Abstract
Automated brain tumor segmentation based on 3D magnetic resonance imaging (MRI) is critical to disease diagnosis. Moreover, robust and accurate achieving automatic extraction of brain tumor is a big challenge because of the inherent heterogeneity of the tumor structure. In this paper, we present an efficient semantic segmentation 3D recurrent multi-fiber network (RMFNet), which is based on encoder–decoder architecture to segment the brain tumor accurately. 3D RMFNet is applied in our paper to solve the problem of brain tumor segmentation, including a 3D recurrent unit and 3D multi-fiber unit. First of all, we propose that recurrent units segment brain tumors by connecting recurrent units and convolutional layers. This quality enhances the model’s ability to integrate contextual information and is of great significance to enhance the contextual information. Then, a 3D multi-fiber unit is added to the overall network to solve the high computational cost caused by the use of a 3D network architecture to capture local features. 3D RMFNet combines both advantages from a 3D recurrent unit and 3D multi-fiber unit. Extensive experiments on the Brain Tumor Segmentation (BraTS) 2018 challenge dataset show that our RMFNet remarkably outperforms state-of-the-art methods, and achieves average Dice scores of 89.62%, 83.65% and 78.72% for the whole tumor, tumor core and enhancing tumor, respectively. The experimental results prove our architecture to be an efficient tool for brain tumor segmentation accurately.
1. Introduction
Glioma originates from glial cells in the brain and has become one of the most terrible diseases that harm human health. Glioma is a high-risk adult brain tumor, with an annual incidence of about 3 to 8 cases per 100,000 people. Medical imaging examinations, such as MRI, play a key part in the diagnosis process of brain tumors [1]. Different forms of MRI images can provide supplementary information for the analysis of different subregions of glioma. Segmentation of brain tumors in multi-modal MRI images has always been a hot research topic. These structural MRI images have positive significance in the majority of cases and the radiologist must take all the four modalities into account to identify each region. There are four standard MRI modalities: fluid attenuated inversion recovery (FLAIR), T1-weighted (T1), T1-weighted contrast-enhanced (T1C) and T2-weighted (T2). Clinically, an accurate segmentation image of brain tumor performs an indispensable role in patient care and evaluation [2]. Automatic segmentation of brain tumors can provide accurate and valuable solutions to further analyze and monitor tumors.
Robust and accurate achieving of automatic segmentation of brain tumors is challenging because of two causes: (1) gliomas have an antenna structure, and they often spread easily and have poor contrast. In addition, their blurred boundaries make it difficult to segment the surrounding glioma tissue. (2) Brain tumors grow anywhere in the brain in almost any shape and size. As mentioned above, regarding the structural heterogeneity of brain tumors, many researchers have proposed solutions to address these problems [3]. Zeineldin et al. [4] applied a novel deep learning method called DeepSeg, which is used to automatically detect and segment brain lesions using FLAIR MRI data. The accurate implementation of automated segmentation technology is very challenging, and it has attracted the attention of many researchers and has become an important research field. Therefore, the realization of automatic segmentation of brain tumors will help to accelerate the large-scale segmentation and assist doctors. According to the above heterogeneity of brain tumor structure, it is significant to propose an automatic brain tumor segmentation method.
Deep learning-based medical image automatic segmentation methods make remarkable advances, convolutional neural networks (CNN) demonstrate impressive segmentation accuracy in 2D natural images [5] and 3D medical image modalities [6]. However, the three-dimensional CNN architecture is usually used in 3D medical images, and it is difficult to put into practice with high computational [7]. In order to deal with this problem, our network leverages 3D multi-fiber units in this article, which greatly reduces the computational cost. Aiming at the problem of blurred boundaries and uncertain positions of brain tumors, a new 3D recurrent multi-fiber network (RMFNet) is proposed in the design of an automatic segmentation method. The RMFNet consists of 3D recurrent units and 3D multi-fiber units. We connect the recurrent units to the convolutional layer, and use the recurrent convolutional layers for feature accumulation to be sure of better feature representation for brain segmentation tasks. In addition, we use 3D multi-fiber unit, which contains a lightweight network integration to reduce computing cost and increase segmentation performance at the same time.
The rest of this article is arranged as follows: Section 2 discusses related work. The architecture of the proposed 3D recurrent multi-fiber network is presented in Section 3. Section 4 presents detailed datasets and exhaustive results. The conclusions of this work are obtained comprehensively in Section 5.
2. Related Work
Deep learning, especially the method based on convolutional neural network, has obtained the most advanced performance in medical image semantic segmentation [8]. Havaei et al. [9] presented a two-pathway CNN model and took a local image block as input in a sliding window manner to predict the label of each pixel. ResNet [10] utilized the efficient bottleneck structure to achieve impressive performance. Ronneberger et al. [11] proposed the earliest and most popular medical image semantic segmentation method called “U-Net”. Since then, U-Net has become very popular and has been used effectively in different forms of medical imaging and computational pathology. Xu et al. [12] proposed a new deep network called LSTM multi-modal UNet, which consists of multi-model UNet and convolution LSTM [13]. Multi-modal UNet includes high density encoders and decoders to take full advantage of exploiting multi-modal data, and convolution LSTM further utilizes sequential information between contiguous slices. Dong et al. [14] adopted a deep atlas network with information consistency constraint to segment the 3D left ventricle for handling high dimensional data and limited annotation data problem. Heinrich et al. [15] designed a novel convolutional architecture called OBELISK-Net to segment 3D multi-organ images. Li et al. [16] used a new type of adversarial model based on a multi-stage learning method to segment three-dimensional multiple spinal structures from multi-modal MRI images [17]. Their results also confirm that deep learning has outstanding performance in resolving 3D medical image segmentation and becomes an indispensable part of medical image processing, it has become the first choice for various medical image segmentation applications. Furthermore, brain tumor segmentation approaches based on deep learning have also attained good segmentation results. We summarize brain tumor segmentation-based methods as follows.
When it comes to brain tumor segmentation, deep learning methods have recently reached state-of-the-art accuracy for segmentation. Ping Liu et al. [18] presented a Deep supervised 3D Squeeze-excitation V-Net (DSSE-V-Net) to automatically segment brain tumors from multi-model MRI images. On the BraTS 2017, Kamnitsas et al. [19] achieved excellent results and proposed a robust segmentation integrating several models called EMMA, which utilized an integrated architecture of several independent training. In particular, EMMA combined DeepMedic [20] and U-Net models and integrated the predictions of their segmentation. In 2018, Myronenko [21] proposed a 3D encoder–decoder model based on ResNet and won the first prize in BraTS18. Zhou et al. [22] ensembled several different networks and used a shared backbone weights to extract multi-scale context information. In [23], the authors used the k-nearest neighbor classifier for the real autism spectrum disorder dataset. They considered the problem of the large amount and complexity of MRI data. For this purpose, they proposed using the adaptive independent subspace analysis (AISA) method to discover meaningful electroencephalogram activity in the MRI scan data. Taking advantage of AISA method, they achieved 94.7% of accuracy. Khan et al. [24] introduced an automated multi-modal classification method using deep learning for brain tumor type classification. They initially employed the linear contrast stretching using edge-based histogram equalization and discrete cosine transform (DCT). Then, they performed deep learning feature extraction using transfer learning. Later on, they adopted a correntropy-based joint learning approach, combined with the extreme learning machine (ELM) for feature selection, and merged robust covariant features based on partial least squares (PLS) into one matrix. The combined matrix was sent to ELM for final multi-modal brain tumor classification.
The perspective of using channel grouping to reduce the computational cost is similar to the idea of group convolution, which was first introduced in AlexNet [25]. Chen et al. [26] designed a high-efficiency 3D CNN to realize real-time dense volumetric segmentation, which built upon the multi-fiber unit [27] for facilitating information flow between groups. These previous works clearly show that deep learning models can be used to segment the brain into anatomical regions. However, these 3D CNN architectures bring high computational overhead due to the use of multi-layer 3D convolutions. As a result of limitations in some of the previous models, these limitations prompted us to propose a new method. In order to make better use of multi-modalities and depth information, we propose a novel network for solving the problem of brain tumors segmentation and adopt several strategies for reducing network parameters.
3. Method
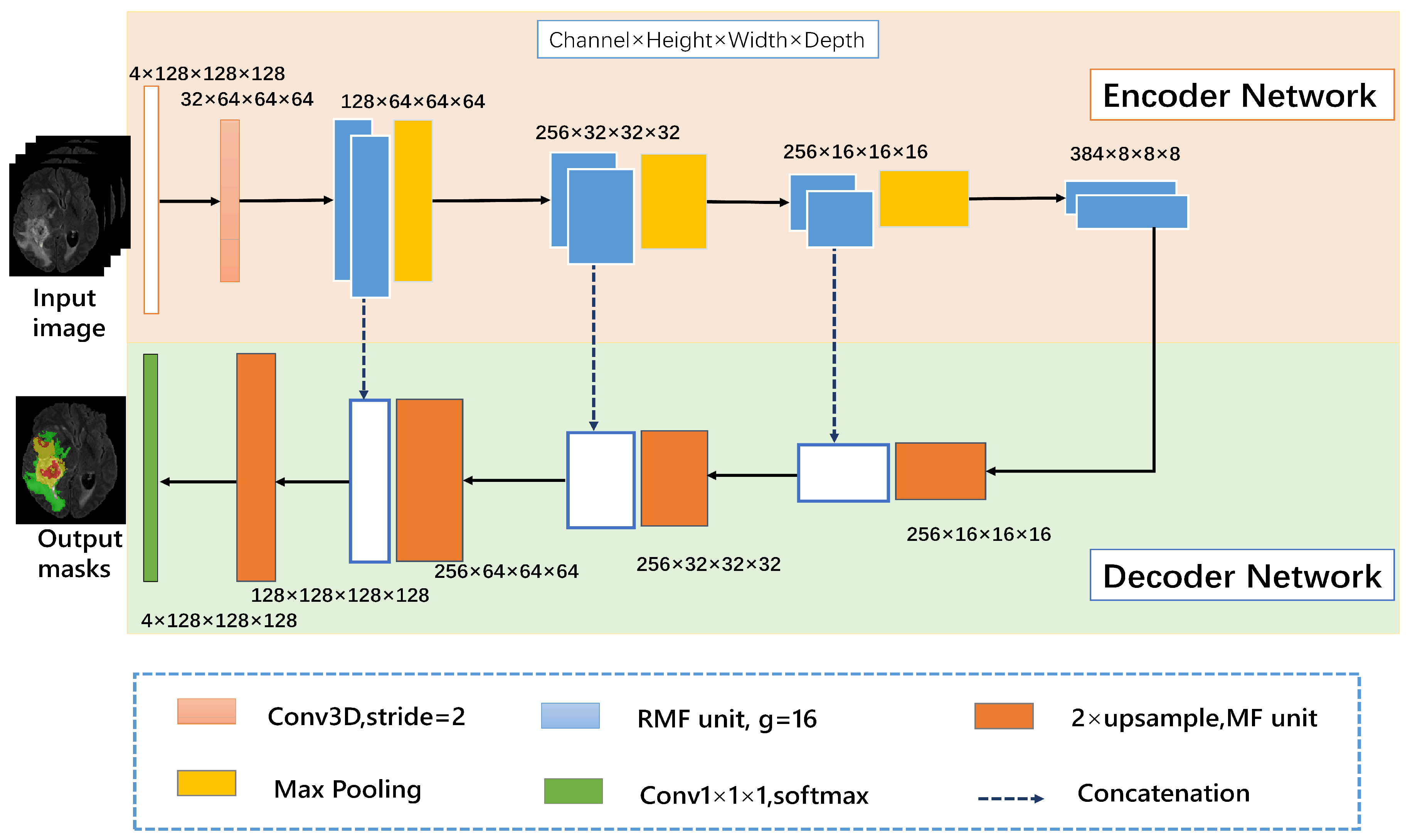
The proposed 3D RMFNet is composed of an encoder network on the pink part and a decoder network on the blue part. Moreover, the number of input channels is set to four since FLAIR, T1, T1C and T2 modalities are exploited to segment the tumor region, and every modality with size 4 × 128 × 128 × 128. 128 × 128 × 128 indicates that each MR sequence has 128 pictures, and the size of each picture is 128 × 128. In the encoding path, the convolution block is composed of a convolution layer with the kernel size of 3 × 3 × 3, a batch normalization layer and ReLU activation function layer. To get more accurate context information, the encoder path contains RMF units to extract rapidly features representation of the input, and a 3D convolution block with a stride of 2 is replaced by the max-pooling operation. The decoder path consists of four upsampling layers and one convolution layer. Four upsampling layers contain MF units to reduce computing costs. We have used concatenation to the feature maps from the encoder network to the decoder network at the same level, which allows capturing multiple contextual information. The trilinear interpolation is used to upsample the feature maps, the final output is generated by the size of 1 × 1 × 1 3D convolution layer. White boxes indicate copied feature maps. These building blocks are explained in detail in the following subsections. Our framework is shown in Figure 1.
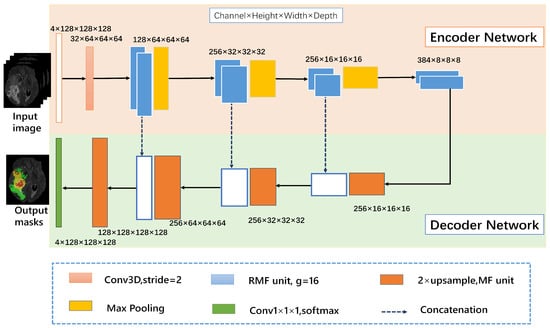
Figure 1.
The 3D recurrent multi-fiber network (RMFNet) comprises an encoder network and a decoder network, the main body of the network consists of RMF units. The input module is a four-channel 3D MRI crop, and the initial 3 × 3 × 3 3D convolution is with 32 filters. In this paper, g represents the number of groups.
3.1. Recurrent Multi-Fiber (RMF) Unit
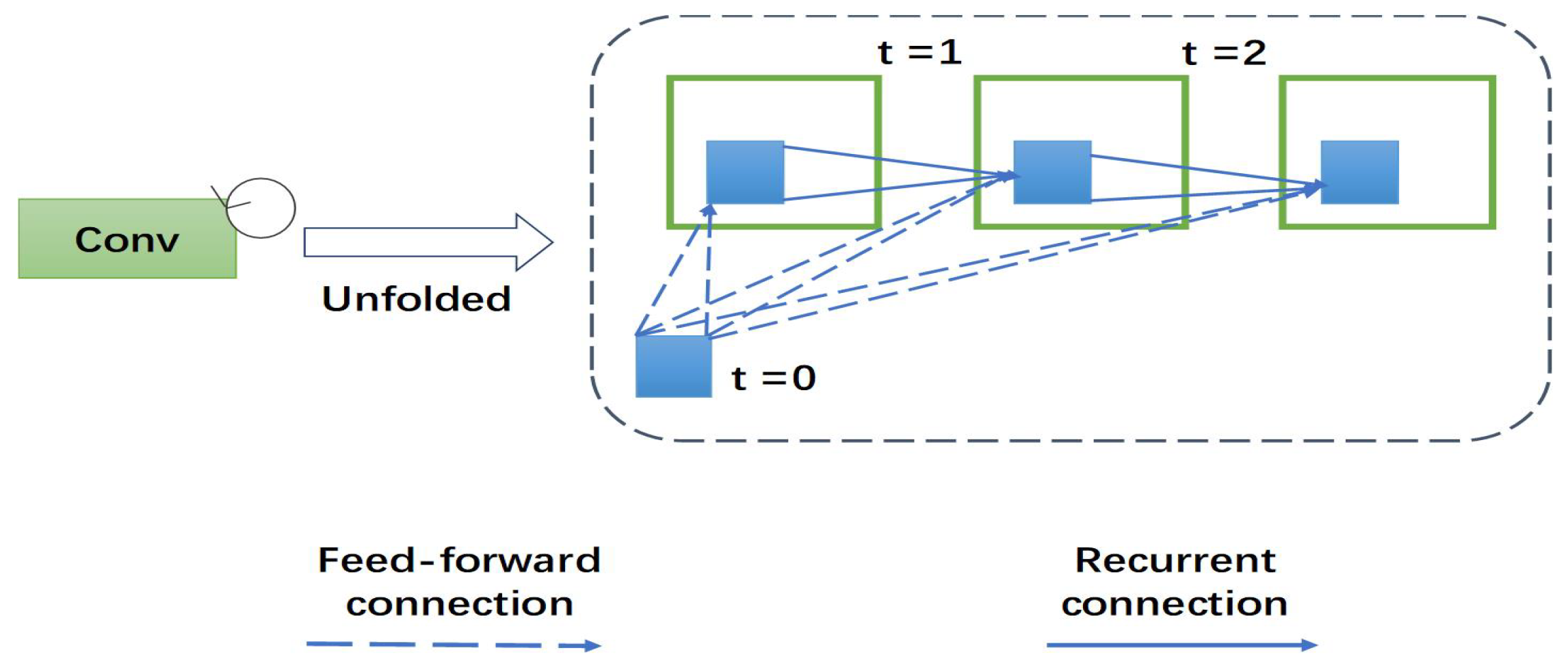
This section presents the recurrent multi-fiber unit. To better understand the role of recurrent multi-fiber unit, we describe the newly-proposed unit as follows: recurrent convolutional neural networks (RCNN) are gradually applied in medical image processing. RCNN and its variants show excellent performance in object recognition tasks using different benchmarks. RCNN is an important part of recurrent multi-fiber (RMF). At each time-step, the RCNN receive a new input and generates an output based on the current input and previous time-step information. Recurrent convolutional layer (RCL) is the key module of RCNN. According to the RCNN, the states of RCL units evolve on discrete time steps. The unfolded RCL layer graphically represents the time-step as shown in Figure 2. When t = 0 only the feedforward input is present. Here, t = 2 (0∼2) refers to the recurrent convolutional operation, including a feed-forward convolution layer and then two sub-sequential recurrent convolutional layers.
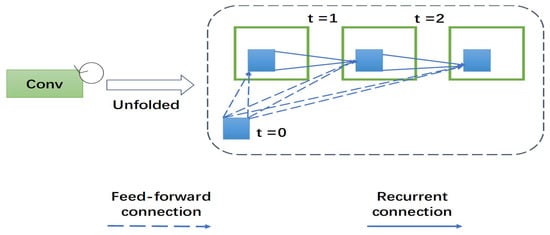
Figure 2.
The overall structure of a 3D recurrent convolutional layer (RCL). An RCL is unfolded for t = 2 time steps.
Let denote the particular input sample in the lth layer of the residual RCNN (RRCNN) block for a pixel located at of a particular input sample on the kth feature map in an RCL. Accordingly, the output of the network at the time step t is presented as follows:
where denotes the inputs to the feed-forward convolutional layer, and represents the inputs to the lth RCL. and are the weights of the feed-forward convolutional layer and the RCL of the kth feature map, respectively, and is the bias. The outputs of the RCL are presented to the rectified linear activation function and are represented as follows:
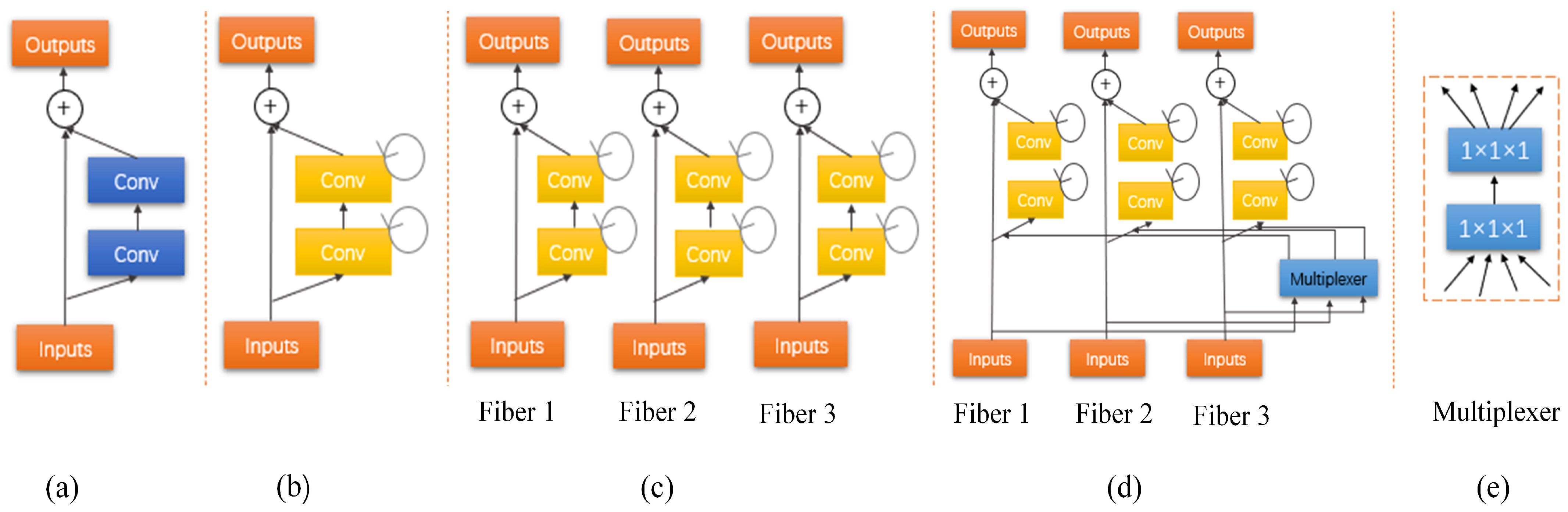
Here, denotes the outputs from of lth layer of the RCNN unit. The basic residual convolutional unit of this architecture is shown in Figure 3a. As showed in Figure 3b, let the output of the RRCNN-block be , so the output is composed of direct mapping parts and residual parts and it can be computed by:
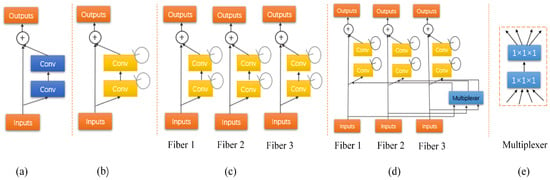
Figure 3.
(a) A residual unit with two 3 × 3 × 3 convolution layers. (b) Recurrent residual convolutional units (RRCU). (c) The developed multi-fiber (MF) design consisting of multiple separated recurrent residual units, called fibers. (d) The recurrent multi-fiber (RMF) unit leverages a multiplexer for transferring information across separated fibers. (e) The multiplexer acts as a router that redirects and amplifies features from all fibers.
3.2. MF Unit
The goal of multi-fiber is to reduce the number of connections between the feature maps and kernels that make sense to calculate the overall cost. As in the examples shown in Figure 3a, let us consider the dimensions of the input feature maps and the kernel size is constant, let represent the number of input channels, represent the number of middle channels and represent the number of output channels. Let Con(b) be the total number of connections as illustrated in Figure 3a. Thus, the amount of connections between these two layers comes to:
Figure 3c shows a multi-fiber grouping strategy. We divide the ordinary residual unit into K parallel and independent residual units that are called fibers. The number of connections can be represented as follows:
Which is K times less than Con(b). The overall width of the unit remains unchanged, but the network parameters are greatly reduced, which significantly reduces the heavy computational burden in 3D convolution.
3.3. Multiplexer Unit
The fibers are independent of each other and prevent them from exchanging information. To facilitate information flow, use two 1 × 1 × 1 convolutions called a multiplexer. The multiplexer acts as a router, redirecting and amplifying the characteristics of all fibers. Details for the multiplexer unit are shown in Figure 3e.
The number of input channels is and then compressed to , and finally enlarged to . The parameters using two convolutions are . However, using a 1 × 1 × 1 convolution layer, params = . Compared with using a 1 × 1 × 1 convolution, we employ two 1 × 1 × 1 convolutions which can reduce half of the parameters. Therefore, the purpose of using two convolutions is to reduce network parameters. Figure 3d describes the full multi-fiber network, where multiplexer is connected at the beginning of the multi-fiber unit, to improve learning ability and extract features without adding additional parameters.
4. Experiments
4.1. Benchmark Dataset and Evaluation Criteria
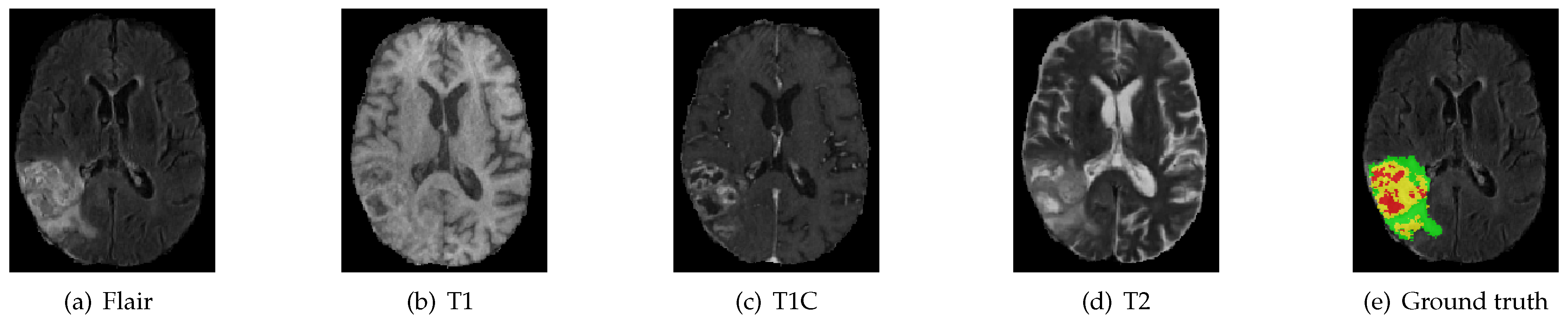
We evaluate our algorithm on the BraTS 2018 challenge dataset against the state-of-the-art methods. We introduce the dataset as follows. The multi-modal 3D MRIs are provided by the BraTS 2018 challenge [3,28,29]. The training data consist of 75 low grade and 210 high grade gliomas. Each subject has FLAIR, T1, T1C and T2 MRI scans, and even ground truth (GT) obtained by hand segmentation from experts. The multi-modal 3D MRIs originated from 19 institutions and employed different protocols, magnetic field strengths and MRI scanners [30]. Annotations include 4 tumor subregions: the normal tissue (label 0), necrotic and non-enhancing tumor (label 1), peritumoral edema (label 2) and active/enhancing tumor (label 4). The annotations are combined into 3 nested subregions. WT, TC and ET refer to the regions of whole tumor (label1, label2, label4), tumor core (label1, label4) and enhancing tumor (label4), respectively, as shown in Figure 4.
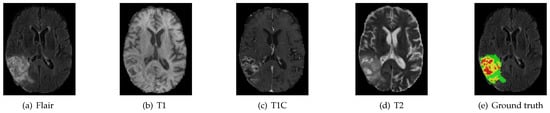
Figure 4.
MRI image modality sequence along with ground truth. The whole tumor (WT) class indicates all visible labels (a combination of green, yellow and red labels), the tumor core (TC) class is a combination of red and yellow, and the enhancing tumor core (ET) class is shown in yellow. The green region indicates peritumoral edema, the yellow region indicates active/enhancing tumor and the red region indicates necrotic and non-enhancing tumor. These are the slices from the axial plane.
The segmentation accuracy has been evaluated using the Dice Similarity Coefficient (DSC) and Hausdorff Distance (HD). Dice coefficient is the most common evaluation metric in medical image segmentation. DSC is used to describe the degree of overlap between the segmentation result and the actual truth label. The higher the Dice score, the better the segmentation performance.
where indicates that the model predicts positive samples, indicates that the model predicts negative samples, indicates that the model predicts positive samples and indicates that the model predicts negative samples.
Hausdorff Distance (HD) are adopted to evaluate the distance between the ground truth boundary and the predicted segmentation boundary. The Hausdorff Distance could be calculated as follows:
where a and b are points of sets A and B, respectively, and d(a, b) is the Euclidean distance between a and b.
4.2. Implementation and Training
We employ standard five-fold cross validation on the dataset for performance analysis and comparison. Specifically, the dataset can be randomly divided into five groups. Four groups of them are used to train deep neural networks, and the last group is used to test performance. This process is repeated five times. The whole experimental procedure is implemented in Pytorch and train RMFNet on NVIDIA Tesla V100 32 GB GPU. Training the RMFNet model for 200 epochs and the batch size is set to 2. In our experiments, the purpose of data augmentation is to increase the sample size of training data and enhance the robustness and generalization ability of deep Learning training algorithm. We apply a random rotation, random intensity shift between and random cropping. During the training, the MRI data are randomly cropped to 128 × 128 × 128, which can ensure that most of the image content is still in the crop area. The probability of random axis mirror flip (for all 3 axes) is 0.5.
To train our model, the Adaptive Moment Estimation (Adam) [31] is used for training, with an initial learning rate of and a l2 weight decay of . In terms of pre-processing, we subtract the average value and divide by the standard deviation of the brain region in order to normalize all input images. The Generalized Dice Loss (GDL) is used to train the network. The formula for GDL is as follows:
The loss function checks each pixel separately, where m is the number of categories, is the standard value of the category i at the j pixel and is the predicted value of the category i at the j pixel. The most critical thing here is w, and w represents the weight of each category, which is determined by the number of pixels in the category as follows:
4.3. Results
To verify the superiority of our presented model, we compare our designed RMFNet model with state-of-the-art models [32,33,34,35,36,37,38]. Among them, 3D U-Net [32] and 3D-ESPNet [33] are the most popular convolutional neural networks and have been a great success in various tasks.
The proposed RMFNet achieves the best results on the brain tumor segmentation from multi-modal 3D MRIs. Table 1 shows the results of our model and comparison model, our results are shown in bold on the last line. The following evaluation results can be used for analysis. In fact, the proposed algorithm achieves a Dice value of 89.62% for whole tumor, 83.65% for tumor core and 78.72% for enhancing tumor. The HD value achieves 5.96 for whole tumor, 7.56 for tumor core and 3.94 for enhancing tumor. According to Dice score, it is observed that, compared with previous models, our model remarkably outperforms 3D U-Net [32] on the BraTS 2018 validation dataset. Compared with the best algorithm [38], we can see that our method only has marginal performance gaps of 5.12% for the enhancing tumor. However, our RMFNet only has 2.65 M parameters and 37.24 G FLOPs, and our model has 10 times less FLOPs than [38]. It is observed that our method can accurately segment different tumor subregions without requiring many parameters, and can outperform the other methods on the BraTS 2018 validation dataset. Thus, our RMFNet is a more efficient algorithm and has the potential of our method in multi-modal 3D MRI image segmentation tasks. The part segmentation results are shown in Figure 5.

Table 1.
Performance comparison with other quantitative results on the BraTS 2018 validation dataset. WT, TC, ET denote whole tumor, tumor core and enhancing tumor, respectively. FLOPs to evaluate the complexity of the model (i.e., multiplication and addition).
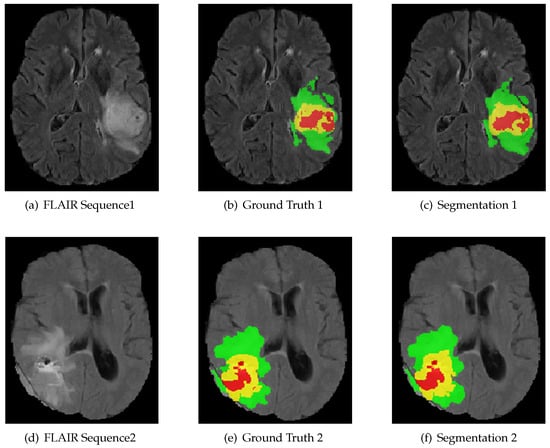
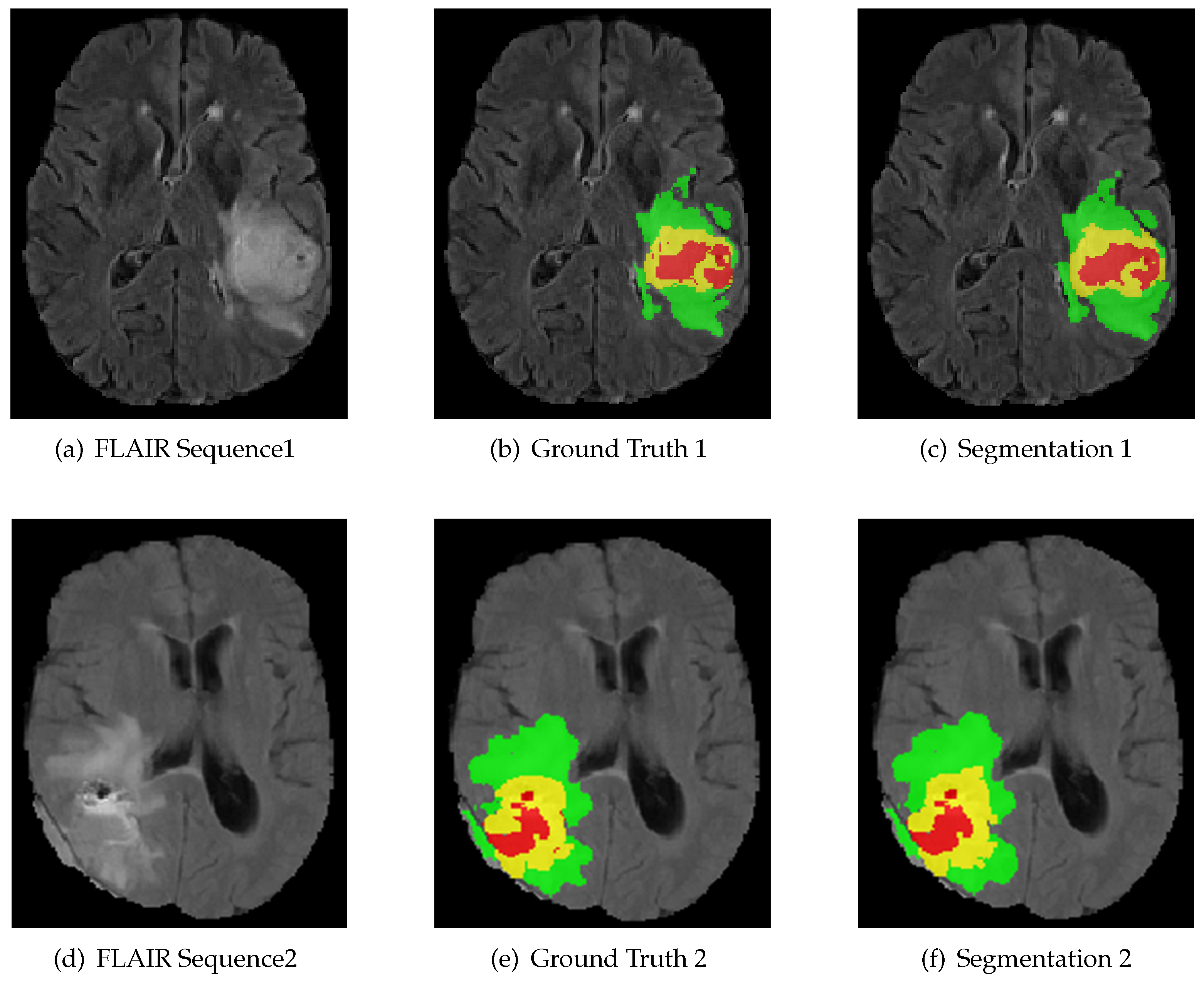
Figure 5.
The result of using RMFNet with corresponding fluid attenuated inversion recovery (FLAIR) slice and ground truth on BraTS 2018 Training dataset. Red: necrotic and non-enhancing tumor (label 1); green: peritumoral edema (label 2); yellow: active/enhancing tumor (label 4).
In this paper, to verify the effectiveness of the backbone established by the recurrent unit, we compare the experimental results of MFNet and RMFNet. The results show that the recurrent unit can improve the performance. Since the recurrent unit has the ability to promote feature accumulation and select multi-scale context information, it can be beneficial to scores. Table 1 clearly shows that our method is superior to complex networks, such as 3D U-Net [32] and 3D-ESPNet [33]. When the recurrent unit is not utilized, the MFNet has only 2.33M parameters. Nevertheless, it also has better segmentation performance. In addition, we estimate the computing time of the network on NVIDIA Tesla V100 32 GB GPU. The input size of RMFNet is 4 × 128 × 128 × 128 with the modalities of FLAIR, T1, T1C and T2. We use the BraTS 2018 benchmarks to estimate the inference time of the method. It takes an average of 2.15 s for RMFNet to segment a patient subject averagely.
4.4. Discussion
In this paper, we come up with a novel architecture for brain tumor segmentation from multi-modal 3D MRIs. Specifically, the tumor is difficult to segment due to its random location, blurred boundaries and irregular shapes. We present a new architecture of 3D recurrent multi-fiber network (RMFNet) that includes RMF units, and we employ RMF units to design a 3D recurrent multi-fiber network for automatic brain tumor segmentation task, to address the huge computational burden in 3D convolution. Our architecture is designed to contain a feature encoding path that encodes the detailed features to abstract features as the network gets deeper and the decoding path with high resolution information from the shallow layers are concatenated with deeper layers. To get as much meaningful feature information as possible, we adopt a recurrent multi-fiber unit as the encoding path in the architecture to better extract more precise detailed information and avoid the degradation as the network grows larger and deeper. Moreover, using a 3D multi-fiber unit can reduce the heavy computational burden in 3D convolution. In the decoder path, we adopt the trilinear interpolation for upsampling the feature information. Meanwhile, we utilize the detailed information and abstract information to segment the tumor accurately. Through a comparative study, the results show that the proposed segmentation algorithm is more effective than the 3D U-net architecture [32]. At the same time, this method has achieved promising results in the extraction of different regions, which is better than other methods. Extensive experiments on the BraTS 2018 challenge dataset verify the effectiveness of our proposed RMFNet. For the whole tumor, the tumor core and the enhancing tumor, we can attain dice scores of 89.62%, 83.65% and 78.72%, respectively. Considering the 3D context, we also compare with the previous state-of-the-art methods. Although the reduction of required parameters affects the training time and execution time, the time is prolonged due to the complexity of the model. Therefore, in the future, we will further modify our method to increase its generalization ability and improve the training speed. Meanwhile, we will adopt different post-processing methods to further improve the segmentation results.
5. Conclusions
Automatic segmentation of a brain tumor has great influence in clinical diagnosis. It reduces the burden of doctors to annotate the lesion area, and presents an accurate contour of the anatomical tumors. In this paper, our model has designed an effective training program to reduce false positives and enhance generalization, and the advantages of the model are intuitive and easy to implement. Our RMFNet only has 2.65 M parameters and 37.24 G FLOPs.
Experimental results show that the performance of our algorithm is obviously better than the previous algorithms. Quantitative evaluation of our method on the 2018 dataset shows that our RMFNet achieved comparable dice sores (89.62%, 83.65% and 78.72% for the whole tumor, the tumor core and the enhancing tumor, respectively) yet with less computational FLOPs and less model parameters, compared with the state-of-the-art brain tumor segmentation approaches, e.g., 3D U-net architecture [32]. The quantitative results show the effectiveness and potential of RMFNet as a clinical tool, reduce the heavy workload of doctors and help to quickly and accurately segment brain tumors. In our experiments, because of conducting 3D segmentation, it takes much more time to train. However, it yields relatively good segmentation results. In the future, we will modify our method to enhance its generalization ability and improve the training speed. Meanwhile, we will apply different post-processing methods to further improve the segmentation results.
Author Contributions
Y.Z., X.R., K.H. and W.L. finished writing this article. Y.Z., X.R. and K.H. designed the methods and concepts of this paper. Y.Z., X.R. and W.L. finished the analysis of the experimental results. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Institute Support Jinan star science and technology plan of Jinan Administration of Science & Technology (No. 20120104).
Acknowledgments
The authors are very grateful to the editors and reviewers for their valuable comments and suggestions on this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the cancer genome atlas glioma mri collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef]
- Zeineldin, R.A.; Karar, M.E.; Coburger, J.; Wirtz, C.R.; Burgert, O. Deepseg: Deep neural network framework for automatic brain tumor segmentation using magnetic resonance flair images. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 909–920. [Google Scholar] [CrossRef]
- Luo, X.; Chen, Z. English text quality analysis based on recurrent neural network and semantic segmentation. Future Gener. Comput. Syst. 2020, 112, 507–511. [Google Scholar] [CrossRef]
- Yuan, W.; Wei, J.; Wang, J.; Ma, Q.; Tasdizen, T. Unified generative adversarial networks for multimodal segmentation from unpaired 3D medical images. Med. Image Anal. 2020, 64, 101731. [Google Scholar] [CrossRef]
- Shuang, Y.; Wang, Z. A novel approach for automatic and robust segmentation of the 3D liver in computed tomography images. Meas. Sci. Technol. 2020, 31, 115701. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; Volume 2016, pp. 770–778. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Lecture Notes in Computer Science, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Xu, F.; Ma, H.; Sun, J.; Wu, R.; Liu, X.; Kong, Y. Lstm multi-modal unet for brain tumor segmentation. In Proceedings of the 2019 IEEE 4th International Conference on Image, Vision and Computing (ICIVC), Xiamen, China, 5–7 July 2019; pp. 236–240. [Google Scholar]
- Akilan, T.; Wu, Q.J.; Safaei, A.; Huo, J.; Yang, Y. A 3D cnn-lstm-based image-to-image foreground segmentation. IEEE Trans. Intell. Transp. Syst. 2019, 21, 959–971. [Google Scholar] [CrossRef]
- Dong, S.; Luo, G.; Tam, C.; Wang, W.; Wang, K.; Cao, S.; Chen, B.; Zhang, H.; Li, S. Deep atlas network for efficient 3D left ventricle segmentation on echocardiography. Med. Image Anal. 2020, 61, 101638. [Google Scholar] [CrossRef]
- Heinrich, M.P.; Oktay, O.; Bouteldja, N. Obelisk-net: Fewer layers to solve 3D multi-organ segmentation with sparse deformable convolutions. Med. Image Anal. 2019, 54, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, G.; Fidon, L.; Ourselin, S.; Cardoso, M.J.; Vercauteren, T. On the compactness, efficiency, and representation of 3D convolutional networks: Brain parcellation as a pretext task. In Lecture Notes in Computer Science, Proceedings of the International Conference on Information Processing in Medical Imaging, Boone, NC, USA, 25–30 June 2017; Springer: Cham, Switzerland, 2017; Volume 10265, pp. 348–360. [Google Scholar]
- Li, T.; Wei, B.; Cong, J.; Li, X.; Li, S. S 3 eganet: 3D spinal structures segmentation via adversarial nets. IEEE Access 2019, 8, 1892–1901. [Google Scholar] [CrossRef]
- Liu, P.; Dou, Q.; Wang, Q.; Heng, P.-A. An encoder-decoder neural network with 3D squeeze-and-excitation and deep supervision for brain tumor segmentation. IEEE Access 2020, 8, 34029–34037. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Bai, W.; Ferrante, E.; McDonagh, S.; Sinclair, M.; Pawlowski, N.; Rajchl, M.; Lee, M.; Kainz, B.; Rueckert, D.; et al. Ensembles of multiple models and architectures for robust brain tumour segmentation. In Lecture Notes in Computer Science, Proceedings of the International MICCAI Brainlesion Workshop, Quebec City, QC, Canada, 14 September 2017; Springer: Cham, Switzerland, 2017; pp. 450–462. [Google Scholar]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D cnn with fully connected crf for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Myronenko, A. 3D mri brain tumor segmentation using autoencoder regularization. In Lecture Notes in Computer Science, Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018; Springer: Cham, Switzerland, 2018; pp. 311–320. [Google Scholar]
- Zhou, C.; Chen, S.; Ding, C.; Tao, D. Learning contextual and attentive information for brain tumor segmentation. In Lecture Notes in Computer Science, Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018; Springer: Cham, Switzerland, 2018; pp. 497–507. [Google Scholar]
- Ke, Q.; Zhang, J.; Wei, W.; Damasevicius, R.; Wozniak, M. Adaptive independent subspace analysis (aisa) of brain magnetic resonance imaging (mri) data. IEEE Access 2019, 7, 12252–12261. [Google Scholar] [CrossRef]
- Khan, M.A.; Ashraf, I.; Alhaisoni, M.; Damaeviius, R.; Scherer, R.; Rehman, A.; Bukhari, S.A.C. Multimodal brain tumor classification using deep learning and robust feature selection: A machine learning application for radiologists. Diagnostics 2020, 10, 565. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Chen, C.; Liu, X.; Ding, M.; Zheng, J.; Li, J. 3D dilated multi-fiber network for real-time brain tumor segmentation in mri. In Lecture Notes in Computer Science, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; Springer: Cham, Switzerland, 2019; Volume 11766, pp. 184–192. [Google Scholar]
- Chen, Y.; Kalantidis, Y.; Li, J.; Yan, S.; Feng, J. Multi-Fiber Networks for Video Recognition; Springer International Publishing: Cham, Switzerland, 2018; Volume 11205, pp. 364–380. [Google Scholar]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. Cancer Imaging Arch. 2017. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. Cancer Imaging Arch. 2017, 286. [Google Scholar] [CrossRef]
- Sharif, M.I.; Li, J.P.; Khan, M.A.; Saleem, M.A. Active deep neural network features selection for segmentation and recognition of brain tumors using mri images. Pattern Recognit. Lett. 2020, 129, 181–189. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation; Springer International Publishing: Cham, Switzerland, 2016; Volume 9901, pp. 424–432. [Google Scholar]
- Nuechterlein, N.; Mehta, S. 3D-ESPNet with Pyramidal Refinement for Volumetric Brain Tumor Image Segmentation; Springer International Publishing: Cham, Switzerland, 2019; Volume 11384, pp. 245–253. [Google Scholar]
- Cheng, J.; Liu, J.; Liu, L.; Pan, Y.; Wang, J. Multi-level glioma segmentation using 3D u-net combined attention mechanism with atrous convolution. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 1031–1036. [Google Scholar]
- Fang, L.; He, H. Three pathways u-net for brain tumor segmentation. In Proceedings of the 7th Medical Image Computing and Computer-Assisted Interventions (MICCAI) BraTS Challenge, Granada, Spain, 16–20 September 2018; Volume 2018, pp. 119–126. [Google Scholar]
- Gates, E.; Pauloski, J.G.; Schellingerhout, D.; Fuentes, D. Glioma Segmentation and a Simple Accurate Model for Overall Survival Prediction; Springer International Publishing: Cham, Switzerland, 2019; Volume 11384, pp. 476–484. [Google Scholar]
- Hu, X.; Li, H.; Zhao, Y.; Dong, C.; Menze, B.H.; Piraud, M. Hierarchical Multi-Class Segmentation of Glioma Images Using Networks with Multi-Level Activation Function; Springer International Publishing: Cham, Switzerland, 2019; Volume 11384, pp. 116–127. [Google Scholar]
- Lyu, C.; Shu, H. A two-stage cascade model with variational autoencoders and attention gates for mri brain tumor segmentation. arXiv 2020, arXiv:2011.02881. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).