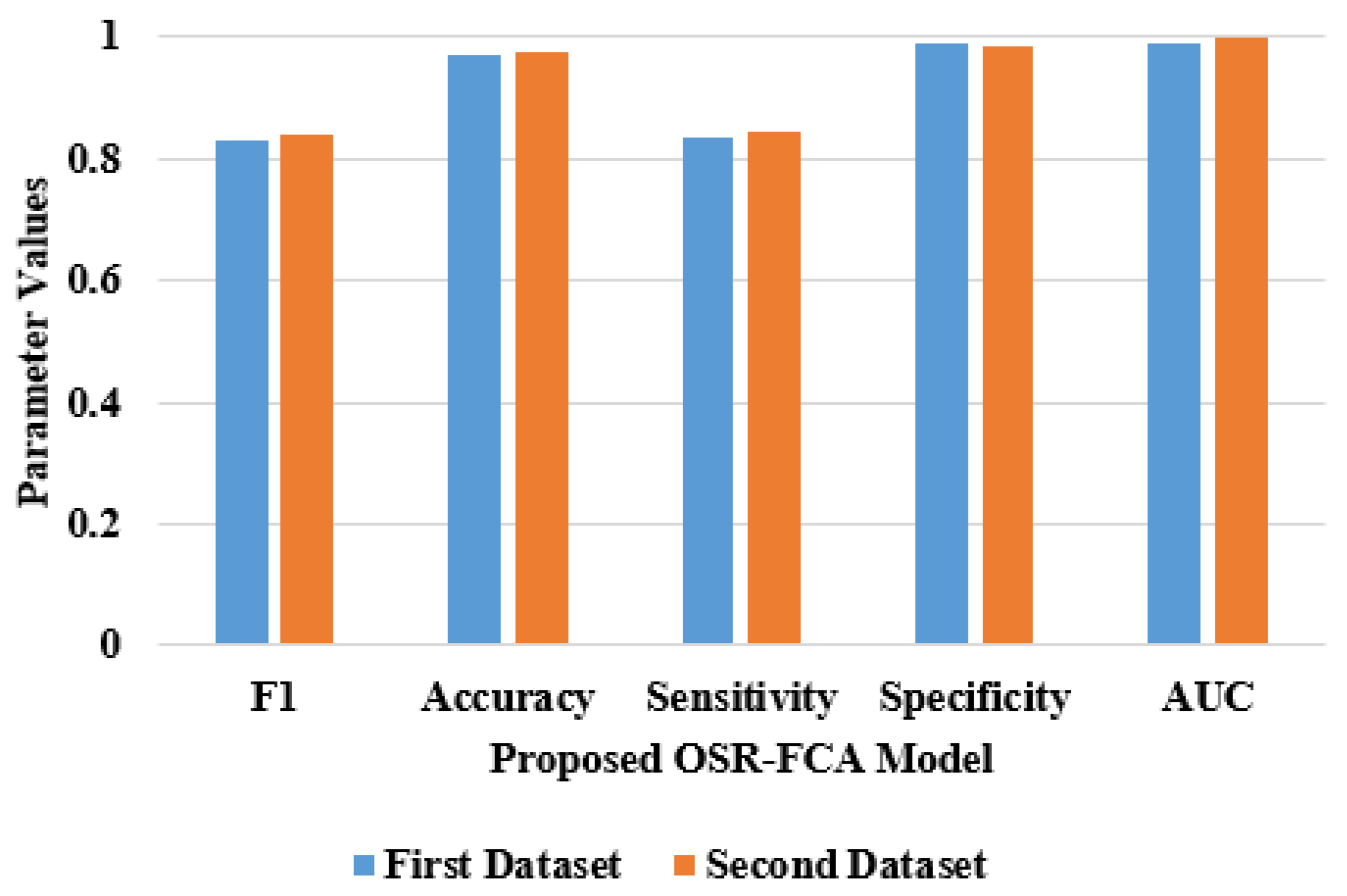
| Proposed OSR-FCA system | 0.8370 | 0.9870 | 0.8321 | 0.9716 | 0.9880 |
| Model Name | F1 | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|
| Basic FCM | 0.8288 | 0.9703 | 0.8198 | 0.9848 | 0.9870 |
| Multi-scale FCM | 0.8288 | 0.9703 | 0.8198 | 0.9848 | 0.9870 |
| Multi-path FCM | 0.8299 | 0.9702 | 0.8298 | 0.9837 | 0.9873 |
| Multi-output fusion FCM | 0.8294 | 0.9702 | 0.8269 | 0.9840 | 0.9873 |
| Multi-scale, multi-path FCM | 0.8242 | 0.9697 | 0.8111 | 0.9849 | 0.9861 |
| Multi-scale, multi-output fusion FCM | 0.8255 | 0.9689 | 0.8392 | 0.9814 | 0.9866 |
| Multi-path, multi-output fusion FCM | 0.8321 | 0.9706 | 0.8325 | 0.9838 | 0.9880 |
| Proposed OSR-FCA system | 0.8420 | 0.9726 | 0.8428 | 0.9852 | 0.9990 |
| Methodologies | Sensitivity | Accuracy | Specificity | Kappa Index |
|---|---|---|---|---|
| % | % | % | % | |
| RNN | 79.33 | 75.03 | 78.45 | 87.60 |
| LSTM | 88.95 | 91.33 | 86 | 81.86 |
| Auto-encoder | 92.77 | 95.17 | 92.24 | 88.45 |
| Proposed CNN | 98.04 | 97.26 | 98.17 | 90.07 |
| Methodologies | Sensitivity | Accuracy | Specificity | Kappa Index |
|---|---|---|---|---|
| % | % | % | % | |
| RNN | 87.43 | 92.02 | 78.14 | 80.14 |
| LSTM | 89.97 | 92.39 | 85.90 | 81.08 |
| Auto-encoder | 95.16 | 94.18 | 92.17 | 86.44 |
| Proposed CNN | 98.62 | 98.70 | 98.83 | 90.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hemamalini, S.; Kumar, V.D.A. Outlier Based Skimpy Regularization Fuzzy Clustering Algorithm for Diabetic Retinopathy Image Segmentation. Symmetry 2022, 14, 2512. https://doi.org/10.3390/sym14122512
Hemamalini S, Kumar VDA. Outlier Based Skimpy Regularization Fuzzy Clustering Algorithm for Diabetic Retinopathy Image Segmentation. Symmetry. 2022; 14(12):2512. https://doi.org/10.3390/sym14122512
Chicago/Turabian StyleHemamalini, Selvamani, and Visvam Devadoss Ambeth Kumar. 2022. "Outlier Based Skimpy Regularization Fuzzy Clustering Algorithm for Diabetic Retinopathy Image Segmentation" Symmetry 14, no. 12: 2512. https://doi.org/10.3390/sym14122512
APA StyleHemamalini, S., & Kumar, V. D. A. (2022). Outlier Based Skimpy Regularization Fuzzy Clustering Algorithm for Diabetic Retinopathy Image Segmentation. Symmetry, 14(12), 2512. https://doi.org/10.3390/sym14122512





