Dynamical Behavior of a Stochastic SIRC Model for Influenza A
Abstract
1. Introduction
2. Preliminaries
3. Global Existence and Uniqueness of the Positive Solution
4. Extinction of the Disease
5. Existence of Ergodic Stationary Distribution
- (i)
- The smallest eigenvalue of diffusion matrix is bounded away from zero in the domain Ξ and some neighborhood thereof;
- (ii)
- If , the mean time τ at which a path issuing from y reaches the set Ξ is finite, and for every compact subset .
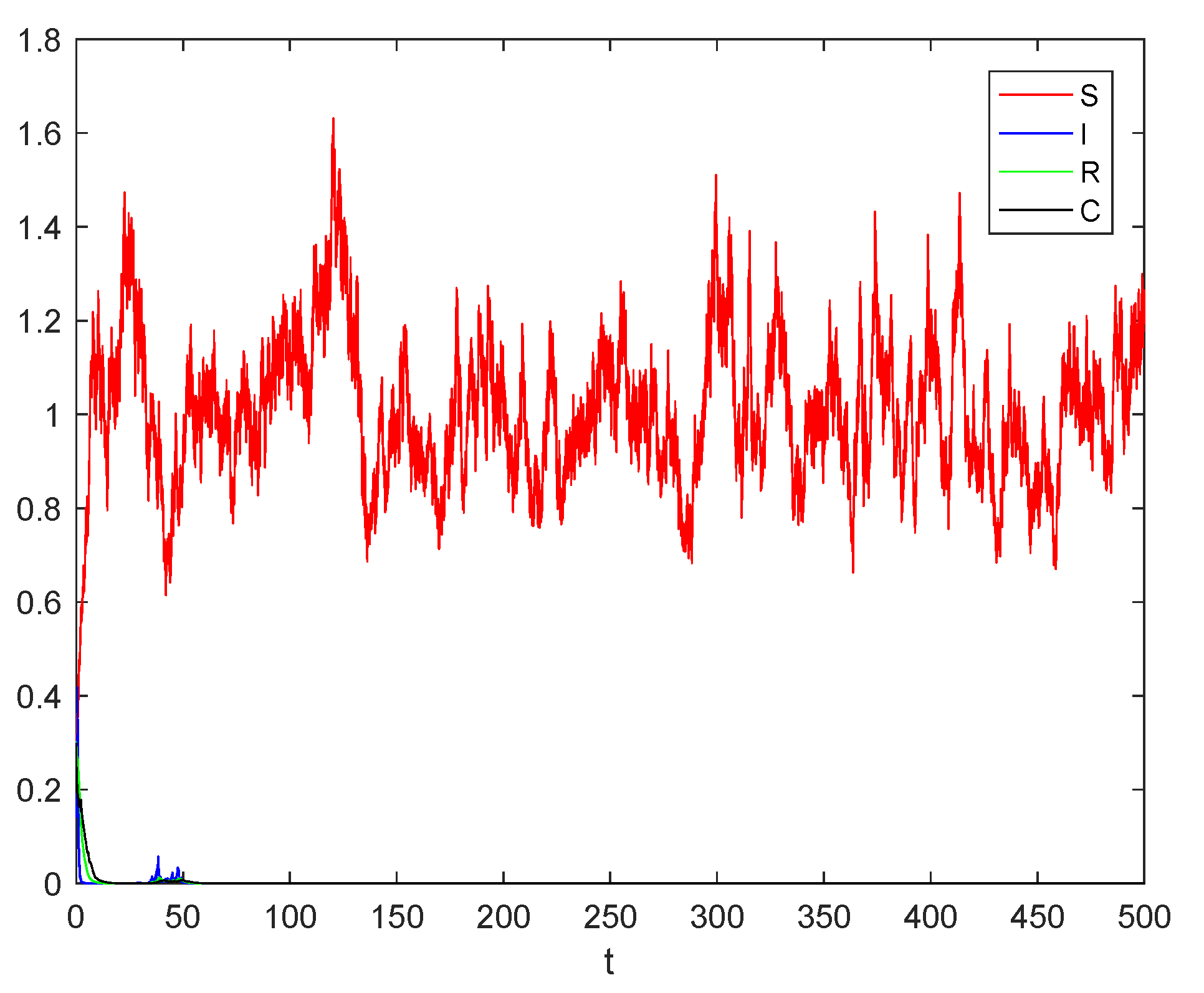
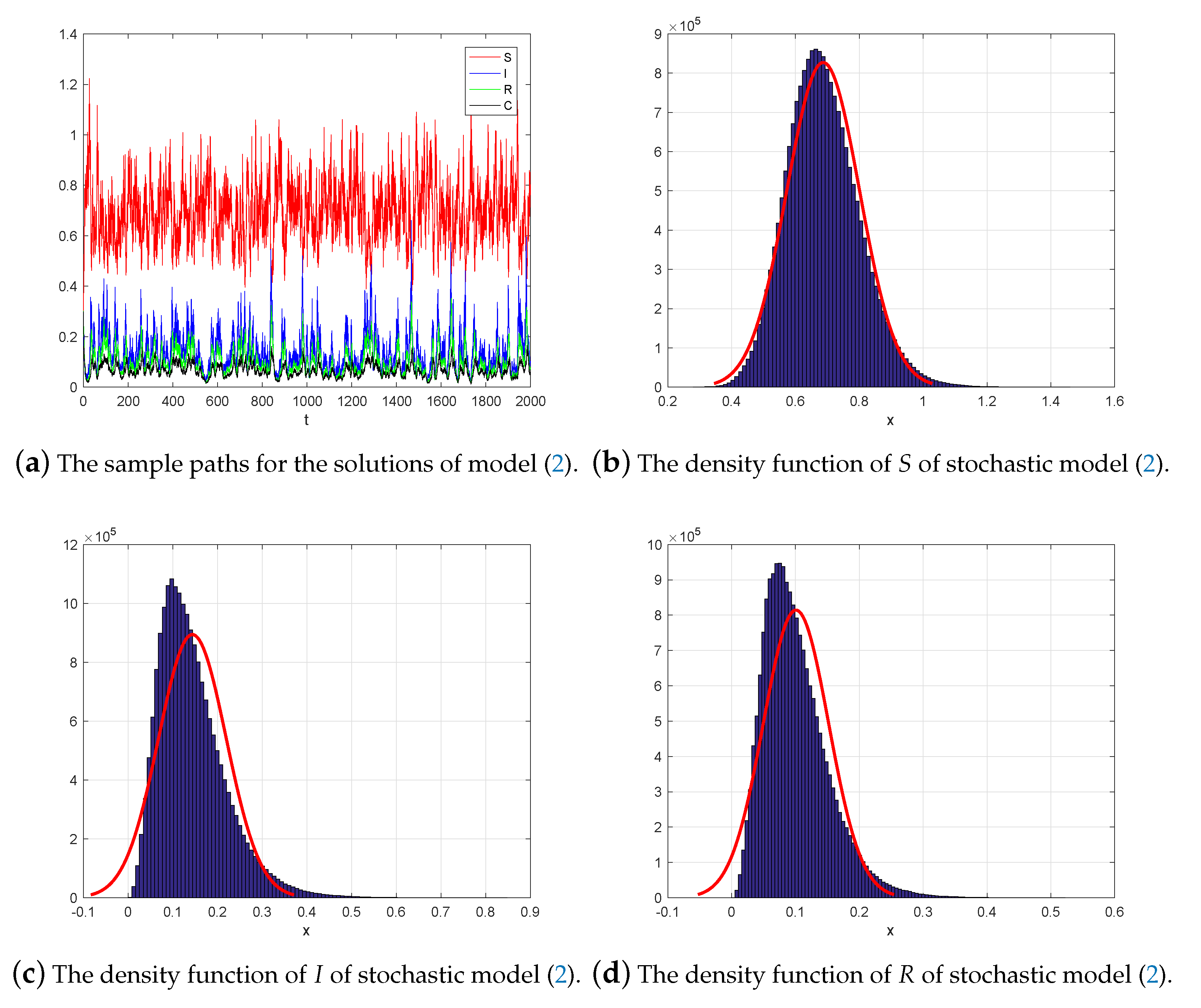
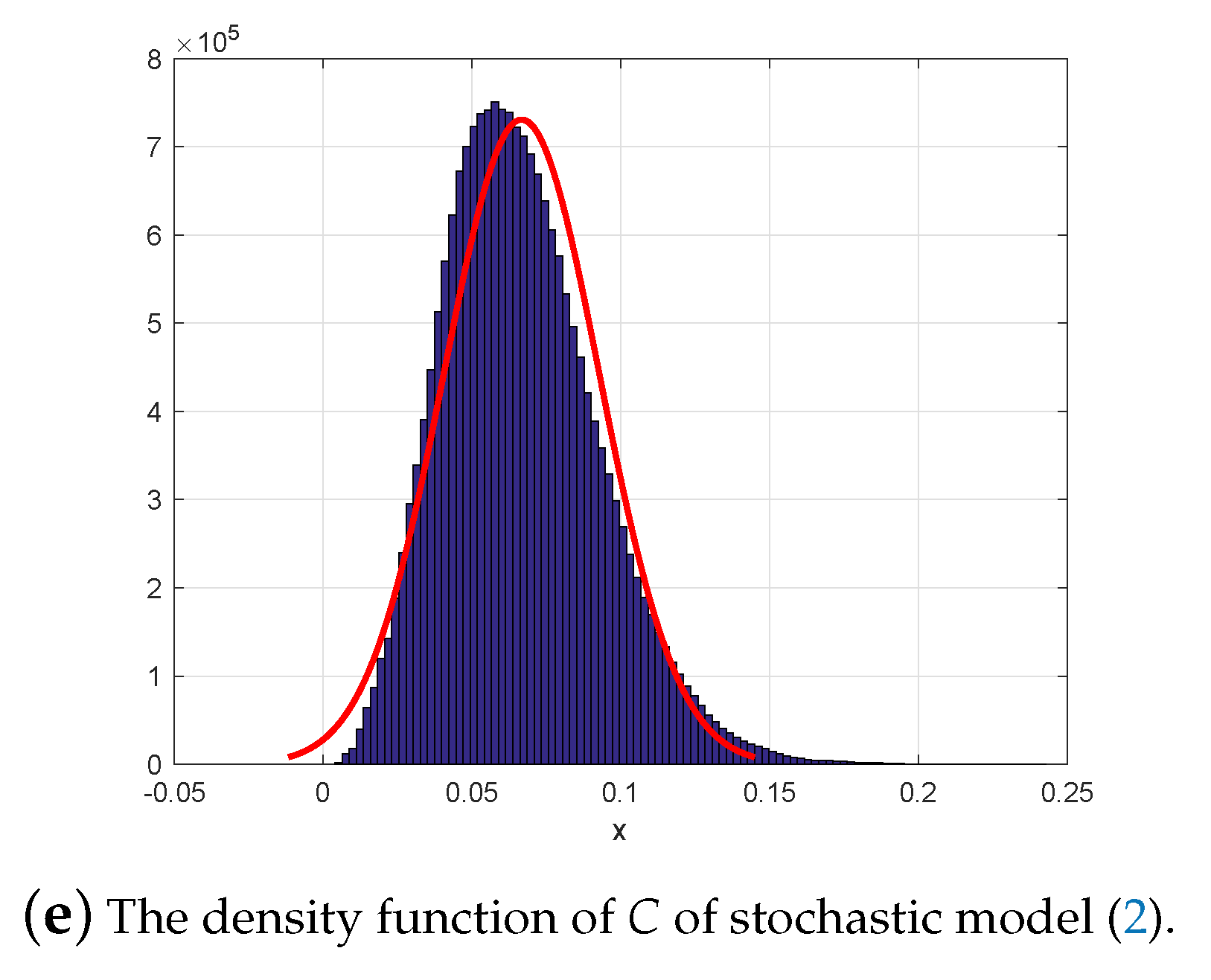
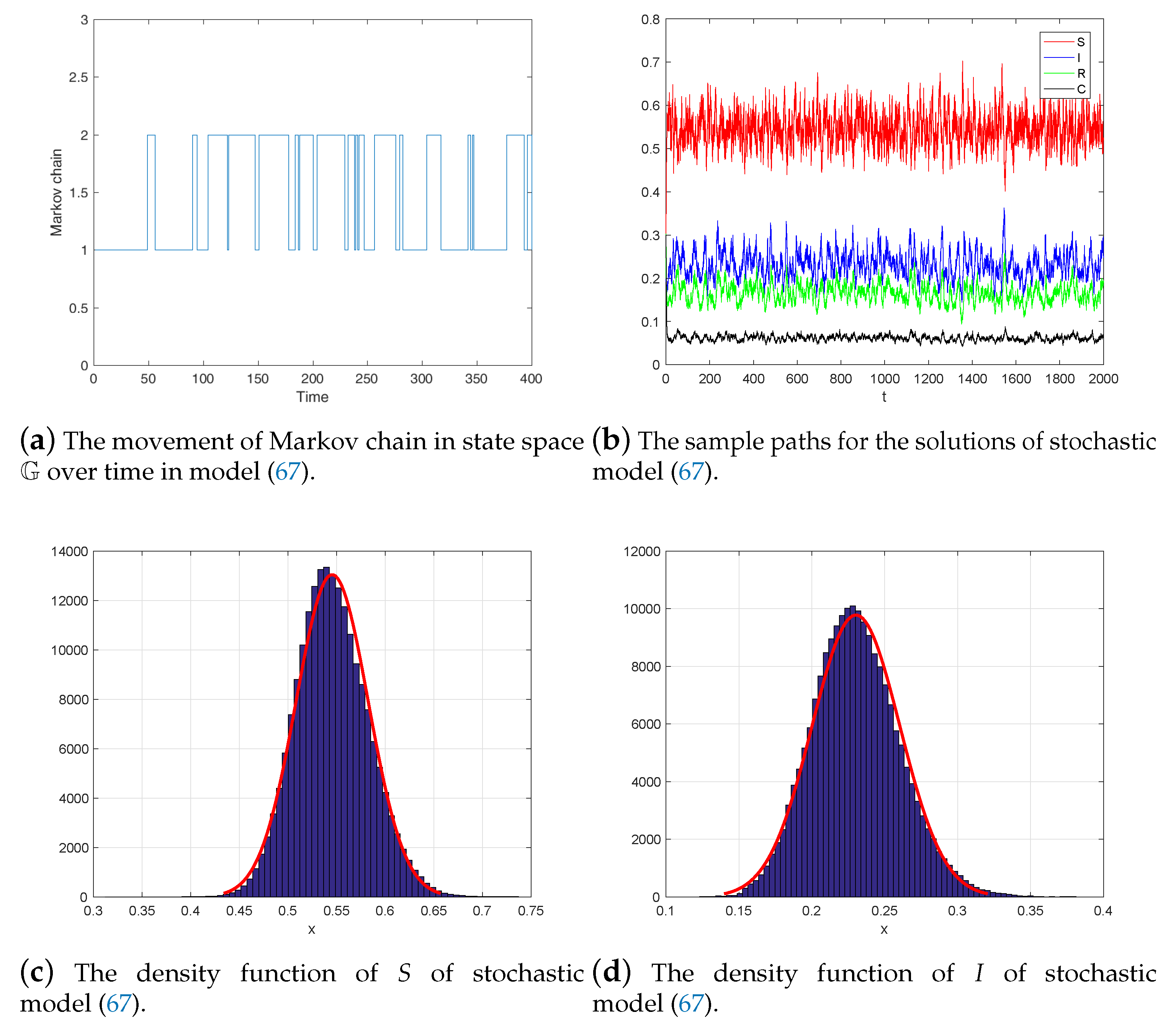
6. Numerical Simulations
7. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Duesberg, P.H. The RNA of influenza virus. Proc. Natl. Acad. Sci. USA 1968, 59, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Ritchey, M.B.; Palese, P.; Kilbourne, E.D. RNAs of influenza A, B, and C viruses. J. Virol. 1976, 18, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, K. Clinical features of influenza. Semin. Respir. Infect. 1992, 7, 26–37. [Google Scholar]
- WHO. World Health Organization Report: Research for Universal Health Coverage. Technical Report, World Health Organization. Available online: http://www.who.int/whr/en/ (accessed on 1 January 2019).
- Palese, P.; Young, J. Variation of influenza A, B, and C viruses. Science 1982, 215, 1468–1474. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.A. Isolation of a Novel Swine Influenza Virus from Oklahoma in 2011 Which Is Distantly Related to Human Influenza C Viruses. PLoS Pathogens 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.M.; Wang, S.C.; Peng, C.; Yu, J.M.; Zhuang, Q.Y.; Hou, G.Y.; Liu, S.; Li, J.P.; Chen, J.M. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes 2014, 49, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Quast, M.; Sreenivasan, C.; Sexton, G.; Nedland, H.; Singrey, A.; Fawcett, L.; Miller, G.; Lauer, D.; Voss, S.; Pollock, S.; et al. Serological evidence for the presence of influenza D virus in small ruminants. Vet. Microbiol. 2015, 180, 281–285. [Google Scholar] [CrossRef]
- Ducatez, M.; Pelletier, C.; Meyer, G. Influenza D Virus in Cattle, France, 2011–2014. Emerg. Infect. Dis. 2015, 21, 368–371. [Google Scholar] [CrossRef]
- Ng, T.F.F.; Kondov, N.O.; Deng, X.; Eenennaam, A.V.; Neibergsa, H.L.; Delwart, E. A Metagenomics and Case–Control Study To Identify Viruses Associated with Bovine Respiratory Disease. J. Virol. 2015, 89, 5340–5349. [Google Scholar] [CrossRef]
- Earn, D.J.; Dushoff, J.; Levin, S.A. Ecology and evolution of the flu. Trends Ecol. Evol. 2002, 17, 334–340. [Google Scholar] [CrossRef]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26, D49–D53. [Google Scholar] [CrossRef] [PubMed]
- Palese, P.; Shaw, M. Orthomyxoviridae: The viruses and their replication. In Fields Virology; Knipe, D., Howley, P., Griffin, D., Lamb, R., Martin, M., Roizman, B., Strauss, E., Eds.; Lippincott Williams & Wilkins: Hiladelphia, PA, USA, 2007. [Google Scholar]
- Feng, T.; Qiu, Z.; Meng, X. Dynamics of a stochastic hepatitis C virus system with host immunity. Discret. Contin. Dyn. Syst.-B 2019, 24, 6367–6385. [Google Scholar] [CrossRef]
- Feng, T.; Qiu, Z.; Meng, X.; Rong, L. Analysis of a stochastic HIV-1 infection model with degenerate diffusion. Appl. Math. Comput. 2019, 348, 437–455. [Google Scholar] [CrossRef]
- Feng, T.; Qiu, Z.; Meng, X. Analysis of a stochastic recovery-relapse epidemic model with periodic parameters and media coverage. J. Appl. Anal. Comput. 2019, 9, 1007–1021. [Google Scholar] [CrossRef]
- Zhang, T.; Gao, N.; Wang, T.; Liu, H.; Jiang, Z. Global dynamics of a model for treating microorganisms in sewage by periodically adding microbial flocculants. Math. Biosci. Eng. 2020, 17, 179–201. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, J.; Li, Y.; Jiang, Z.; Han, X. Dynamics analysis of a delayed virus model with two different transmission methods and treatments. Adv. Differ. Equ. 2020, 2020, 1. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Mol. Biol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef]
- Webster, R.; Laver, W.; Air, G.; Schild, G. Molecular mechanisms of variation in influenza viruses. Nature 1982, 296, 115–121. [Google Scholar] [CrossRef]
- Larson, H.; Tyrrell, D.; Bowker, C.; Potter, C.; Schild, G. Immunity to challenge in volunteers vaccinated with an inactivated current or earlier strain of influenza A(H3N2). J. Hyg. 1978, 80, 243–248. [Google Scholar] [CrossRef]
- Davies, J.; Grilli, E.; Smith, A. Influenza A: Infection and reinfection. J. Hyg. 1984, 92, 125–127. [Google Scholar] [CrossRef]
- Levine, A. Viruses; W.H. Freeman & Company: New York, NY, USA, 1992. [Google Scholar]
- Casagrandi, R.; Bolzoni, L.; Levin, S.A.; Andreasen, V. The SIRC model and influenza A. Math. Biosci. 2006, 200, 152–169. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guo, S. Dynamics of a SIRC epidemiological model. Electron. J. Differ. Equ. 2017, 2017, 1–18. [Google Scholar]
- Zhu, F.; Meng, X.; Zhang, T. Optimal harvesting of a competitive n-species stochastic model with delayed diffusions. Math. Biosci. Eng. 2019, 16, 1554–1574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, T. The stationary distribution of a microorganism flocculation model with stochastic perturbation. Appl. Math. Lett. 2020, 103, 106217. [Google Scholar] [CrossRef]
- Liu, G.; Qi, H.; Chang, Z.; Meng, X. Asymptotic stability of a stochastic May mutualism system. Comput. Math. Appl. 2020, 79, 735–745. [Google Scholar] [CrossRef]
- Chi, M.; Zhao, W. Dynamical analysis of two-microorganism and single nutrient stochastic chemostat model with monod-haldane response function. Complexity 2019, 2019, 8719067. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, J.; Chi, M.; Bian, F. Dynamics analysis of stochastic epidemic models with standard incidence. Adv. Differ. Equ. 2019, 2019, 22. [Google Scholar] [CrossRef]
- Jiang, D.; Ji, C.; Shi, N.; Yu, J. The long time behavior of DI SIR epidemic model with stochastic perturbation. J. Math. Anal. Appl. 2010, 372, 162–180. [Google Scholar] [CrossRef]
- Ji, C.; Jiang, D. Threshold behaviour of a stochastic SIR model. Appl. Math. Model. 2014, 38, 5067–5079. [Google Scholar] [CrossRef]
- Tuckwell, H.C.; Williams, R.J. Some properties of a simple stochastic epidemic model of SIR type. Math. Biosci. 2007, 208, 76–97. [Google Scholar] [CrossRef]
- Dieu, N.T.; Nguyen, D.H.; Du, N.H.; Yin, G. Classification of Asymptotic Behavior in a Stochastic SIR Model. SIAM J. Appl. Dyn. Syst. 2016, 15, 1062–1084. [Google Scholar] [CrossRef]
- Miao, A.; Zhang, T.; Zhang, J.; Wang, C. Dynamics of a stochastic SIR model with both horizontal and vertical transmission. J. Appl. Anal. Comput. 2018, 8, 1108–1121. [Google Scholar]
- Beretta, E.; Kolmanovskii, V.; Shaikhet, L. Stability of epidemic model with time delays influenced by stochastic perturbations. Math. Comput. Simul. 1998, 45, 269–277. [Google Scholar] [CrossRef]
- Bacaër, N. On the stochastic SIS epidemic model in a periodic environment. J. Math. Biol. 2014, 71, 491–511. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gray, A.; Greenhalgh, D.; Mao, X.; Pan, J. The SIS epidemic model with Markovian switching. J. Math. Anal. Appl. 2012, 394, 496–516. [Google Scholar] [CrossRef]
- Cai, Y.; Kang, Y.; Banerjee, M.; Wang, W. A stochastic epidemic model incorporating media coverage. Commun. Math. Sci. 2016, 14, 893–910. [Google Scholar] [CrossRef]
- Gao, N.; Song, Y.; Wang, X.; Liu, J. Dynamics of a stochastic SIS epidemic model with nonlinear incidence rates. Adv. Differ. Equ. 2019, 2019, 41. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, L.; Yuan, S. The effect of media coverage on threshold dynamics for a stochastic SIS epidemic model. Phys. A Stat. Mech. Appl. 2018, 512, 248–260. [Google Scholar] [CrossRef]
- Zhou, Y.; Yuan, S.; Zhao, D. Threshold behavior of a stochastic SIS model with jumps. Appl. Math. Comput. 2016, 275, 255–267. [Google Scholar]
- Song, Y.; Miao, A.; Zhang, T.; Wang, X.; Liu, J. Extinction and persistence of a stochastic SIRS epidemic model with saturated incidence rate and transfer from infectious to susceptible. Adv. Differ. Equ. 2018, 2018, 293. [Google Scholar] [CrossRef]
- Lahrouz, A.; Settati, A. Qualitative Study of a Nonlinear Stochastic SIRS Epidemic System. Stoch. Anal. Appl. 2014, 32, 992–1008. [Google Scholar] [CrossRef]
- Liu, Q.; Jiang, D.; Shi, N.; Hayat, T.; Alsaedi, A. Stationary distribution and extinction of a stochastic SIRS epidemic model with standard incidence. Phys. A Stat. Mech. Appl. 2017, 469, 510–517. [Google Scholar] [CrossRef]
- Liu, M.; Bai, C.; Wang, K. Asymptotic stability of a two-group stochastic SEIR model with infinite delays. Commun. Nonlinear Sci. Numer. Simul. 2014, 19, 3444–3453. [Google Scholar] [CrossRef]
- Mao, X. Stochastic Differential Equations and Applications, 2nd ed.; Horwood Publishing: Chichester, UK, 2007. [Google Scholar]
- Khasminskii, R. Stochastic Stability of Differential Equations, 2nd ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2012; Volume 66. [Google Scholar]
- Li, F.; Zhang, S.; Meng, X. Dynamics analysis and numerical simulations of a delayed stochastic epidemic model subject to a general response function. Comput. Appl. Math. 2019, 38, 95. [Google Scholar] [CrossRef]
- Mao, X.; Marion, G.; Renshaw, E. Environmental Brownian noise suppresses explosions in population dynamics. Stoch. Proces. Appl. 2002, 97, 95–110. [Google Scholar] [CrossRef]
- Liptser, R. A strong law of large numbers for local martingales. Stochastics 1980, 3, 217–228. [Google Scholar] [CrossRef]
- Gard, T.C. Introduction to Stochastic Differential Equations(Pure and Applied Mathematics); Marcel Dekker Inc.: New York, NY, USA, 1988. [Google Scholar]
- Zhu, C.; Yin, G. Asymptotic Properties of Hybrid Diffusion Systems. SIAM J. Control Optim. 2007, 46, 1155–1179. [Google Scholar] [CrossRef]
- Gao, M.; Jiang, D. Ergodic stationary distribution of a stochastic chemostat model with regime switching. Phys. A Stat. Mech. Appl. 2019, 524, 491–502. [Google Scholar] [CrossRef]
- Settati, A.; Lahrouz, A. Stationary distribution of stochastic population systems under regime switching. Appl. Math. Comput. 2014, 244, 235–243. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Ding, T.; Gao, N.; Song, Y. Dynamical Behavior of a Stochastic SIRC Model for Influenza A. Symmetry 2020, 12, 745. https://doi.org/10.3390/sym12050745
Zhang T, Ding T, Gao N, Song Y. Dynamical Behavior of a Stochastic SIRC Model for Influenza A. Symmetry. 2020; 12(5):745. https://doi.org/10.3390/sym12050745
Chicago/Turabian StyleZhang, Tongqian, Tingting Ding, Ning Gao, and Yi Song. 2020. "Dynamical Behavior of a Stochastic SIRC Model for Influenza A" Symmetry 12, no. 5: 745. https://doi.org/10.3390/sym12050745
APA StyleZhang, T., Ding, T., Gao, N., & Song, Y. (2020). Dynamical Behavior of a Stochastic SIRC Model for Influenza A. Symmetry, 12(5), 745. https://doi.org/10.3390/sym12050745




