An Improved Sub-Model PLSR Quantitative Analysis Method Based on SVM Classifier for ChemCam Laser-Induced Breakdown Spectroscopy
Abstract
1. Introduction
2. Dataset
3. SVM and Partial Least Squares Regression (PLSR) Algorithm
3.1. Support Vector Machine
3.2. Partial Least Square Regression
- 1.
- Extracting a PLS components (t, Equation (9)) of X.
- 2.
- Obtaining the parameter vectors (p and q) of regression equations (Equations (10) and (11)).
- 3.
- Building regression equations (Equations (12) and (13)).
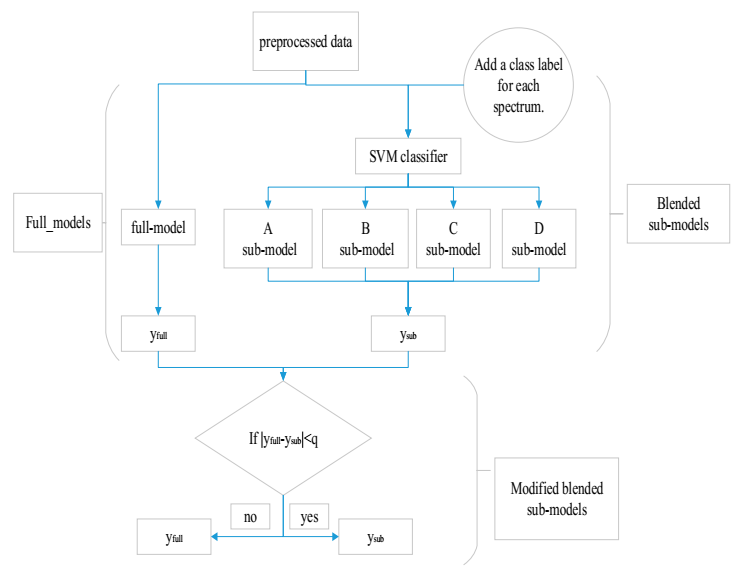
4. Implementation
4.1. Selecting a Sub-Model by the SVM Classifier
4.2. The Optimized Method for Final Outputs of Blended Sub-Models a Sub-Model
4.3. Model Evaluation
5. Results
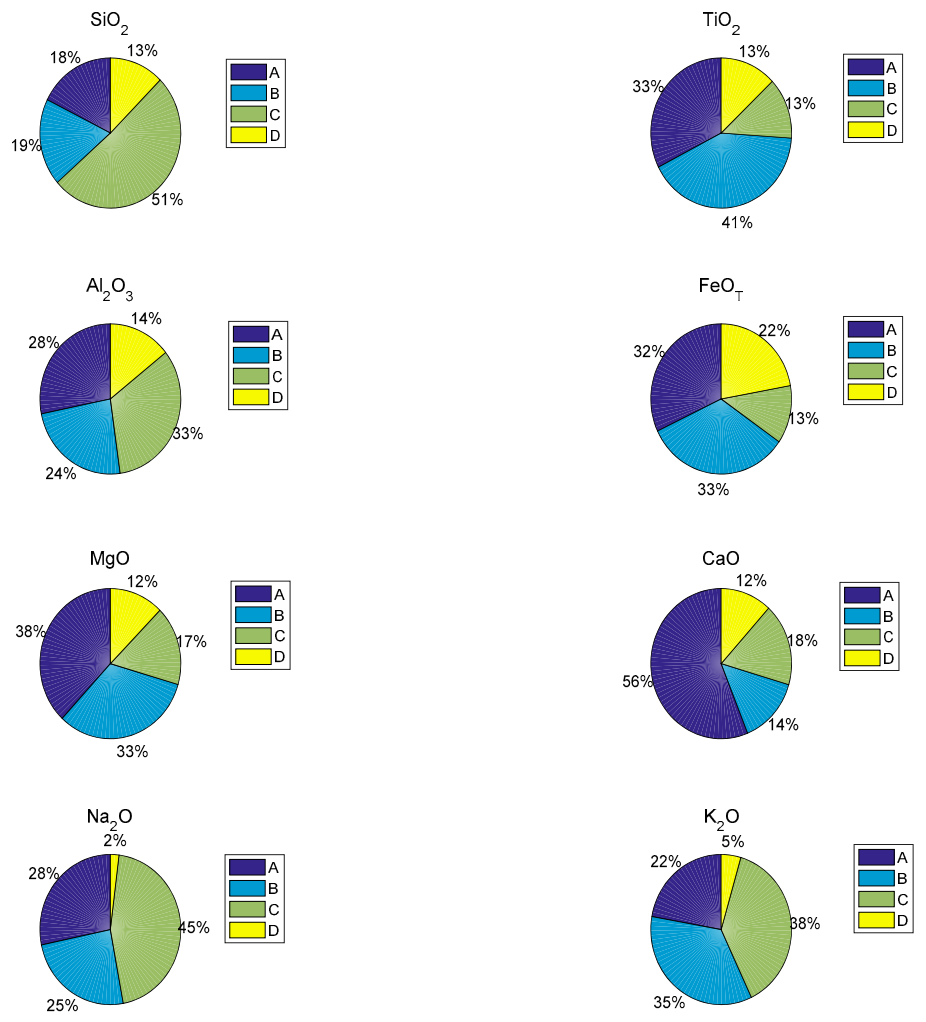
5.1. Performances of Regression Sub-Models and SVM Classifiers
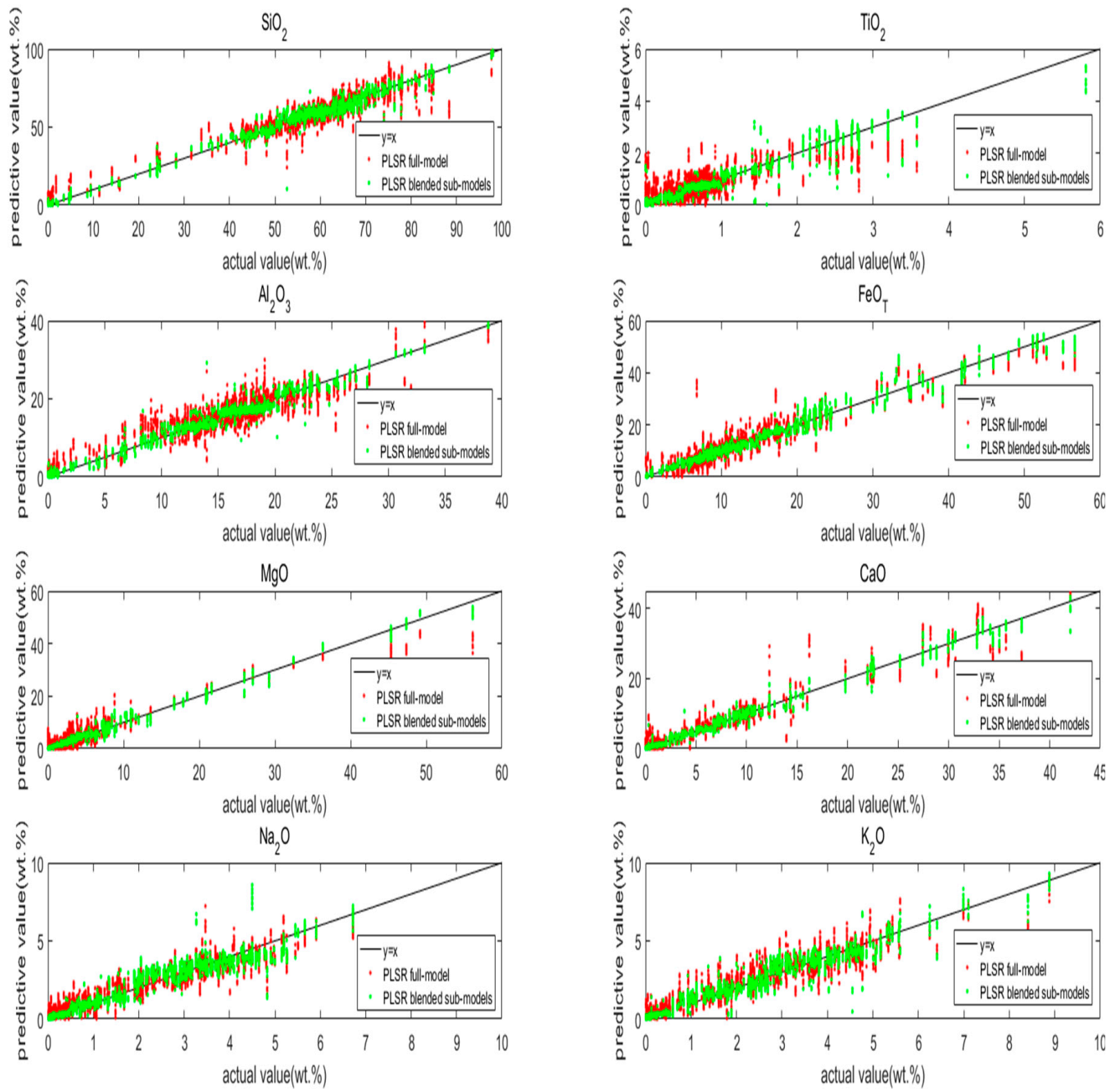
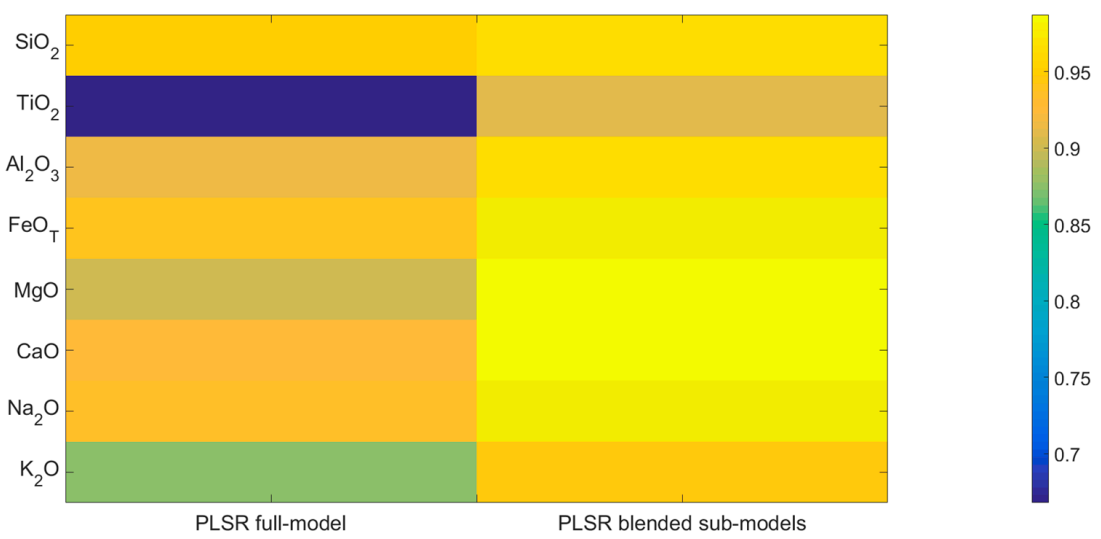
5.2. PLSR Models
5.3. Effect of Optimized Idea
5.4. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, J.; Li, X.; Lu, H.; Xu, J.; Li, H. An LIBS quantitative analysis method for alloy steel at high temperature based on transfer learning. J. Anal. At. Spectrom. 2018, 33, 1184–1195. [Google Scholar] [CrossRef]
- Cousin, A.; Sautter, V.; Payré, V.; Forni, O.; Mangold, N.; Gasnault, O.; Le Deit, L.; Johnson, J.; Maurice, S.; Salvatore, M.; et al. Classification of igneous rocks analyzed by ChemCam at Gale crater, Mars. Icarus 2017, 288, 265–283. [Google Scholar] [CrossRef]
- Roux, C.P.M.; Rakovský, J.; Musset, O.; Monna, F.; Buoncristiani, J.F.; Pellenard, P.; Thomazo, C. In situ Laser Induced Breakdown Spectroscopy as a tool to discriminate volcanic rocks and magmatic series, Iceland. Spectrochim. Acta Part B At. Spectrosc. 2015, 103–104, 63–69. [Google Scholar] [CrossRef]
- Sautter, V.; Fabre, C.; Forni, O.; Toplis, M.J.; Cousin, A.; Ollila, A.M.; Meslin, P.Y.; Maurice, S.; Wiens, R.C.; Baratoux, D.; et al. Igneous mineralogy at Bradbury Rise: The first ChemCam campaign at Gale crater. J. Geophys. Res. Planets 2014, 119, 30–46. [Google Scholar] [CrossRef]
- Rossi, M.; Aglio, M.D.; De Giacomo, A.; Gaudiuso, R.; Senesi, G.S.; De Pascale, O.; Capitelli, F.; Nestola, F.; Ghiara, M.R. Multi-methodological investigation of kunzite, hiddenite, alexandrite, elbaite and topaz, based on laser-induced breakdown spectroscopy and conventional analytical techniques for supporting mineralogical characterization. Phys. Chem. Miner. 2014, 41, 127–140. [Google Scholar] [CrossRef]
- Clegg, S.M.; Wiens, R.C.; Anderson, R.; Forni, O.; Frydenvang, J.; Lasue, J.; Cousin, A.; Payré, V.; Boucher, T.; Dyar, M.D.; et al. Recalibration of the Mars Science Laboratory ChemCam instrument with an expanded geochemical database. Spectrochim. Acta Part B At. Spectrosc. 2017, 129, 64–85. [Google Scholar] [CrossRef]
- Wiens, R.C.; Maurice, S.; Barraclough, B.; Saccoccio, M.; Barkley, W.C.; Bell, J.F., III; Bender, S.; Bernardin, J.; Blaney, D.; Blank, J.; et al. The ChemCam Instrument Suite on the Mars Science Laboratory (MSL) Rover: Body Unit and Combined System Tests. Space Sci. Rev. 2012, 170, 167–227. [Google Scholar] [CrossRef]
- Fu, H.; Ni, Z.; Wang, H.; Jia, J.; Dong, F. Accuracy improvement of calibration-free laser-induced breakdown spectoscopy. Plasma Sci. Technol. 2019, 21, 034001. [Google Scholar] [CrossRef]
- Yang, J.; Li, X.; Xu, J.; Ma, X. A Calibration-Free Laser-Induced Breakdown Spectroscopy (CF-LIBS) Quantitative Analysis Method Based on the Auto-Selection of an Internal Reference Line and Optimized Estimation of Plasma Temperature. Appl. Spectrosc. 2018, 72, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wang, B.; Jiang, X.; Zhang, J.; Niu, K.; Yuan, Y. Detection of K in soil using time-resolved laser-induced breakdown spectroscopy based on convolutional neural networks. Plasma Sci. Technol. 2019, 21, 34014. [Google Scholar] [CrossRef]
- Guezenoc, J.; Payré, V.; Fabre, C.; Syvilay, D.; Cousin, A.; Gallet-Budynek, A.; Bousquet, B. Variable selection in laser-induced breakdown spectroscopy assisted by multivariate analysis: An alternative to multi-peak fitting. Spectrochim. Acta Part B At. Spectrosc. 2019, 152, 6–13. [Google Scholar] [CrossRef]
- Anderson, R.B.; Clegg, S.M.; Frydenvang, J.; Wiens, R.C.; McLennan, S.; Morris, R.V.; Ehlmann, B.; Dyar, M.D. Improved accuracy in quantitative laser-induced breakdown spectroscopy using sub-models. Spectrochim. Acta Part B At. Spectrosc. 2017, 129, 49–57. [Google Scholar] [CrossRef]
- Yuanyuan, C.; Zhibin, W. Quantitative analysis modeling of infrared spectroscopy based on ensemble convolutional neural networks. Chemom. Intell. Lab. Syst. 2018, 181, 1–10. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, T.; Xu, J.; Luo, X.; Ying, Y. DeepSpectra: An end-to-end deep learning approach for quantitative spectral analysis. Anal. Chim. Acta 2019, 1058, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Bjerrum, E.J.; Glahder, M.; Skov, T. Data Augmentation of Spectral Data for Convolutional Neural Network (CNN) Based Deep Chemometrics. arXiv 2017, arXiv:1710.01927. [Google Scholar]
- Padarian, J.; Minasny, B.; McBratney, A.B. Using deep learning to predict soil properties from regional spectral data. Geoderma Reg. 2019, 16, e00198. [Google Scholar] [CrossRef]
- Fabre, C.; Maurice, S.; Cousin, A.; Wiens, R.C.; Forni, O.; Sautter, V.; Guillaume, D. Onboard calibration igneous targets for the Mars Science Laboratory Curiosity rover and the Chemistry Camera laser induced breakdown spectroscopy instrument. Spectrochim. Acta Part B At. Spectrosc. 2011, 66, 280–289. [Google Scholar] [CrossRef]
- Boser, E.; Guyon, M.; Vapnik, V.N. A Training Algorithm for Optimal Margin Classifiers. In Proceedings of the 5th Annual Workshop on Computational Learning Theory (COLT’ 92), Pittsburgh, PA, USA, 27–29 July 1992; pp. 144–152. [Google Scholar]

| A Class | B Class | C Class | D Class | |
|---|---|---|---|---|
| SiO2 | 0–30% | 30–50% | 50–70% | 70–100% |
| TiO2 | 0–0.5% | 0.5–1% | 1–2% | 2–100% |
| AL2O3 | 0–10% | 10–15% | 15–20% | 20–100% |
| FeOT | 0–8% | 8–15% | 15–20% | 20–100% |
| MgO | 0–2% | 2–3.5% | 3.5–7% | 7–100% |
| CaO | 0–2% | 2–7% | 7–19% | 19–100% |
| Na2O | 0–0.5% | 0.5–1.9% | 1.9–5.4% | 5.4–100% |
| K2O | 0–0.6% | 0.6–2.7% | 2.7–5.3% | 5.3–100% |
| Sub-Model | Training Spectra | Testing Spectra | RMSEP of Each PLSR Sub-Model | |
|---|---|---|---|---|
| SiO2 | A sub-model | 600 | 300 | 0.9004 |
| B sub-model | 1614 | 810 | 0.9227 | |
| C sub-model | 4314 | 2160 | 1.3831 | |
| D sub-model | 959 | 480 | 1.0504 | |
| TiO2 | A sub-model | 2200 | 1100 | 0.0755 |
| B sub-model | 3240 | 1620 | 0.0737 | |
| C sub-model | 1074 | 540 | 0.1203 | |
| D sub-model | 714 | 360 | 0.1843 | |
| AL2O3 | A sub-model | 1500 | 750 | 0.2758 |
| B sub-model | 1948 | 980 | 0.3574 | |
| C sub-model | 2760 | 1380 | 0.4761 | |
| D sub-model | 1260 | 630 | 0.6580 | |
| FeOT | A sub-model | 2080 | 1040 | 0.3455 |
| B sub-model | 2680 | 1340 | 0.4479 | |
| C sub-model | 1294 | 650 | 0.4370 | |
| D sub-model | 1434 | 720 | 0.7856 | |
| MgO | A sub-model | 2500 | 1250 | 0.1240 |
| B sub-model | 2480 | 1240 | 0.1469 | |
| C sub-model | 1468 | 739 | 0.3703 | |
| D sub-model | 1000 | 500 | 0.6468 | |
| CaO | A sub-model | 4000 | 2000 | 0.1166 |
| B sub-model | 1220 | 610 | 0.4008 | |
| C sub-model | 1648 | 830 | 0.5528 | |
| D sub-model | 600 | 300 | 1.0975 | |
| Na2O | A sub-model | 1740 | 870 | 0.0833 |
| B sub-model | 1780 | 890 | 0.1163 | |
| C sub-model | 3608 | 1810 | 0.2592 | |
| D sub-model | 200 | 100 | 0.7203 | |
| K2O | A sub-model | 1754 | 880 | 0.0792 |
| B sub-model | 2514 | 1260 | 0.1836 | |
| C sub-model | 2620 | 1309 | 0.2240 | |
| D sub-model | 320 | 160 | 0.4929 |
| Training Set | Testing Set | |||
|---|---|---|---|---|
| Optimal Parameters of SVM Classifiers | CV | CA | ||
| C | γ | |||
| SiO2 | 97.0059 | 0.0010 | 99.6261% | 97.8667% (3670/3750) |
| TiO2 | 73.5167 | 0.0030 | 99.5849% | 97.2652% (3521/3620) |
| AL2O3 | 48.5029 | 0.0052 | 99.2100% | 96.6310% (3614/3740) |
| FeOT | 97.0059 | 0.0017 | 99.4658% | 98.0533% (3677/3750) |
| MgO | 168.8970 | 0.0017 | 99.2213% | 97.7480% (3646/3730) |
| CaO | 97.0059 | 0.0010 | 99.8527% | 98.9037% (3699/3740) |
| Na2O | 55.7152 | 0.0017 | 99.5497% | 97.8474% (3591/3670) |
| K2O | 97.0059 | 0.0052 | 99.0844% | 96.8975% (3498/3610) |
| Average | __ | 97.65% | ||
| Oxide | Training Spectra | Testing Spectra | Modified PLSR Blended Sub-Models | PLSR Blended Sub-Models | PLSR Full Model |
|---|---|---|---|---|---|
| SiO2 | 7488 | 3750 | 3.7936 | 3.1837 | 5.6280 |
| TiO2 | 7228 | 3620 | 0.4378 | 0.2309 | 0.4797 |
| AL2O3 | 7468 | 3740 | 2.0075 | 1.2711 | 2.7404 |
| FeOT | 7488 | 3750 | 2.6758 | 1.7602 | 3.5476 |
| MgO | 7448 | 3730 | 2.1826 | 0.8550 | 2.2724 |
| CaO | 7468 | 3740 | 2.3404 | 0.9731 | 2.4510 |
| Na2O | 7328 | 3670 | 0.7549 | 0.4948 | 0.7834 |
| K2O | 7208 | 3610 | 0.6760 | 0.4568 | 0.7009 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, L.; Liu, F.; Zhang, L. An Improved Sub-Model PLSR Quantitative Analysis Method Based on SVM Classifier for ChemCam Laser-Induced Breakdown Spectroscopy. Symmetry 2021, 13, 319. https://doi.org/10.3390/sym13020319
Han L, Liu F, Zhang L. An Improved Sub-Model PLSR Quantitative Analysis Method Based on SVM Classifier for ChemCam Laser-Induced Breakdown Spectroscopy. Symmetry. 2021; 13(2):319. https://doi.org/10.3390/sym13020319
Chicago/Turabian StyleHan, Liang, Feng Liu, and Li Zhang. 2021. "An Improved Sub-Model PLSR Quantitative Analysis Method Based on SVM Classifier for ChemCam Laser-Induced Breakdown Spectroscopy" Symmetry 13, no. 2: 319. https://doi.org/10.3390/sym13020319
APA StyleHan, L., Liu, F., & Zhang, L. (2021). An Improved Sub-Model PLSR Quantitative Analysis Method Based on SVM Classifier for ChemCam Laser-Induced Breakdown Spectroscopy. Symmetry, 13(2), 319. https://doi.org/10.3390/sym13020319





