1. Introduction
In this paper, an automatic DR diagnosis system is proposed. Pointing to the ‘fact sheet’ of the world health organisation (WHO), over 2.2 billion people all over the world have a vision impairment or blindness. One billion of these cases can be prevented or at least controlled if these cases are detected at an early stage. Diabetic retinopathy (DR) is the fifth leading cause of vision impairment and is estimated at 4.5 million out of the one billion cases [
1,
2,
3,
4]. Currently, the need for eye disease screening and check ups has increased rapidly. Despite that, there is a notable shortfall of ophthalmologists, particularly in developing countries. Hence, the automation detection of the DR process by supervised procedures is a benefit and can significantly assist in working out this issue [
5,
6].
In the last couple of decades, the clinical importance of the segmentation of retinal blood vessels from fundus images has attracted researchers to diagnose many diseases. Consequently, the segmentation of retinal blood vessels is an attractive field of research as it helps in retinal image analysis and computer-assisted diagnosis [
7,
8].
Retinal vessel investigation is noninvasive, as it can be observed directly without any surgical intervention [
9]. Additionally, it is an indispensable element to any automatic diagnosis system for ophthalmic disorders. The variation in the structure of retinal blood vessels can serve as an indicator of serious diseases such as stroke, glaucoma, hypertension, arteriosclerosis, cardiovascular, age-related macular degeneration (AMD), refractive error and DR [
7,
10]. It is commonly accepted by the retinal medical community that uses
colour fundus images and 3D Optic Coherent Tomography (OCT) images modality as a clinical indicator for DR diagnosis because of its low cost and the simplicity in computation [
11,
12]. Fundus image modality is more often used than OCT images in both screening and clinical diagnosis systems. The lesions structure of the retina such as micro-aneurysms (MA), neovascularisation, haemorrhages, hard exudates, and cotton-wool-spots can be more prominent in fundus retinal images. In retinal fundus image diagnosis, there are two objects to be segmented: the anatomical structure (optic disc, blood vessels, macula, and fovea) and the lesions caused by diseases. The identification of the anatomical structure (main region) of the retinal image can assist in the analysis of conditions that affect these regions. For example, glaucoma affects the optic disc by making it more dilated, hypertension affects the vessels by tortuosity, and proliferative DR affects the number of new vessels [
13]. The latter is the focal point of the present work. The anatomical attributes of retinal blood vessels, such as width, length, tortuosity, local branching pattern, and angles, play an important role in diagnostic results. They are widely used not only to predict ophthalmological disorders (i.e., DR, glaucoma, and age-related macular degeneration), but also to non-ophthalmological disorders (i.e., hypertension, arteriosclerosis, obesity, and cardiovascular disease) [
14,
15]. Therefore, the analysis of the vasculature structure in retinal fundus images allows both monitoring and evaluating the progress of these disorders.
Manual segmentation of the vascular tree in retinal fundus images is a tedious task for many reasons [
16]. First, the variation in the contrast between blood vessel pixels and background in fundus images can be very tiny. Second, the noise presence and the difference in illumination conditions can cause problems. Third, the blood vessel variations, such as width, shape, branching angles, and brightness, may not be readily observable. Finally, the presence of lesions and haemorrhage in the background of retinal images can introduce confusion. Hence, automatic segmentation is a necessity.
A substantial amount of work is reported in the literature for detecting blood vessels in colour retinal fundus images [
9,
10,
17]. Since retinal image analysis is complicated, it usually fails when a stand-alone image processing technique is used [
18]. According to [
19], the most classical segmentation techniques on the fundus images encompass six domains: (i) pattern recognition, (ii) mathematical morphology, (iii) matched filter, (iv) vessel tracking, (v) model-based, (vi) parallel-hardware. Pattern recognition is considered the most accurate and robust [
18]. Parallel-hardware methods can accelerate the speed of execution, but the accuracy may be relatively lowered [
20]. Hybrid methods, which combine one or more different domains to improve system performance, are in common use. Mathematical morphology, matched filter, vessel tracking and model-based techniques can contribute to each other or be applied to pattern recognition techniques and/or parallel-hardware methods to build an efficient, automated, comprehensive blood vessel segmentation system.
Machine learning, supervised and unsupervised, can be used in retinal blood vessel segmentation. In supervised learning, where for each input data, there is a corresponding output label, one can use a feature vector and a gold standard (GS) dataset. The latter includes manual markings for blood vessels, to train a classifier to divide retinal colour image pixels into vessel pixels and non-vessel pixels. On the other hand, where labels cannot exist, unsupervised methods explore interesting patterns of input data. In unsupervised methods, no prior knowledge is required upon the pattern before segmenting; moreover, the run is faster than the supervised methods. Although the supervised approaches are more accurate than the unsupervised method, it requires a craft, features engineering and lots of extracted features to obtain good results in the segmentation of the retinal blood vessel [
21,
22,
23,
24].
In the last five years, a new dawn of an era has shone based on deep learning as an automatic features extraction such as Deep convolutional neural networks(CNN). CNN shows the highest robustness and effectiveness in segmenting the blood vessels. Unlike any conventional supervised approach, the deep learning neural network (DNN) methods are intelligent in automatically learning the features by using a predefined filter, which is used to extract features from the input image. Additionally to this, Transfer learning, an advanced approach of Deep learning, is recently used. Rather than building and training the network to learn features from scratch, Transfer learning concepts are uses as a pre-trained model on vast numbers of different images. So the model is already pre-learned by the tremendous number of different features and only need to fine-tune for the more suitable and relevant application. Since the proposed work belongs to the former category, previous work using supervised learning is reviewed next.
A supervised learning method as an ensemble system based on feature-based AdaBoost classifier (FABC) using a collection of a 41-D feature vector is constructed for segmentation, but a high relative number of features is necessary to get the accurate results [
25]. A 7-D feature vector comprises a grey level and moment invariant based feature segments. An artificial neural network (ANN) is used as a classifier. The method approaches are unable to achieve the desired results without sophisticated preprocessing steps [
26]. An ensemble system of bagged and boosting analysis composed of the 9-D feature vector is constructed from the gradient vector field, morphological transformation, line strength measures, and Gabor filter to handle healthy and pathological colour retinal images. The methods use a number of features, and a decision tree applies where the effects of weak learners are combined using bootstrap aggregation to increase the accuracy of the segmentation [
27]. The retinal blood vessel morphology is exploited to identify the successive stage of the severity of the DR disease. A technique based on an ANN is used to automate the screening of DR. An ANN that contains one input layer, five hidden layers, and one output layer is used. The input to the ANN is derived from the Gabor filter and moment invariant based features. The input image is processed using a mathematical morphology operator, mean, and Gaussian filter. This technique is used to diagnose diseases like hypertension and diabetes mellitus, for which a change in the retinal vasculature is a primary symptom. The method is applied only on the one dataset while it fails on another dataset [
17]. The retinal vessel is uprooted using Lattice Neural Networks, and dendritic processing introduced relatively better performance in vessels. Nevertheless, the final results are low in DRIVE, STARE datasets [
28]. Extensive preprocessing steps, such as zero-phase whitening, global contrast normalisation and gamma correlation on fundus image patches, are used as inputs to CNN. Although the prediction on the DRIVE, STARE, and CHASE_DB1, showed reduced vessels misclassification and central vessels reflex problem, the results are mainly based on preprocessing steps [
29]. Five stages of DNN with the autoencoder transform the selected RGB patches of the retinal image into a vessel map image. Even though the prediction is made at every pixel in a patch, final results are based on the boundless cross-modality training on datasets [
24]. A supervised hierarchical retinal blood vessel segmentation method is presented to diagnose several ailments, such as cardiovascular diseases, DR, and hypertension. The proposed method uses two superior layers: A convolution neural network, as a deep hierarchical feature extractor, and a random forest, which works as a trainable classifier. This method is tested only on two datasets. Although it outperforms the aforementioned methods, the time cost of this method is relatively high [
30]. A supervised method based on simple extreme machine learning uses a 39-D feature vector constructed from local features (29 features), morphological transformation operators (6 features), phase congruency (1 feature), Hessian components (2 features) and divergence of a vector field (1 feature). Regardless of the fact that the method tries only one public dataset and another unknown private dataset, the obtained results are relatively low concerning the other publicly available datasets [
12]. Information obtained from a stationary wavelet transform with a multi-scale convolution neural network can catch the tiny vessel pixel. This method uses a rotation operator as a basis of a joint strategy for data augmentation and prediction [
31]. A hybrid feature set and hierarchical classification used to segment retinal blood vessels from a retinal colour image. Six groups of discerning features were used to construct the feature vector compose of local intensity variation features, the local binary pattern features, a histogram of gradient features, vector field divergence features, morphological transformation, and high-order-local auto-correlation features [
32]. Moreover, a random forest-based technique was used as a feature selection in addition to using two hierarchical trainers, comprised of a support vector machine (SVM) and random forest, used as weak classifiers. At the same time, the final results are from a Bayesian network. The final output results completely depend on the random forest technique, as a feature selection in addition to hierarchical classification. However, it was so low when it tested for each classifier separately. A hybrid segmentation method with a self-mask generated schema was used to extract the retinal blood vessel from the fundus retinal image. However, its performance was very low on the DRIVE dataset [
4]. A transfer learning model, VGG16, as a pre-trained model with multilevel/multi-scale deep supervision layers was incorporated to segment the retinal blood vessel. The VGG16 model was improved by convolving a vessel-specific Gaussian kernel with two pre-initialised scales. The final output is an activation map with well-learned vessel-specific feature at multiple scales. The activation map is increasingly obtained by the symmetric feature map. Regardless of using a pre-trained model, VGG16, which trained on ImageNet (15 million of high-resolution labelled images), the method required better post-processing steps [
6]. Despite the increasing use of deep learning, there are some drawbacks that face it. First, the need for copious numbers of images to train and test, which does not exist in our case, and second, a high amount of power in processing using a graphical processing unit (GPU). Furthermore, it has a complicated, long structure, which needs a lot of experience, training and effort.
For fulfilling the retinal blood vessel segmentation, this paper uses hand-crafted features and a multi-layer perceptron neural network (MLP), with the following contribution.
Carefully, 24 hand-crafted features are outlined.
Special multi-layer perceptron neural network structure for full gauge fundus retinal evaluation.
Cross entropy loss function.
Transformed green channel image.
Morphological transformation are applied twice. First, white top-hat and black bottom-hat are used as filters to extract features and to suppress noise during the extraction. Second, a closing operator is used to connect the isolated vessels and to fill the gap areas in the segmented images during the post-processing step.
Model for automatic feature extraction and classification of blood vessel in retinal fundus images.
Maximum and minimum moments of phase congruency are used for the first time.
Retinal vessel segmentation with improved generalisation on multiple datasets.
In-depth visual analysis plus comparison with the state-of-the-art methods.
This paper is organised as follows.
Section 2 outlines the datasets and the methodology, where feature extracting steps are discussed in detail. In
Section 3, the results of the proposed method are presented. The discussion of the results is in
Section 4. The conclusions of the paper are described in
Section 5.
2. Materials and Methods
Three datasets are commonly in use for vessel segmentation research. These are the Digital Retinal Images for Vessel Extraction (DRIVE) dataset [
33], the Structure Analysis of Retina (STARE) dataset [
34] and CHASE_DB1 dataset [
35]. Because we do some early processing on STARE and CHASE_DB datasets in creating mask images related to each one, the difference between the three datasets must be explained as follows.
- 1.
DRIVE dataset
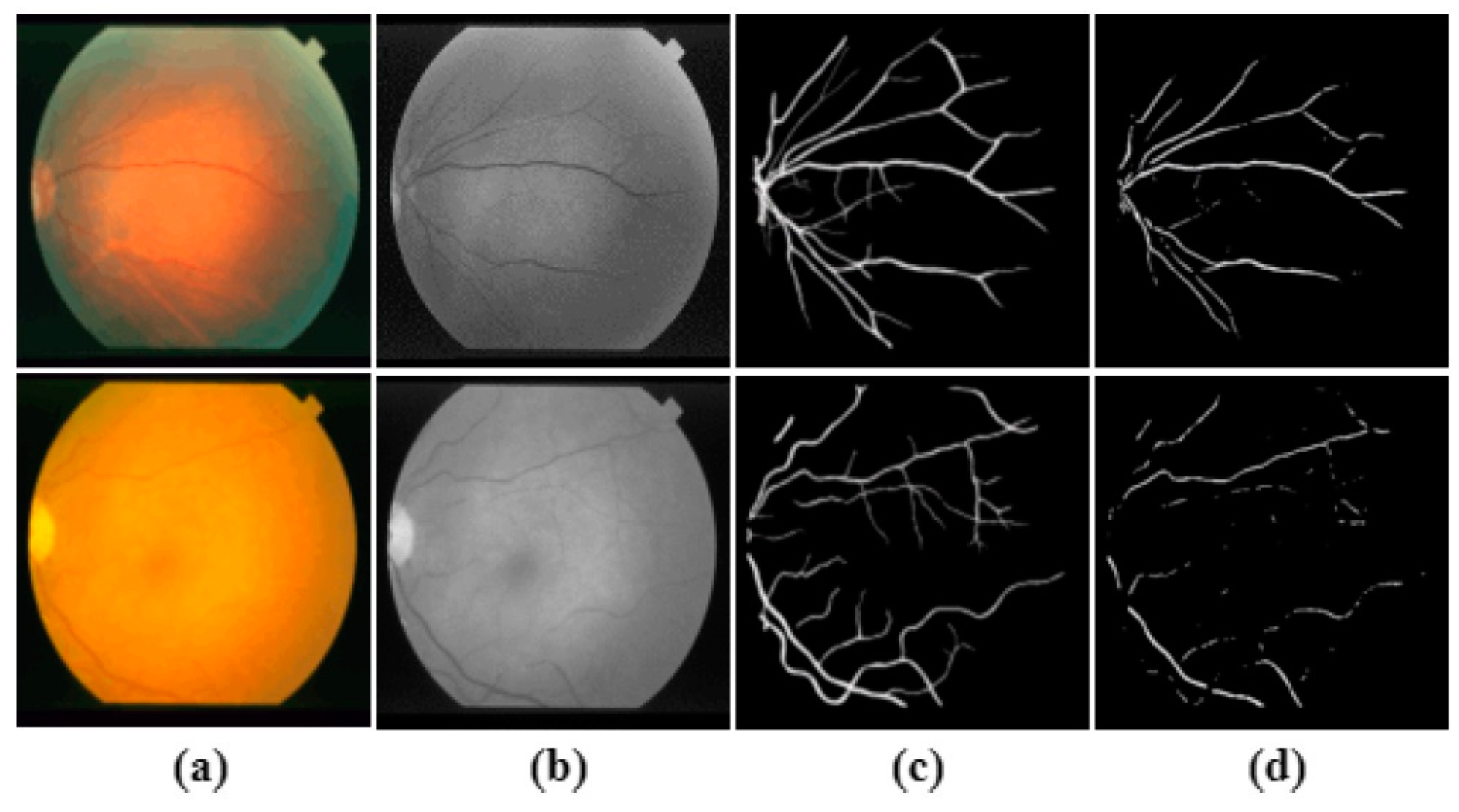
The DRIVE dataset consists of 40 images (7 of which are pathology) divided into 20 images for training and 20 for testing (3 of which are pathology). The image size is 565 × 584 pixels captured by a Canon CR5 non-mydriatic 3 charge-coupled-device (CCD) cameras at a 45° field of view (FOV). The image has a FOV approximately 540 pixels in diameter. The DRIVE dataset provides a mask for every image to facilitate the identification of the field of view (FOV) area, as shown in
Figure 1.
Every image is accompanied by a manual segmentation label, manually created by three experts (observers) and validated by an experienced ophthalmologist (annotator). The training set is segmented only once and twice for the testing set, resulting in 577,649 pixels (
) marked as a vessel in the first set (first observer), against 556,532 pixels (
) in the second set (second observer). In this paper, the first set (
first observer) is used as the ground truth (label) for the performance evaluation, while the second set is serving as a human observer reference for the performance comparison. The DRIVE dataset is depicted in
Figure 2 and
Figure 3.
- 2.
STARE dataset
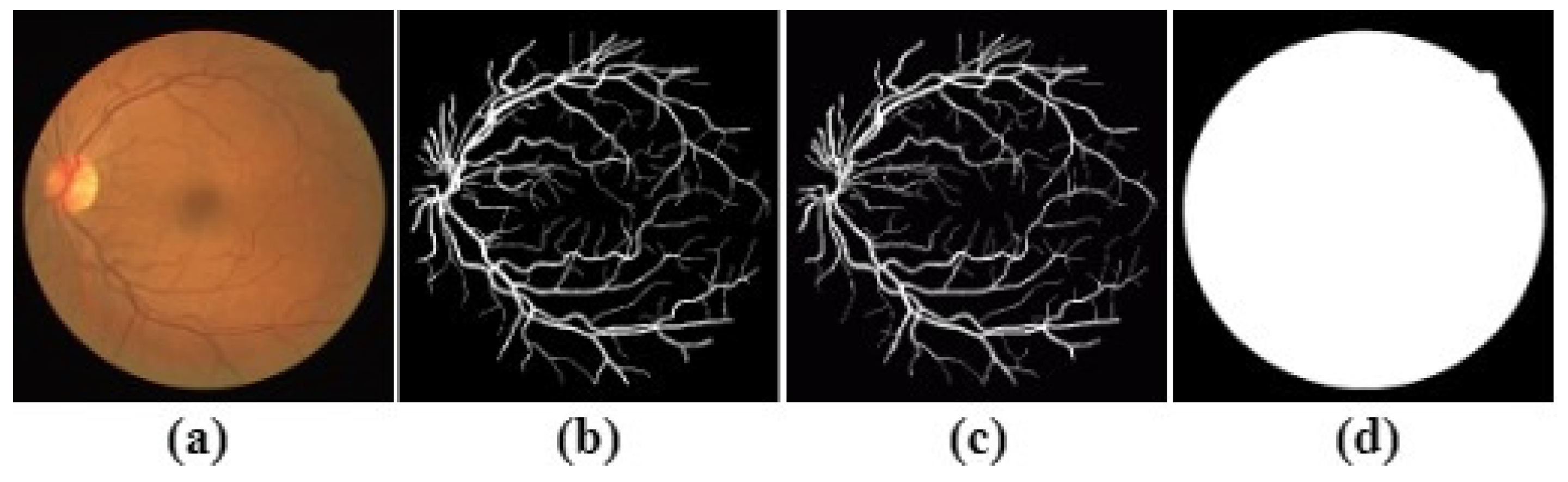
The STARE (Structure Analysis of Retina) dataset comprises 20 images (10 of which present pathology) of size
pixels. The dataset is captured by a TopCon TRV-50 fundus camera at 35° FOV. The dataset is manually segmented twice by two observers; the first one segments
of pixels as vessels and
for the other. Since the blood vessels of the second observer are much thinner than the first, the
first is considered the ground truth.
Figure 4 illustrates a sample of the STARE dataset. Unlike DRIVE, STARE does not offer masks to determine the FOV, so the method described in [
11,
34] is leveraged to create them.
- 3.
The CHASE_DB1 Dataset
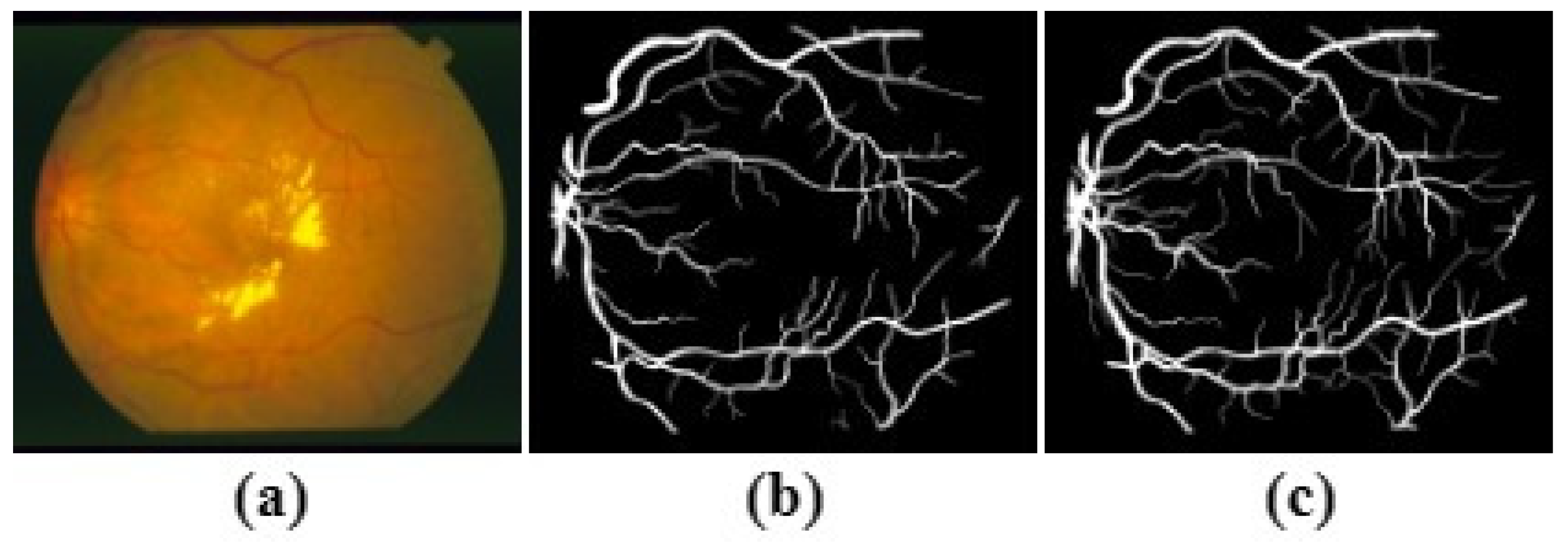
The CHASE_DB1 Dataset includes 28 images of size
pixels, captured from 14 school children for both left and right eyes by a hand-held NidekNM-200-D fundus camera at 30° FOV. The segmentation results of the
first observer are deployed as the ground truth. This dataset is characterised by nonuniform background illumination. Thus, the contrast of blood vessels is inferior, with the presence of central vessel reflexes. The CHASE_DB1 is pictured in
Figure 5. Like STARE, CHASE_DB1 does not offer mask images, so as in the above, the method described in [
11,
34] is also leveraged to create them.
The proposed model to segment retinal blood vessels from a colour retinal image contains four stages: (i) green channel extraction, (ii) features extraction, (iii) evaluation of segmentation, (iv) post-processing. In the first stage, the green channel is extracted from the RGB retinal fundus image. The choice of the green channel, in particular, is due to the fact that in this channel, the blood vessel tree is more vivid than in other channels. In the second stage, a 24 feature vector is constructed for pixel representation. The features of this vector encode information on the local intensity value (1 feature), morphological white top-hat value at six different scales (6 features), morphology black-bottom-hat values at six different scales (6 features), maximum-moment of phase congruency (1 feature), minimum-moment of phase congruency (1 feature), Hessian components values (4 features) and difference of Gaussian features at five different scales (5 features). At the end of this stage, a features matrix based on the feature vector and the correspondence manual label is constructed for each pixel in the green channel image. Later, a training sample from this feature matrix is randomly selected and used as input to the classifier to perform the segmentation. In the third stage, a supervised learning approach based on a multi-layer perceptron neural network is adopted. The rule for blood vessel extraction is to learn by the algorithm through the provided training sample. The segmented reference images are usually termed label or gold standard images. The last stage, post-processing, is used to enhance the classifier performance and acts to reduce two types of artefacts. The proposed method is tested on the three publicly available datasets, DRIVE, STARE, and CHASE_DB1, which contain digital colour retinal images with 8 bits per colour channel. A complete block diagram for the proposed model is shown in
Figure 6.
The remainder of this section is dedicated to explaining the method in detail.
2.1. Feature Extraction
As mentioned above, in the second stage of our proposed method, a 24 feature vector is constructed for pixel representation. The features composing the vector are as follows.
2.1.1. Local Intensity Feature
In a retinal fundus image, blood vessel pixels appear much darker than the background pixels. So, the intensity value for each pixel in the retinal green channel is considered as a local intensity feature.
Figure 7 depicts an image after a local intensity feature is extracted from the colour image.
2.1.2. Morphological White Top-Hat Transformation
In general, mathematical morphology transformation in image processing can combine (merge) two sets of inputs, the first related to the image and the second a predefined linear structuring element. One type of such transformation, white top-hat, is an operator used for lighting objects on a dark background. In the beginning, the opening operator removes objects smaller than the structuring element. Then the difference between the original image and the output of the opening operator is eliminated. Specifically, the opening operator uses the predefined structuring element, which is oriented at a particular angle
, to eradicate a vessel or part of it when the vessel’s width is shorter than the element and, both are orthogonal [
15].
Let
I be the green image to be processed, and
S be the linear structuring element. Then the white top-hat transformation feature of
I is given by
where
is the opening operator,
the length of the structuring element and
the rotation angle of
S, spanning
in steps of
. The result of the white top-hat transformation in all direction is given by
where
, and set
A is a set that contains the orientation angles.
The length of
S is critical to extract the vessel with the largest diameter. Therefore, a set of different lengths is used to formulate six different features. In case the opening operator
along a class of
S is considered, the summation of white top-hat transformation at each direction will brighten the vessels regardless of its direction. The effectiveness of the summation on the entire retinal image will enhance all vessels regardless of their orientation. That includes the small (tortuous) vessels in the bright zone.
Figure 8 shows the image after applying the white top-hat operator.
Following the white top-hat feature extractor computation proposed in [
15], six different features are extracted. Not only do vessel-like structures become much lighter but they also acquire intensity values relative to the local background. So, the intensity values of extracted blood vessels are relative to the local intensity of neighbourhood pixels in the original image [
36]. The final response of the white top-hat transformation operator is a corrected uniform image with enhanced edge information.
2.1.3. Morphological Black Bottom-Hat Transformation
The counterpart of morphological white top-hat is the morphological black bottom-hat transformation. While the former is used for light objects on a dark background, the latter is used for the opposite. Black bottom-hat transformation is defined as the residue of the closing operator and the original image. In other words, it is a closing • of the image minus the image [
12]. The black bottom-hat transformation feature can be defined as follows
Let
I be the green image to be processed,
S is the structuring element, • is the closing operator and
is an orientation angle at a given scale
c. Then the black bottom-hat transformation feature on image
I is given by
Now,
where
is the summation of the black bottom-hat transformation in all directions of
S at a different
. Both
c and
are defined as in the white top-hat transformation.
The image is simplified by both of the above morphological filters. So the cross-curvature computation is easy, and the vessel segments are linearly coherent.
Figure 9 shows the image after applying black top-hat operators.
2.1.4. The Principal Moments of Phase Congruency
Instead of extracting features by using intensity gradient-based methods in the spatial domain, the phase congruency method is based on Fourier components. Its importance stems from its ability to detect edges and corner points in an image effectively and invariantly to change in contrast and illumination conditions. Furthermore, phase congruency has a significant advantage over all other feature detectors as it can mark features that discriminate like edges (blood vessels) correctly. It is based on the local energy model, reported by Morrone et al. [
37], which suggests that features as line and edges are perceived at points where the Fourier components are maximal in phase. The model is defined as a ratio of local energy to the overall path length used up by Fourier components from the head to tail.
Let
x be some location at a signal, the
nth Fourier component of which at point
x has amplitude
, and let
be the magnitude at that point of the vector from the head to tail (considered as the local energy). Then the phase congruency, (
, of the signal is given by
In case all the Fourier components are in phase, then all the complex vectors will be aligned, and the
will be 1; otherwise, it will be 0. While the model is not based on gradient intensity for detecting features, the construction of a dimensionless measure
at any point in the image is possible. The most notable drawback of this model is the bad localisation and its sensitivity to noise. A modified model is proposed by Kovesi [
38,
39], to compensate for the image noise. It provides proper localisation since
is represented as the phase difference between the amplitudes of the weighted mean of the local phase energy angle
and the local phase of
nth Fourier component
, both evaluated at point
x. As such, the modified model can be expressed as follows.
where (
) is the (green channel) pixel coordinate,
I is the total number of orientation.
K is the total number of scales,
is a weighted factor for frequency spread in orientation
i (because
from many frequencies is significantly better than
over a few frequencies),
T is the estimated noise influence (recommended value is 3), and
is a constant incorporated to avoid division by zero if the local energy vanishes. In Equation (
6), a
cosine of the phase difference between the amplitude of
and
at a particular point is used instead of a
sine because when the frequency components are aligned (in phase) with the “local energy”, the phase difference value can be expressed as
, with the
cosine being 1, resulting in a maximum
. The weighting function
w used to compute frequencies in ranges is defined as a sigmoid function
where
c is the filter response cut-off value, below which
values are ignored,
is the value of the gain factor that controls the sharpness of the cut-off (its value set to 10), and the fractional value of
of the frequencies is calculated by taking the amplitude of the sum of the filter response and dividing by the maximum amplitude
at each point (pixel) (x,y) of the images as follows.
Herein, Equation (
6) represents
for all orientation from
overall scales
K in a 2D image. Unlike the proposed method in [
12], the modified model in [
39,
40] is used by calculating
independently for each orientation using Equation (
6), then the moment of
covariance is computed. Hence, the variation of the moment within an orientation is taken into account. In such a case, the principal axis is corresponding to the axis about which the magnitude of the moment is minimum (minimum moment), which provides a good indicator for the orientation of the feature. On the other side, the magnitude of the maximum moment corresponding to the moment about the axis perpendicular to the principal axis is considered as an indicator of the significance of the feature. Accordingly, the maximum moment of
is used directly to establish whether the point in the retinal image has a significant edge feature (blood vessel), where the magnitude has a large value. Meanwhile, if the minimum moment of
is also large then it is considered as a strong indicator that the feature point has a strong 2D component and can be considered a corner.
The calculation of the maximum and minimum moment of
is represented in [
39,
40] based on a classical moment analysis equation. Firstly, the
information is combined in an orientation matrix (
) given by
evaluated over all multiple orientations formulated from the
covariance matrix (
) given by
where
is the amplitude of
, which is determined at orientation
o, and the sum is performed overall six orientations.
Figure 10 vividly depicts the phase congruency filter response at the six different directions. According to the above illustration of the PC model, in case all Fourier components are in phase, all the complex vectors would be aligned, indicating that the feature point has a robust 2D component and can be considered an edge like feature.
A singular value decomposition on a
covariance matrix is performed, and the maximum moment, (
M), and minimum moment, (
m), are obtained as follows.
where
Figure 11 visualises the extracted features image of the two different moments (M) and (m) features.
2.1.5. Multi-Scale Second-Order of Local Image Structure (Hessian matrix)
The Hessian matrix
for a point
in the 2D retinal image is used for vessel detection and is given by
where the second partial derivatives components are presented as
,
,
and
for the neighbours of the point
. The Hessian matrix is exploited to describe the second-order structure of intensity variation value around a pixel on the retinal image. The information obtained from the
describes the local curvature of the data in a small neighbourhood surrounding each pixel. The often employed approach to the retinal image is inspired by the theory of linear scale-space [
36]. Accordingly, a multi-scale algorithm is designed to identify vascular structure by investigating the signs and magnitudes of the eigenvalues of
, which reflect specific shapes of structures in the retinal image, besides their brightness.
Let
be the intensity of the a
green retinal image at a particular point
in an image, and
is a
Gaussian kernel. Then,
Hessian matrix
components of the second order derivatives of
multiplied by
at a scale
are defined as:
where the
Gaussian kernel is given by
with
the standard deviation of the Gaussian distribution, referring to a scale of the filter, and ∗ the convolution operator. Different values of
are experimentally tested, and the optimal values of
are detected as
, 1, 2, and
. The response of second-order derivative of a Gaussian function at a scale
represents a probe kernel that can be a measure of the contrast between regions inside and outside the given range (
) in the derivative direction in the retinal image [
41,
42]. In this respect, the blood vessel pixels gain the highest response at a suitable scale of
, while the background pixels get a low response. It is noteworthy that the nature of the second-order derivative of the Hessian matrix is invariant to linear grey variation of the retinal image pixels, because of the smooth nature of the Gaussian kernel. So only local contrast is assessed along with the actual greyscale values.
As soon as the elements of the Hessian matrix are obtained, the corresponding eigenvalues and eigenvectors are determined. The eigenvalues,
, have an essential role in the discrimination of the local orientation pattern. Eigenvalues not only represent the magnitude of maximal local contrast change but also the change of the magnitude of the local intensity in the other perpendicular direction. Consequently, the eigenvalue decomposition of Hessian can be exploited to distinguish between the edge-like feature in a fundus retinal image and the background. Concerning blood vessel skeleton, darker features are considered. We are interested in only the case where one eigenvalue has a positive and high magnitude.
Table 1 illustrates all possible variants of the structure’s shape and brightness that can be identified through the analysis of Hessian eigenvalues in any 2D medical image. We are only interested in the fundus retinal image modality (in our case, the blob-like structures are
dark).
Geometric information for the blob-like structure is obtained through a dissimilarity measure based on the second-ordered derivative that is called blobness ratio,
, given by
This ratio is maximal for a blob-like structure and invariant to the grey level. Since the intensity value of the blood vessel pixels is lower (darker) than the background pixels, the magnitude of the second-derivative (eigenvalues) will be high at the blob-like structure and small at the background. So the ratio will be higher at the blood vessel pixels and lower at the background pixels.
The magnitude of the second-order derivatives at the background is small, distinguishing background structure from the blood vessels. The norm of Hessian is used to quantify this property in terms of the structureness ratio,
S, and is given by
where
is the Forbenius norm of the components of Hessian matrix. Intuitively, the structureness ratio
S is small for the background pixels mostly because of their lack of structure and higher amount of blood vessel pixels.
Finally, a vessel function
combines the components of the blobness ratio
, and structureness ratio,
, to map these features into a probability-like estimates
of being a vessel given by
where
is equal to
and
c is half of the maximum Frobenius norm
S. Both
V and
S are computed at a different scale
. Herein,
accounts for the eccentricity of the second-order ellipse. Then the maximum value for both
V,
S are taken as a Hessian feature for each pixel and they are given by:
Both , , and are used as a features for every pixel of a green image.
The visualisation of the four features of the Hessian matrix components are delineated in
Figure 12.
2.1.6. Difference of Gaussian
Difference of Gaussian (DoG) kernels are exploited as an edge detector [
16]. A DoG kernel is much closer to the second-order of Gaussian function, which is described in detail in
Section 2.1.5. To characterise a pixels features regarding its neighbours, a DoG filter is utilised at five different smoothing scales,
, with the base scale set to
. Using DoG as a band path filter not only serves to remove noise in an image but also vividly depicts the homogeneous area in an image. The visualisation of the five DoG features are depicted in
Figure 13.
In this paper, a 24-D feature vector is constructed for each pixel in a retinal image and is used to differentiate vessel pixels, lesions, and background. Five groups of features are used. The RGB image is transformed into a green channel image because the contrast between blood vessel pixels and background is more precise and prominent than the other channels. So, the intensity value of each pixel in the green channel image is used as a feature. The moments of phase congruency emphasise the edges of the retinal blood vessel and it is the first time to be used as a feature in the fundus retinal image. The Hessian matrix components emphasise the zero crossing. The white top-hat transformation using multi-scales and multi-orientation structuring elements are efficient in making a corrected uniform image. Black bottom-hat transformation using multi-scales and a multi-orientation structuring element is used as a filter to suppress noise. The difference of Gaussian is used, and the response of the filter is a homogeneous image.
2.2. Segmentation
The procedure can be distinguished in three steps: first, scaling the features by normalisation; second, design of the neural network, in which the configuration is decided and trained (applied what the NN learned); third, testing, to identify each pixel as a vessel or a non-vessel to get the final binary image.
2.2.1. Normalisation
In the end, in the feature extraction process, each pixel in the green channel image is characterised by a vector
(in a 24-D feature space) given by
where the features
have distinct ranges and values. Normalisation, by narrowing down the range between the different features is necessary and leads to an easier classification procedure of assigning the candidate pixel to the suitable class: class
of blood vessel pixel or class
of non-vessel pixel. One can obtain a normalised version
of features
by
where
is the mean value of the
jth feature and
their standard deviation. Finally, the features are normalised with a zero mean and unit variance for each pixel independently. The resulting features are used in the classification procedure.
According to [
26], using the results of linear classifiers to separate the two classes of vessel segmentation gives poor results because of the nature of the distribution of the training dataset at the feature space, which is nonlinear. Consequently, segmentation can be made more accurately and efficiently using a nonlinear classifier, such as support vector machine (SVM) [
32], neural network [
24], a multi-layer feed forward neural network [
17], decision tree [
43], random forest [
32], extreme learning machine [
12] and convolution neural network(CNN) [
44].
2.2.2. Design of the Neural Network
In the present work, we use a modified MLP neural network of one input layer (with 24 input units), three hidden layers, and one output layer as a binary classifier. In the purpose of getting the optimal topology of hidden layers, several topologies and different numbers of neurons were tested. The test results showed that excellent results are obtained when each of the three hidden layers contains 27 units. A rectified linear unit (Relu) activation function is selected for the hidden layers [
45]. Further, in classification, cross-entropy (CE) was found better to use as a loss function than a Minimum Mean Square Error (MSE) loss function, because of the unbalanced distribution of vessel and non-vessel pixels [
45], where the CE measures the nearness of two probability distributions.
2.2.3. Training
To train the classifier, a set
of
M candidates features for which the feature vector [
(
22)], and the classification result,
(vessel pixel) or
(non-vessel) are known. The sample that forms
is collected randomly from the manually labelled pixels, vessel and non-vessel, from the training images as proposed in [
26,
43] for each dataset. Once the classifier passes the training steps, it becomes ready to work and classify images of which the results are not known a priori.
2.2.4. Application of Neural Network (Testing)
The trained neural network is used to test “unseen” dataset images. Specifically, a binary image is generated in which blood vessel pixels are distinguished from background pixels. The mathematical descriptions of pixels are individually input to MLP. In particular, the input layer units receive a vector
V of the normalised features, as in Equations (
22) and (
23). The Relu function has a tremendous out-of-sample prediction ability due to its simplicity. It also alleviates the problem of a vanishing gradient in the learning phase of the ANN [
46]. Finally, we obtained a binary image that contains only extracted blood vessel and background pixels.
In this paper, we use a multi-layer neural network (MLP) as a binary classifier, since there are only two classes (blood vessel) and (non-vessel). MLP is an extension of an artificial neural network (ANN), considered to be more nonlinear when mapping a set of the input variable to a set of output variables [
47]. Also, it does not make any assumption regarding the probabilistic information about the pattern under consideration.
2.3. Post-Processing
In the post-processing stage, two steps are used to reduce two types of classification artefacts. First, the discontinuous vessels pixels, which are surrounded by vessel neighbours and are misclassified as background pixels (
). Second, the non-vessel pixels, which appear as small isolated regions and are misclassified as vessels (
). For the sake of overcoming both artefacts, a mathematical operator is used. A mathematical closing operator is applied for the first artefact to fill any holes smaller than a predefined structure (a disk of radius 9 in our experiments) in the vessel map. For the other artefact, the acreage of isolated pixels are connected in eight connected areas, then any area below 100 pixels is removed, as proposed by [
9]. Once both artefacts are corrected, they will be reclassified correctly as vessel pixels for the first artefact and background pixels for the second.
The experimental results in
Section 3 show that the combination of the five groups of features, 24D features, which are encoded to the modified MLP, yields great performance. Our experiments are tested in the environment of 2.4 GHz Intel7-77ooQH CPU and 16 GB memory using Scikit-learn: open source machine learning libraries [
48].
5. Conclusions
In this paper, we have proposed an integrated method to segment blood vessels in retinal images. The experimental results prove that the method succeeds both absolutely and in comparison with seven other state-of-the-art similar methods using three well known publicly available datasets. The proposed method encompasses many elements that wholly contribute to its success. The first element is a feature choice, where 24 carefully selected features are employed. The second element is the choice of the classifier, namely an MLP neural network, and the design of this classifier. The third element is the way the classifier is trained, where randomly chosen images from all three datasets participate in the training. The fourth element is post-processing, where classification artefacts can be mitigated. Classification results are provided both visually and quantitatively, wherein the latter case, five standard classification metrics are employed. Both the visual and quantitative results show the good performance of the proposed method vividly, despite the wide variation of vessel sizes. Actually, the method detects both wide and tiny blood vessel with the same efficiency. We avoid using preprocessing to preserve the blood vessel structure and avoid losing any information. Furthermore, our results are only based on the crafty choice of features and the correcting of the outlier values by normalisation. So concentrating on the handling of the outlier values by the feature engineer may help in decreasing the reliability of using feature selection techniques or hierarchical classification as a way to increase system performance. Our experimental results contradict previous thought that the only advantage is in using feature selection and/or hierarchy classification techniques to improve performance. On the contrary, it suggests that using features that are selected craftily or by using feature engineers can give promising results. In the retinal disease diagnosis community, the relevant dataset is scarce. In other words, the availability for investigating and researching using real images from patients who suffer from an eye ailment is a hard task, particularly in a developing country. So, the only available choice is to work on the most publicity available dataset. The average accuracy, sensitivity and specificity values accomplished by the proposed method are 0.9607, 0.7542, 0.9843 on DRIVE, 0.9634, 0.7806, and 0.9825 on STARE, and 0.9577, 0.7585, and 0.9846 on CHASE_DB1, which are comparable with the current blood vessel segmentation techniques. Although 24-D feature vectors are constructed for every pixel in the image, the ensuing space and time complexity is offset by the simplicity of the method, compared to the cutting edge methods like deep learning. The fast implementation and simplicity of the proposed method make it a handy early prediction tool of DR.