On the Hierarchical Bernoulli Mixture Model Using Bayesian Hamiltonian Monte Carlo
Abstract
1. Introduction
2. Materials and Methods
2.1. Bernoulli Mixture Model
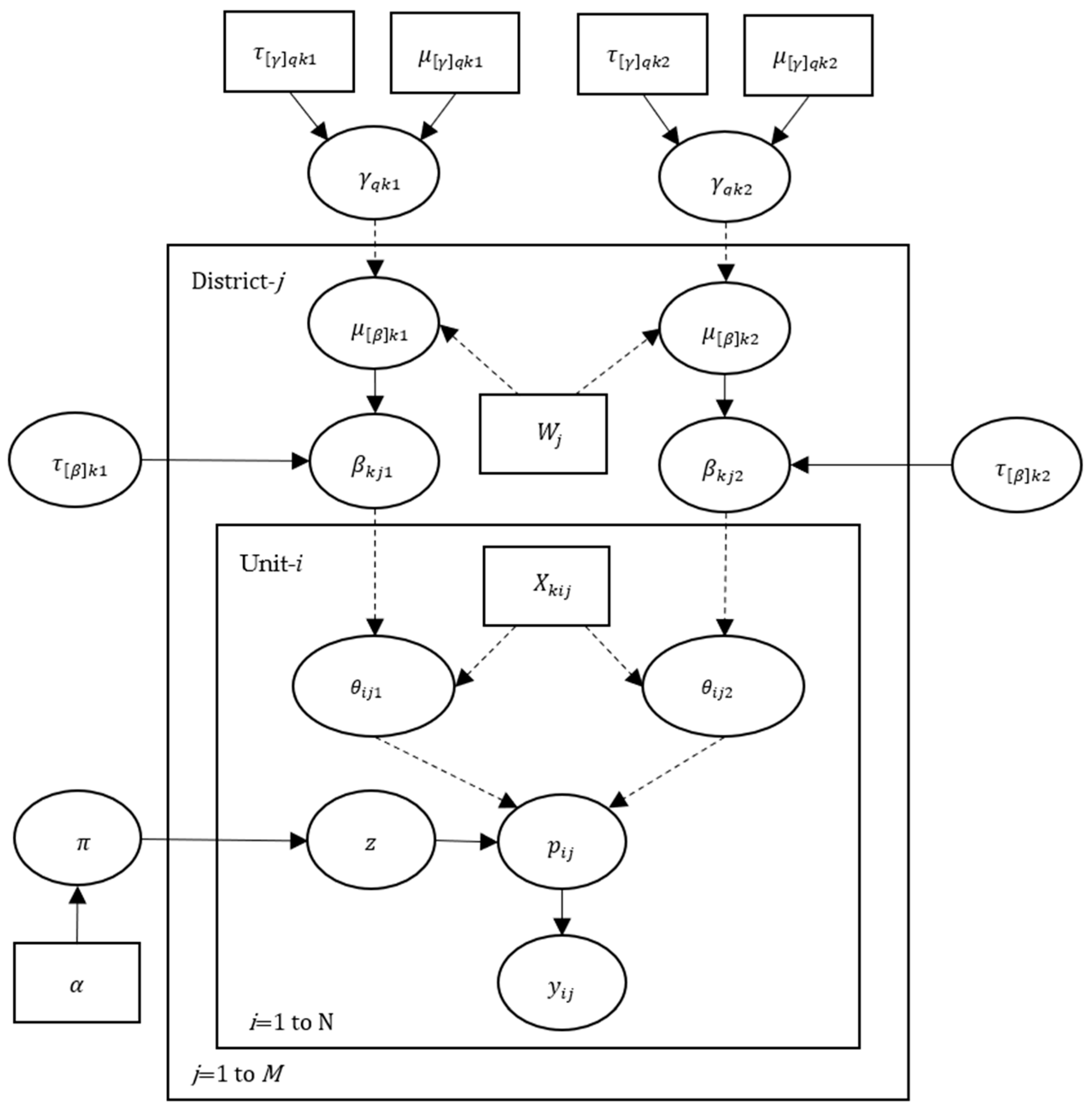
2.2. Directed Acyclic Graph of Hibermimo
2.3. Prior Distribution of Hibermimo
2.4. Posterior Distribution of Hibermimo
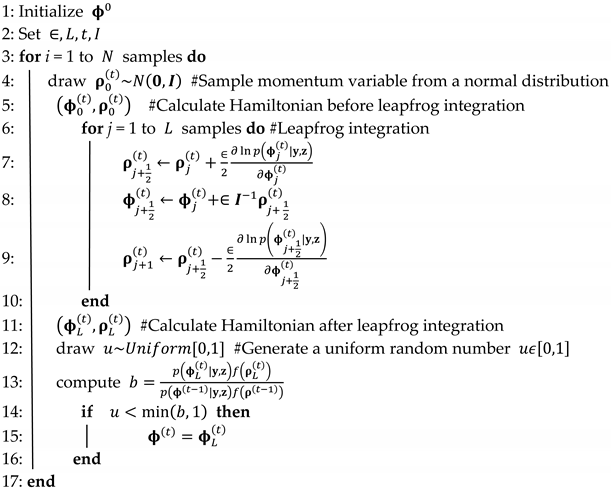
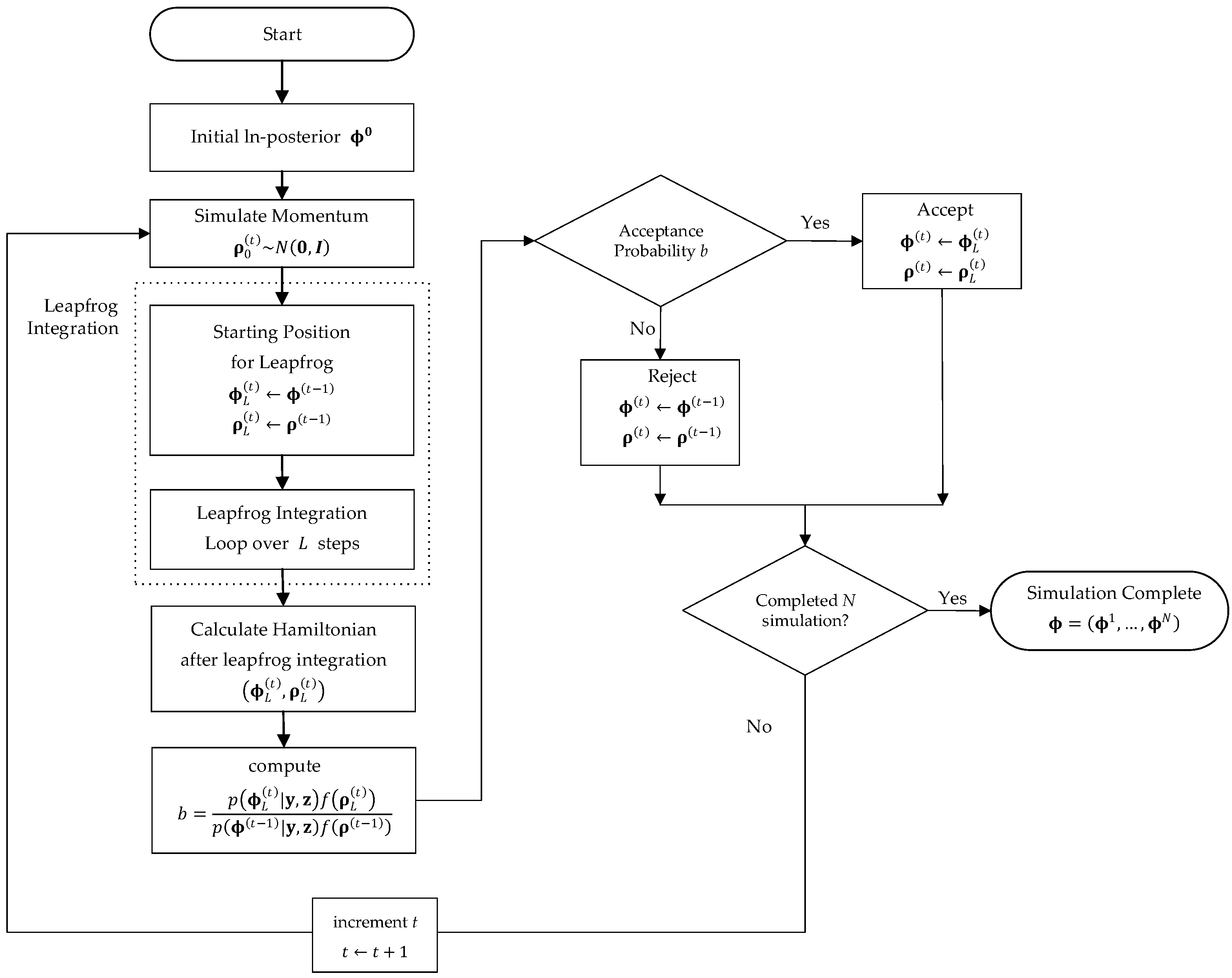
2.5. Hamiltonian Monte Carlo (HMC)
- Step 1.
- Specify the likelihood function of the Bernoulli Mixture Model .
- Step 2.
- Determine the prior distributions of Hibermimo: , , and .
- Step 3.
- Perform the first derivative of the ln-posterior for each Hibermimo parameter ; .
- Step 4.
- Set the initial value of the parameter , the diagonal mass matrix , the leapfrog integration step size (indicating the leapfrog step jumps), the number of leapfrog integration steps , and the number of iterations t.
- Step 5.
- Perform the parameter estimation of Hibermimo using the HMC algorithm;Algorithm 1 contains a pseudo-code for an implementation of the Hamiltonian algorithm for Hibermimo.
Algorithm 1 The Hamiltonian Monte Carlo for Hibermimo. - Step 6.
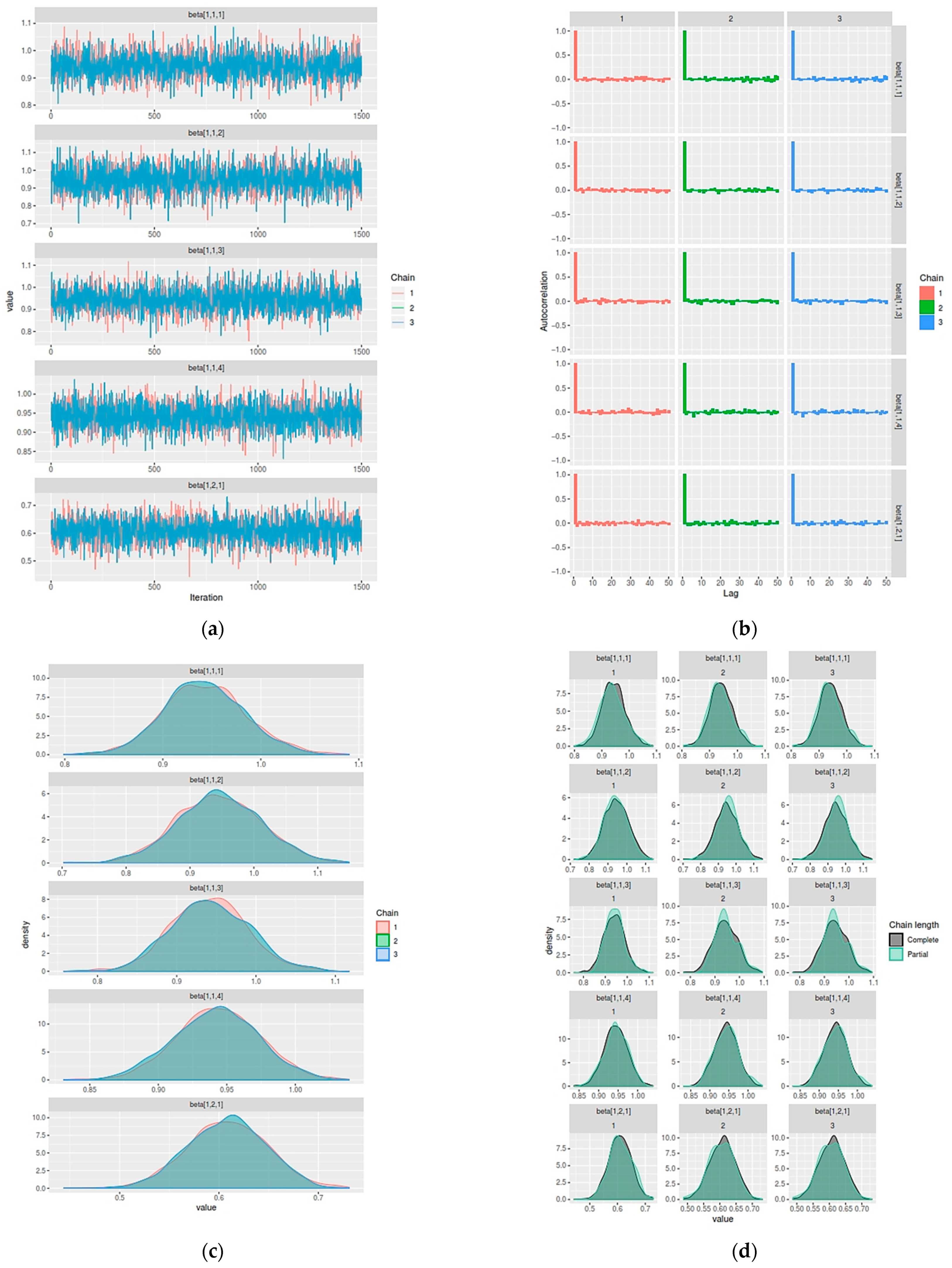
- Monitor and evaluate the convergence of the algorithm.
- Step 7.
- Plot the posterior distribution of Hibermimo.
- Step 8.
- Obtain a summary of the posterior distribution of Hibermimo.
3. Results
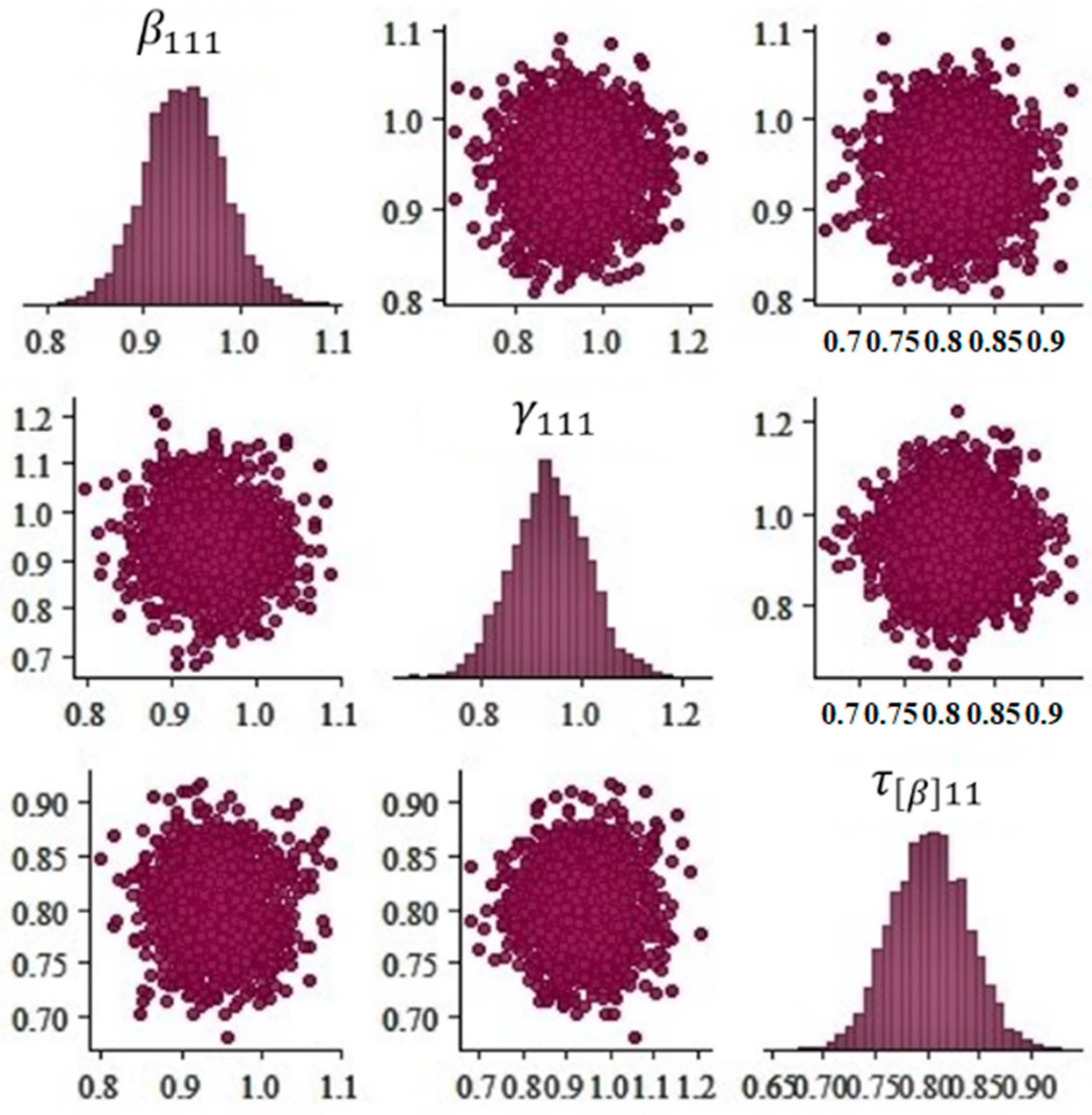
3.1. Parameter Estimation of Hibermimo
3.2. Application
3.2.1. Bayesian Bernoulli Mixture Aggregate Regression Model
3.2.2. Hierarchical Bernoulli Mixture Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification, 2nd ed.; Wiley-Interscience: Hoboken, NJ, USA, 2000; ISBN 0-471-05669-3. [Google Scholar]
- Iriawan, N.; Fithriasari, K.; Ulama, B.S.S.; Suryaningtyas, W.; Pangastuti, S.S.; Cahyani, N.; Qadrini, L. On The Comparison: Random Forest, SMOTE-Bagging, and Bernoulli Mixture to Classify Bidikmisi Dataset in East Java. In Proceedings of the 2018 International Conference on Computer Engineering, Network and Intelligent Multimedia (CENIM), Surabaya, Indonesia, 26–27 November 2018; pp. 137–141. [Google Scholar]
- Grim, J.; Pudil, P.; Somol, P. Multivariate Structural Bernoulli Mixtures for Recognition of Handwritten Numerals. In Proceedings of the Proceedings 15th International Conference on Pattern Recognition. ICPR-2000, Barcelona, Spain, 3–7 September 2000; Volume 2, pp. 585–589. [Google Scholar]
- González, J.; Juan, A.; Dupont, P.; Vidal, E.; Casacuberta, F. Pattern Recognition and Image Analysis. In Proceedings of the Pattern Recognition and Image Analysis, Universitat Jaume I, Servei de Comunicació i Publicacions, Benicasim, Spain, 16–18 May 2001. [Google Scholar]
- Juan, A.; Vidal, E. On the Use of Bernoulli Mixture Models for Text Classification. Pattern Recognit. 2002, 35, 2705–2710. [Google Scholar] [CrossRef]
- Juan, A.; Vidal, E. Bernoulli Mixture Models for Binary Images. In Proceedings of the 17th International Conference on Pattern Recognition, 2004, ICPR 2004, Cambridge, UK, 26 August 2004; Volume 3, pp. 367–370. [Google Scholar]
- Patrikainen, A.; Mannila, H. Subspace Clustering of High-Dimensional Binary Data—A Probabilistic Approach. In Proceedings of the In Workshop on Clustering High Dimensional Data and its Applications, SIAM International Conference on Data Mining, Lake Buena Vista, FL, USA, 24 April 2004. [Google Scholar]
- Bouguila, N. On Multivariate Binary Data Clustering and Feature Weighting. Comput. Stat. Data Anal. 2010, 54, 120–134. [Google Scholar] [CrossRef]
- Zhu, S.; Takigawa, I.; Zhang, S.; Mamitsuka, H. A Probabilistic Model for Clustering Text Documents with Multiple Fields. In Advances in Information Retrieval; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4425, pp. 331–342. ISBN 978-3-540-71494-1. [Google Scholar]
- Sun, Z.; Rosen, O.; Sampson, A.R. Multivariate Bernoulli Mixture Models with Application to Postmortem Tissue Studies in Schizophrenia. Biometrics 2007, 63, 901–909. [Google Scholar] [CrossRef]
- Tikka, J.; Hollmén, J.; Myllykangas, S. Mixture Modeling of DNA Copy Number Amplification Patterns in Cancer. In Computational and Ambient Intelligence; Sandoval, F., Prieto, A., Cabestany, J., Graña, M., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4507, pp. 972–979. ISBN 978-3-540-73006-4. [Google Scholar]
- Myllykangas, S.; Tikka, J.; Böhling, T.; Knuutila, S.; Hollmén, J. Classification of Human Cancers Based on DNA Copy Number Amplification Modeling. BMC Med. Genom. 2008, 1, 15. [Google Scholar] [CrossRef]
- Saeed, M.; Javed, K.; Atique Babri, H. Machine Learning Using Bernoulli Mixture Models: Clustering, Rule Extraction and Dimensionality Reduction. Neurocomputing 2013, 119, 366–374. [Google Scholar] [CrossRef]
- Hox, J.J. Multilevel Analysis: Techniques and Applications, 2nd ed.; Quantitative Methodology Series; Routledge, Taylor & Francis: New York, NY, USA, 2010; ISBN 978-1-84872-845-5. [Google Scholar]
- Goldstein, H. Multilevel Statistical Models, 4th ed.; Wiley Series in Probability and Statistics; Wiley: Chichester, UK, 2011; ISBN 978-0-470-74865-7. [Google Scholar]
- Hox, J.J. Applied Multilevel Analysis; TT-Publikaties: Amsterdam, The Netherlands, 1995; ISBN 978-90-801073-2-8. [Google Scholar]
- Ismartini, P.; Iriawan, N.; Setiawan; Ulama, B.S.S. Toward a Hierarchical Bayesian Framework for Modelling the Effect of Regional Diversity on Household Expenditure. J. Math. Stat. 2012, 8, 283–291. [Google Scholar] [CrossRef][Green Version]
- Ringdal, K. Recent Developments in: Methods for Multilevel Analysis. Acta Sociol. 1992, 35, 235–243. [Google Scholar] [CrossRef]
- Suryaningtyas, W.; Iriawan, N.; Fithriasari, K.; Ulama, B.; Susanto, I.; Pravitasari, A. On The Bernoulli Mixture Model for Bidikmisi Scholarship Classification with Bayesian MCMC. J. Phys. Conf. Ser. 2018, 1090, 012072. [Google Scholar] [CrossRef]
- Carpenter, B.; Gelman, A.; Hoffman, M.D.; Lee, D.; Goodrich, B.; Betancourt, M.; Brubaker, M.; Guo, J.; Li, P.; Riddell, A. Stan: A Probabilistic Programming Language. J. Stat. Soft. 2017, 76, 1–32. [Google Scholar] [CrossRef]
- McLachlan, G.; Peel, D. Finite Mixture Models; John Wiley and Sons: New York, NY, USA, 2000; ISBN 0-471-00626-2. [Google Scholar]
- King, R.; Morgan, B.J.T.; Gimenez, O.; Brooks, S.P. Bayesian Analysis for Population Ecology; Interdisciplinary Statistics Series; Chapman & Hall/CRC: Boca Raton, FL, USA, 2010; ISBN 978-1-4398-1187-0. [Google Scholar]
- Carlin, B.P.; Chib, S. Bayesian Model Choice via Markov Chain Monte Carlo Methods. J. R. Stat. Soc. Ser. B 1995, 57, 473–484. [Google Scholar] [CrossRef]
- Box, G.E.P.; Tiao, G.C. Bayesian Inference in Statistical Analysis; Addison-Wesley Series in Behavioral Science; Addison-Wesley: Reading, MA, USA, 1973; ISBN 978-0-201-00622-3. [Google Scholar]
- Guo, G.; Zhao, H. Multilevel Modeling for Binary Data. Annu. Rev. Sociol. 2000, 26, 441–462. [Google Scholar] [CrossRef]
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Dunson, D.B.; Vehtari, A.; Rubin, D.B. Bayesian Data Analysis, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2014; ISBN 978-1-4398-9820-8. [Google Scholar]
- Solikhah, A.; Kuswanto, H.; Iriawan, N.; Fithriasari, K. Fisher’s z Distribution-Based Mixture Autoregressive Model. Econometrics 2021, 9, 27. [Google Scholar] [CrossRef]
- Gamerman, D. Markov Chain Monte Carlo for Dynamic Generalised Linear Models. Biometrika 1998, 85, 215–227. [Google Scholar] [CrossRef]
- Iriawan, N.; Fithriasari, K.; Ulama, B.S.S.; Susanto, I.; Suryaningtyas, W.; Pravitasari, A.A. On the Markov Chain Monte Carlo Convergence Diagnostic of Bayesian Bernoulli Mixture Regression Model for Bidikmisi Scholarship Classification. In Proceedings of the Third International Conference on Computing, Mathematics and Statistics (iCMS2017), Langkawi, Malaysia, 7–8 November 2017; Kor, L.-K., Ahmad, A.-R., Idrus, Z., Mansor, K.A., Eds.; Springer: Singapore, 2019; pp. 397–403, ISBN 978-981-13-7279-7. [Google Scholar]
- Wang, Z.; Mohamed, S.; De Freitas, N. Adaptive Hamiltonian and Riemann Manifold Monte Carlo Samplers. In Proceedings of the 30th International Conference on Machine Learning, Proceedings of Machine Learning Research (PMLR), Atlanta, GA, USA, 17–19 June 2013; Volume 28, pp. 1462–1470. [Google Scholar]
- Hoffman, M.D.; Gelman, A. The No-U-Turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo. J. Mach. Learn. Res. 2014, 15, 1593–1623. [Google Scholar]
- Grantham, N.S. Clustering Binary Data with Bernoulli Mixture Models. In Unpublished Written Preliminary Exam; NC State University: Raleigh, NC, USA, 2014. [Google Scholar]
- Hanson, K.M. Markov Chain Monte Carlo Posterior Sampling with The Hamiltonian Method. Proc. SPIE—Int. Soc. Opt. Eng. 2001, 4322, 456–467. [Google Scholar] [CrossRef]
- Metropolis, N.; Rosenbluth, A.W.; Rosenbluth, M.N.; Teller, A.H.; Teller, E. Equation of State Calculations by Fast Computing Machines. J. Chem. Phys. 1953, 21, 1087. [Google Scholar] [CrossRef]
- Rossberg, K. A First Course in Analytical Mechanics. Am. J. Phys. 1984, 52, 1155. [Google Scholar] [CrossRef]
- Andersen, H.C. Molecular Dynamics Simulations at Constant Pressure and/or Temperature. J. Chem. Phys. 1980, 72, 2384–2393. [Google Scholar] [CrossRef]
- Stan Development Team. Stan User’s Guide, Version 2.18.0. 2018. Available online: https://mc-stan.org/docs/2_18/stan-users-guide/index.html (accessed on 21 October 2020).
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Rubin, D.B. Bayesian Data Analysis; Chapman and Hall/CRC: Boca Raton, FL, USA, 2004; ISBN 1-58488-388-X. [Google Scholar]
- Koop, G. Bayesian Econometrics; J. Wiley: Hoboken, NJ, USA, 2003; ISBN 978-0-470-84567-7. [Google Scholar]
- Gelfand, A.E.; Smith, A.F.M. Sampling-Based Approaches to Calculating Marginal Densities. J. Am. Stat. Assoc. 1990, 85, 398–409. [Google Scholar] [CrossRef]
- Arnold, B.C.; Castillo, E.; Sarabia, J.-M. Conditional Specification of Statistical Models; Springer Series in Statistics; Springer: New York, NY, USA, 1999; ISBN 978-0-387-98761-3. [Google Scholar]
- Kay, R.; Little, S. Transformations of the Explanatory Variables in the Logistic Regression Model for Binary Data. Biometrika 1987, 74, 495–501. [Google Scholar] [CrossRef]
- Mason, W.M.; Wong, G.Y.; Entwisle, B. Contextual Analysis through the Multilevel Linear Model. Sociol. Methodol. 1983, 14, 72–103. [Google Scholar] [CrossRef]
- Goldstein, H. Multilevel Mixed Linear Model Analysis Using Iterative Generalized Least Squares. Biometrika 1986, 73, 43–56. [Google Scholar] [CrossRef]
- Longford, N. A Fast Scoring Algorithm for Maximum Likelihood Estimation in Unbalanced Mixed Models with Nested Random Effects. ETS Res. Rep. Ser. 1987, 74, 817–827. [Google Scholar] [CrossRef]
- Bryk, A.S.; Raudenbush, S.W. Toward a More Appropriate Conceptualization of Research on School Effects: A Three-Level Hierarchical Linear Model. Am. J. Educ. 1988, 97, 65–108. [Google Scholar] [CrossRef]
- Goldstein, H.; Rasbash, J. Improved Approximations for Multilevel Models with Binary Responses. J. R. Stat. Soc. Ser. A 1996, 159, 505–513. [Google Scholar] [CrossRef]
- Rodriguez, G.; Goldman, N. An Assessment of Estimation Procedures for Multilevel Models with Binary Responses. J. R. Stat. Soc. Ser. A (Stat. Soc.) 1995, 158, 73–89. [Google Scholar] [CrossRef]
- Taylor, H.M.; Karlin, S. An Introduction to Stochastic Modelling; Academic Press: New York, NY, USA, 1994; ISBN 978-0-12-684887-4. [Google Scholar]
- Bolstad, W.M. Understanding Computational Bayesian Statistics, 1st ed.; Wiley Series in Computational Statistics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; ISBN 0-470-04609-0. [Google Scholar]
- Wilkinson, L.; Wills, G. The Grammar of Graphics, Statistics and Computing, 2nd ed.; Springer: New York, NY, USA, 2005; ISBN 978-0-387-24544-7. [Google Scholar]
- Gelman, A.; Rubin, D.B. Inference from Iterative Simulation Using Multiple Sequences. Stat. Sci. 1992, 7, 457–472. [Google Scholar] [CrossRef]
- Raftery, A.E.; Lewis, S. How Many Iterations in the Gibbs Sampler? Department of Statistics, University of Washington: Seattle, WA, USA, 1991. [Google Scholar]
- Geweke, J. Evaluating the Accuracy of Sampling-Based Approaches to the Calculation of Posterior Moments; Federal Reserve Bank of Minneapolis: Minneapolis, MN, USA, 1991. [Google Scholar]
- Heidelberger, P.; Welch, P.D. Simulation Run Length Control in the Presence of an Initial Transient. Oper. Res. 1983, 31, 1109–1144. [Google Scholar] [CrossRef]
- Fernández-i-Marín, X. Ggmcmc: Analysis of MCMC Samples and Bayesian Inference. J. Stat. Soft. 2016, 70, 1–20. [Google Scholar] [CrossRef]
- Watanabe, S. Asymptotic Equivalence of Bayes Cross Validation and Widely Applicable Information Criterion in Singular Learning Theory. J. Mach. Learn. Res. 2010, 11, 3571–3594. [Google Scholar]
- Prasetyo, R.B.; Kuswanto, H.; Iriawan, N.; Ulama, B.S.S. Binomial Regression Models with a Flexible Generalized Logit Link Function. Symmetry 2020, 12, 221. [Google Scholar] [CrossRef]
| Variable | Description | Data Scale |
|---|---|---|
| Percentage of the poverty population | Ratio | |
| The average extent of school | Ratio | |
| Percentage of population aged 19–24 out of school | Ratio | |
| Percentage of households with roofs made from asbestos/zinc + bamboo/wood + straw/fiber/leaves/other | Ratio | |
| Percentage of households with wooden walls | Ratio | |
| Percentage of households receiving subsidies | Ratio | |
| Percentage of households receiving insufficient student aid for high school students | Ratio | |
| Percentage of households whose members have accessed the internet in the last 3 months | Ratio |
| Parameters | Mean | 2.5% | 50% | 97.5% | n_eff | Rhat |
|---|---|---|---|---|---|---|
| 0.704 | 0.698 | 0.711 | 0.724 | 8717 | 1 | |
| 0.296 | 0.276 | 0.295 | 0.315 | 8717 | 1 | |
| 0.983 | 0.976 | 0.983 | 0.990 | 9511 | 1 | |
| 0.995 | 0.988 | 0.995 | 0.998 | 10,123 | 1 | |
| 0.040 | 0.028 | 0.040 | 0.051 | 2970 | 1 | |
| 0.025 | 0.015 | 0.025 | 0.034 | 4253 | 1 | |
| 0.025 | 0.015 | 0.025 | 0.035 | 3983 | 1 | |
| −0.026 | −0.035 | −0.026 | −0.016 | 3649 | 1 | |
| … | … | … | … | … | … | … |
| 0.176 | 0.162 | 0.176 | 0.190 | 2755 | 1 | |
| 0.018 | 0.005 | 0.018 | 0.030 | 2404 | 1 | |
| 0.095 | 0.082 | 0.095 | 0.108 | 3246 | 1 | |
| −0.016 | −0.042 | −0.016 | 0.011 | 3896 | 1 |
| Parameter | Districts of Micro-Level Mix-1 | Districts of Micro-Level Mix-2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Bangkalan | Sampang | Pamekasan | Sumenep | Bangkalan | Sampang | Pamekasan | Sumenep | |
| 0.943 (0.042) | 0.944 (0.065) | 0.942 (0.048) | 0.943 (0.030) | 0.607 (0.040) | 0.610 (0.046) | 0.608 (0.093) | 0.608 (0.087) | |
| 0.358 (0.012) | 0.358 (0.019) | 0.359 (0.036) | 0.359 (0.012) | 0.129 (0.076) | 0.129 (0.033) | 0.128 (0.072) | 0.129 (0.040) | |
| 0.052 (0.031) | 0.051 (0.030) | 0.051 (0.037) | 0.051 (0.013) | 0.284 (0.024) | 0.286 (0.018) | 0.285 (0.035) | 0.285 (0.077) | |
| 0.144 (0.047) | 0.145 (0.018) | 0.146 (0.010) | 0.145 (0.017) | 0.364 (0.052) | 0.363 (0.019) | 0.367 (0.018) | 0.366 (0.083) | |
| 0.283 (0.059) | 0.284 (0.038) | 0.284 (0.011) | 0.283 (0.074) | 0.425 (0.018) | 0.429 (0.025) | 0.428 (0.024) | 0.427 (0.017) | |
| 0.243 (0.090) | 0.246 (0.020) | 0.243 (0.019) | 0.243 (0.037) | 0.390 (0.013) | 0.391 (0.085) | 0.390 (0.017) | 0.391 (0.013) | |
| 0.444 (0.029) | 0.447 (0.046) | 0.447 (0.048) | 0.446 (0.046) | 0.355 (0.027) | 0.357 (0.051) | 0.355 (0.018) | 0.355 (0.074) | |
| 0.161 (0.068) | 0.162 (0.033) | 0.162 (0.010) | 0.161 (0.014) | 0.331 (0.020) | 0.332 (0.014) | 0.331 (0.014) | 0.331 (0.014) | |
| 0.208 (0.035) | 0.209 (0.014) | 0.209 (0.093) | 0.208 (0.011) | 0.286 (0.063) | 0.287 (0.013) | 0.285 (0.016) | 0.286 (0.013) | |
| 0.241 (0.093) | 0.242 (0.035) | 0.242 (0.021) | 0.242 (0.013) | 0.037 (0.023) | 0.038 (0.006) | 0.037 (0.028) | 0.037 (0.010) | |
| 0.424 (0.010) | 0.427 (0.034) | 0.424 (0.087) | 0.424 (0.015) | 0.082 (0.001) | 0.083 (0.001) | 0.080 (0.002) | 0.082 (0.004) | |
| 0.104 (0.077) | 0.106 (0.014) | 0.103 (0.075) | 0.103 (0.094) | 0.116 (0.059) | 0.117 (0.067) | 0.115 (0.078) | 0.116 (0.028) | |
| 0.084 (0.007) | 0.086 (0.005) | 0.086 (0.008) | 0.085 (0.005) | 0.391 (0.012) | 0.393 (0.015) | 0.390 (0.026) | 0.390 (0.014) | |
| 0.092 (0.004) | 0.095 (0.004) | 0.090 (0.002) | 0.092 (0.002) | 0.490 (0.021) | 0.494 (0.031) | 0.493 (0.020) | 0.492 (0.015) | |
| 0.352 (0.010) | 0.354 (0.017) | 0.351 (0.030) | 0.352 (0.017) | 0.256 (0.013) | 0.256 (0.019) | 0.255 (0.007) | 0.255 (0.022) | |
| 0.181 (0.001) | 0.183 (0.008) | 0.181 (0.007) | 0.181 (0.009) | 0.320 (0.009) | 0.323 (0.007) | 0.322 (0.016) | 0.321 (0.091) | |
| 0.583 (0.017) | 0.587 (0.013) | 0.584 (0.028) | 0.584 (0.020) | 0.108 (0.071) | 0.108 (0.090) | 0.110 (0.023) | 0.109 (0.086) | |
| 0.231 (0.011) | 0.233 (0.010) | 0.232 (0.063) | 0.232 (0.089) | 0.415 (0.065) | 0.420 (0.016) | 0.407 (0.013) | 0.411 (0.011) | |
| 0.083 (0.001) | 0.084 (0.002) | 0.084 (0.004) | 0.083 (0.003) | 0.169 (0.043) | 0.166 (0.013) | 0.170 (0.039) | 0.169 (0.011) | |
| 0.185 (0.013) | 0.187 (0.015) | 0.187 (0.020) | 0.186 (0.016) | 0.121 (0.069) | 0.123 (0.013) | 0.122 (0.066) | 0.122 (0.068) | |
| 0.083 (0.004) | 0.068 (0.004) | 0.059 (0.004) | 0.071 (0.002) | 0.141 (0.046) | 0.140 (0.029) | 0.144 (0.067) | 0.143 (0.046) | |
| 0.412 (0.019) | 0.414 (0.020) | 0.414 (0.013) | 0.413 (0.014) | 0.214 (0.014) | 0.218 (0.024) | 0.211 (0.032) | 0.212 (0.071) | |
| 0.621 (0.058) | 0.163 (0.029) | 0.621 (0.053) | 0.162 (0.060) | 0.096 (0.010) | 0.096 (0.003) | 0.099 (0.013) | 0.097 (0.069) | |
| −0.398 (0.042) | −0.401 (0.013) | −0.398 (0.008) | −0.398 (0.012) | 0.285 (0.052) | 0.286 (0.011) | 0.285 (0.015) | 0.285 (0.016) | |
| 0.526 (0.009) | 0.252 (0.017) | 0.540 (0.016) | 0.254 (0.097) | 0.564 (0.072) | 0.568 (0.098) | 0.567 (0.085) | 0.566 (0.090) | |
| 0.262 * (0.011) | 0.267 * (0.046) | 0.265 * (0.011) | 0.266 * (0.077) | 0.326 * (0.010) | 0.328 * (0.047) | 0.328 * (0.023) | 0.327 * (0.018) | |
| 0.596 * (0.065) | 0.581 * (0.034) | 0.606 * (0.019) | 0.599 * (0.029) | 0.881 * (0.064) | 0.885 * (0.012) | 0.879 * (0.016) | 0.881 * (0.019) | |
| Macro-Level Parameters () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.936 (0.076) | 0.002 (0.008) | 0.003 (0.003) | 0.008 (0.001) | 0.001 (0.003) | 0.001 (0.002) | 0.002 (0.002) | 0.010 (0.002) | 0.005 (0.004) | |
| 0.356 (0.011) | 0.002 (0.006) | 0.001 (0.007) | 0.002 (0.005) | 0.007 (0.005) | 0.001 (0.007) | 0.002 (0.009) | 0.004 (0.002) | 0.007 (0.002) | |
| 0.053 (0.004) | 0.003 (0.008) | 0.001 (0.003) | 0.002 (0.001) | 0.003 (0.001) | 0.0001 (0.001) | 0.001 (0.002) | 0.008 (0.001) | 0.002 (0.001) | |
| 0.142 (0.020) | 0.001 (0.001) | 0.007 (0.010) | 0.001 (0.002) | 0.002 (0.001) | 0.001 (0.002) | 0.006 (0.001) | 0.001 (0.001) | 0.009 (0.012) | |
| 0.278 (0.030) | 0.002 (0.003) | 0.009 (0.002) | 0.003 (0.005) | 0.003 (0.004) | 0.002 (0.003) | 0.006 (0.009) | 0.003 (0.004) | 0.007 (0.002) | |
| 0.239 (0.027) | 0.001 (0.001) | 0.003 (0.003) | 0.002 (0.002) | 0.001 (0.002) | 0.009 (0.001) | 0.005 (0.007) | 0.004 (0.004) | 0.006 (0.007) | |
| 0.444 (0.032) | 0.004 (0.005) | 0.010 (0.018) | 0.004 (0.004) | 0.015 (0.011) | 0.002 (0.006) | 0.010 (0.013) | 0.010 (0.009) | 0.020 (0.023) | |
| 0.159 (0.014) | 0.001 (0.003) | 0.007 (0.001) | 0.001 (0.001) | 0.005 (0.003) | 0.002 (0.002) | 0.003 (0.003) | 0.002 (0.001) | 0.002 (0.005) | |
| 0.203 (0.062) | 0.004 (0.003) | 0.004 (0.008) | 0.002 (0.001) | 0.001 (0.001) | 0.002 (0.001) | 0.006 (0.003) | 0.002 (0.001) | 0.001 (0.002) | |
| 0.239 (0.036) | 0.003 (0.004) | 0.002 (0.002) | 0.002 (0.002) | 0.005 (0.006) | 0.002 (0.002) | 0.005 (0.005) | 0.004 (0.001) | 0.002 (0.002) | |
| 0.414 (0.087) | 0.018 (0.002) | 0.011 (0.002) | 0.003 (0.003) | 0.007 (0.001) | 0.007 (0.007) | 0.009 (0.003) | 0.003 (0.003) | 0.008 (0.010) | |
| 0.100 (0.015) | 0.007 (0.001) | 0.004 (0.001) | 0.001 (0.002) | 0.002 (0.003) | 0.008 (0.0004) | 0.003 (0.004) | 0.001 (0.001) | 0.004 (0.005) | |
| 0.092 (0.013) | 0.002 (0.001) | 0.003 (0.005) | 0.010 (0.001) | 0.005 (0.001) | 0.007 (0.001) | 0.004 (0.006) | 0.002 (0.001) | 0.008 (0.012) | |
| 0.080 (0.009) | 0.051 (0.007) | 0.005 (0.001) | 0.0002 (0.001) | 0.001 (0.002) | 0.002 (0.002) | 0.002 (0.002) | 0.008 (0.001) | 0.008 (0.001) | |
| 0.338 (0.026) | 0.006 (0.007) | 0.005 (0.009) | 0.012 (0.015) | 0.011 (0.017) | 0.005 (0.009) | 0.005 (0.004) | 0.004 (0.003) | 0.002 (0.011) | |
| 0.177 (0.021) | 0.002 (0.003) | 0.004 (0.004) | 0.002 (0.003) | 0.005 (0.006) | 0.007 (0.004) | 0.003 (0.005) | 0.002 (0.001) | 0.009 (0.013) | |
| 0.577 (0.015) | 0.004 (0.002) | 0.005 (0.004) | 0.004 (0.003) | 0.010 (0.006) | 0.007 (0.004) | 0.006 (0.006) | 0.011 (0.010) | 0.020 (0.016) | |
| 0.226 (0.013) | 0.002 (0.005) | 0.008 (0.012) | 0.004 (0.002) | 0.002 (0.003) | 0.002 (0.005) | 0.007 (0.005) | 0.004 (0.004) | 0.003 (0.007) | |
| 0.080 (0.006) | 0.001 (0.001) | 0.006 (0.001) | 0.001 (0.001) | 0.003 (0.003) | 0.002 (0.001) | 0.002 (0.002) | 0.009 (0.002) | 0.001 (0.002) | |
| 0.181 (0.023) | 0.002 (0.003) | 0.006 (0.005) | 0.002 (0.002) | 0.008 (0.009) | 0.002 (0.004) | 0.009 (0.011) | 0.003 (0.004) | 0.011 (0.013) | |
| 0.092 (0.008) | 0.006 (0.001) | 0.001 (0.002) | 0.002 (0.001) | 0.002 (0.002) | 0.005 (0.001) | 0.088 (0.002) | 0.009 (0.001) | 0.003 (0.004) | |
| 0.409 (0.029) | 0.004 (0.003) | 0.012 (0.011) | 0.001 (0.002) | 0.005 (0.010) | 0.002 (0.002) | 0.012 (0.010) | 0.003 (0.007) | 0.004 (0.004) | |
| 0.158 (0.072) | 0.003 (0.004) | 0.003 (0.002) | 0.003 (0.001) | 0.002 (0.002) | 0.002 (0.002) | 0.002 (0.002) | 0.002 (0.001) | 0.008 (0.006) | |
| −0.390 (0.013) | −0.005 (0.004) | −0.004 (0.001) | −0.007 (0.0003) | −0.0004 (0.001) | −0.004 (0.0003) | −0.006 (0.001) | −0.001 (0.000) | −0.0003 (0.001) | |
| 0.250 (0.035) | 0.003 (0.004) | 0.009 (0.003) | 0.002 (0.003) | 0.006 (0.008) | 0.008 (0.002) | 0.001 (0.002) | 0.008 (0.002) | 0.004 (0.007) | |
| 0.254 (0.019) | 0.008 (0.001) | 0.010 (0.007) | 0.004 (0.003) | 0.005 (0.005) | 0.003 (0.003) | 0.003 (0.003) | 0.040 (0.004) | 0.011 (0.009) | |
| 0.657 (0.024) | 0.056 (0.011) | 0.053 (0.016) | 0.044 (0.009) | 0.057 (0.006) | 0.045 (0.031) | 0.001 (0.049) | 0.036 (0.017) | 0.092 (0.032) | |
| Model | WAIC |
|---|---|
| Bayesian Bernoulli Mixture aggregate regression model (BBMARM) | 2392.3 |
| Hierarchical Bernoulli mixture model (Hibermimo) | 1218.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suryaningtyas, W.; Iriawan, N.; Kuswanto, H.; Zain, I. On the Hierarchical Bernoulli Mixture Model Using Bayesian Hamiltonian Monte Carlo. Symmetry 2021, 13, 2404. https://doi.org/10.3390/sym13122404
Suryaningtyas W, Iriawan N, Kuswanto H, Zain I. On the Hierarchical Bernoulli Mixture Model Using Bayesian Hamiltonian Monte Carlo. Symmetry. 2021; 13(12):2404. https://doi.org/10.3390/sym13122404
Chicago/Turabian StyleSuryaningtyas, Wahyuni, Nur Iriawan, Heri Kuswanto, and Ismaini Zain. 2021. "On the Hierarchical Bernoulli Mixture Model Using Bayesian Hamiltonian Monte Carlo" Symmetry 13, no. 12: 2404. https://doi.org/10.3390/sym13122404
APA StyleSuryaningtyas, W., Iriawan, N., Kuswanto, H., & Zain, I. (2021). On the Hierarchical Bernoulli Mixture Model Using Bayesian Hamiltonian Monte Carlo. Symmetry, 13(12), 2404. https://doi.org/10.3390/sym13122404